Expression of the Z Variant of α1-Antitrypsin Suppresses Hepatic Cholesterol Biosynthesis in Transgenic Zebrafish
Abstract
1. Introduction
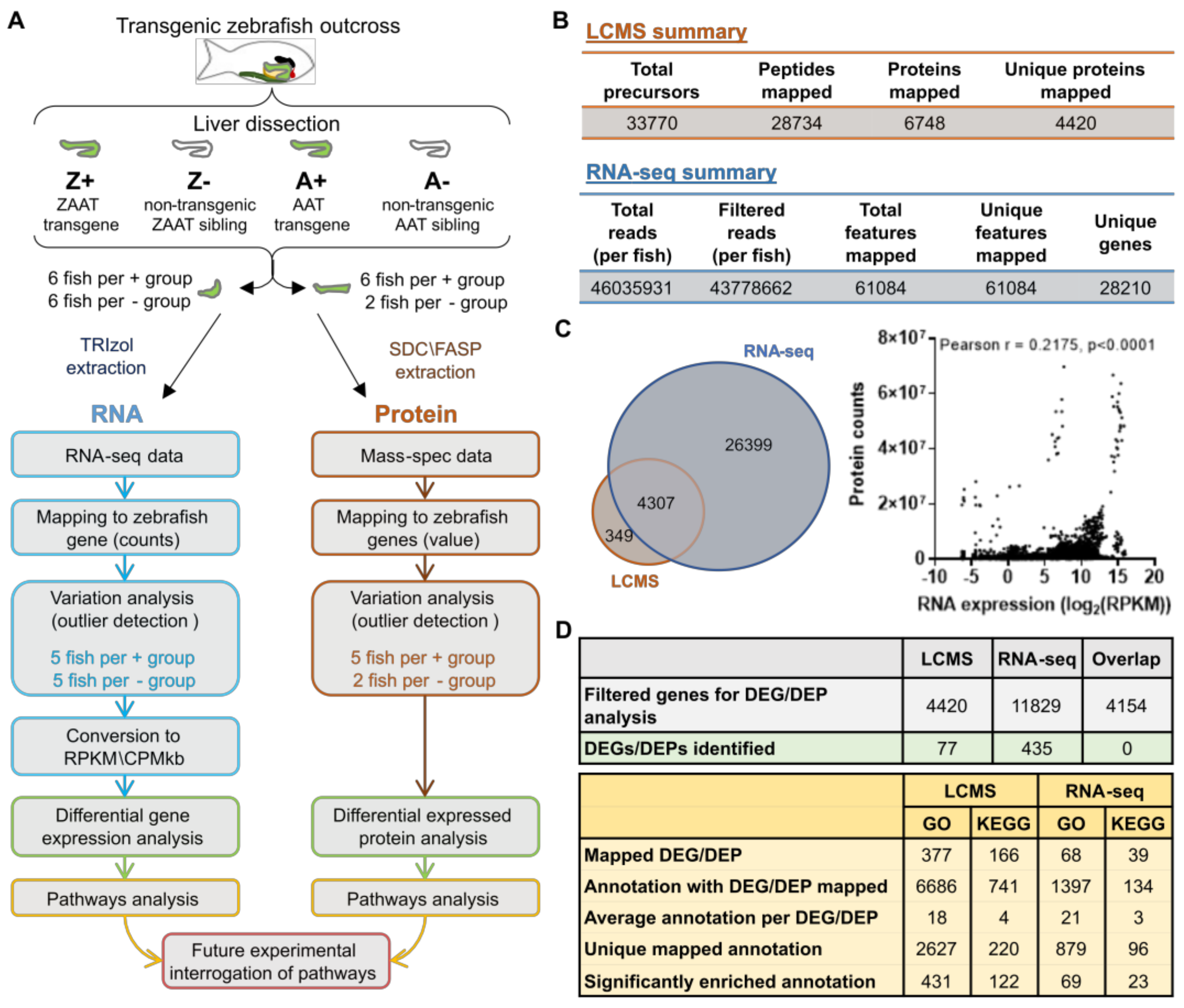
2. Results
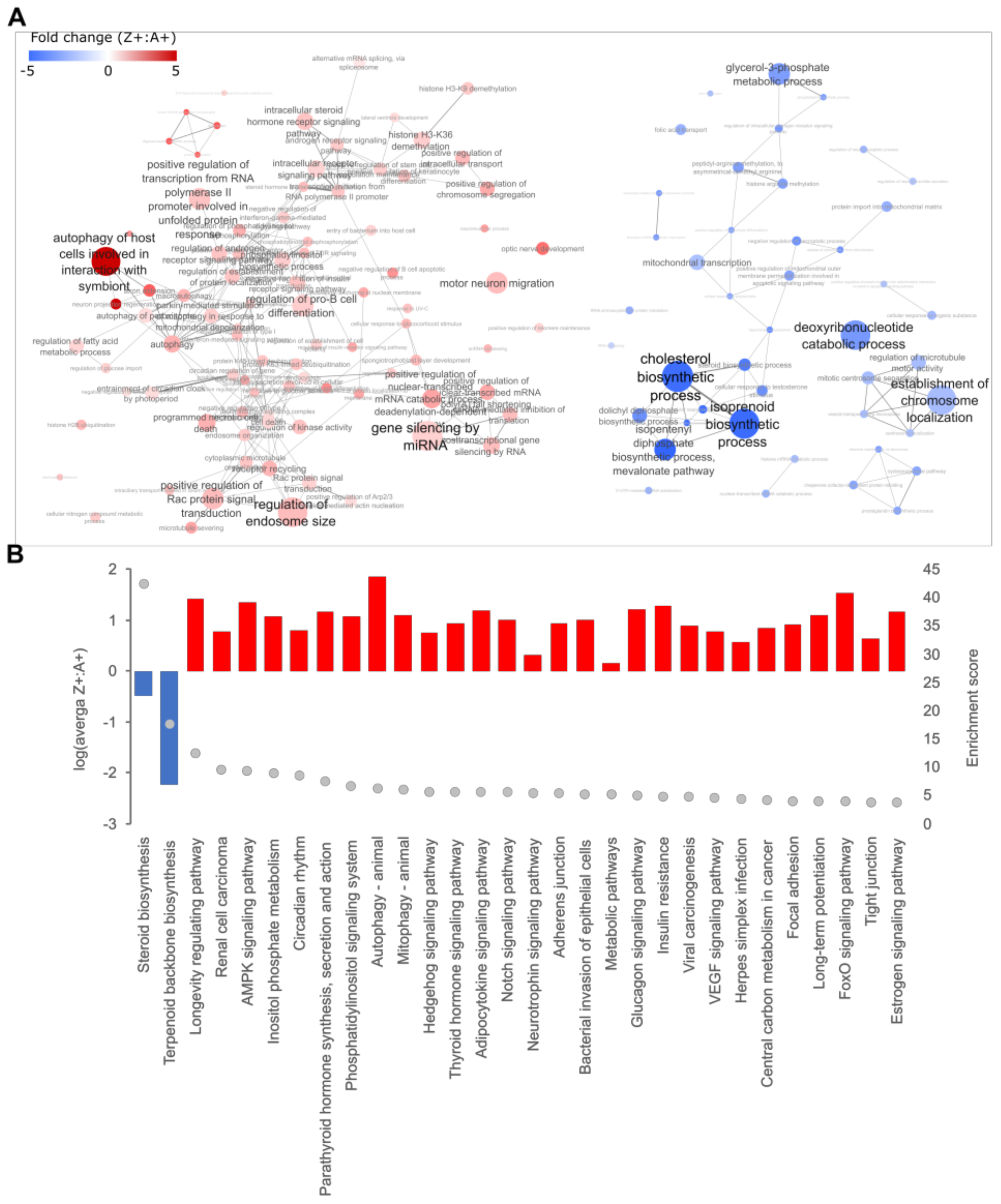
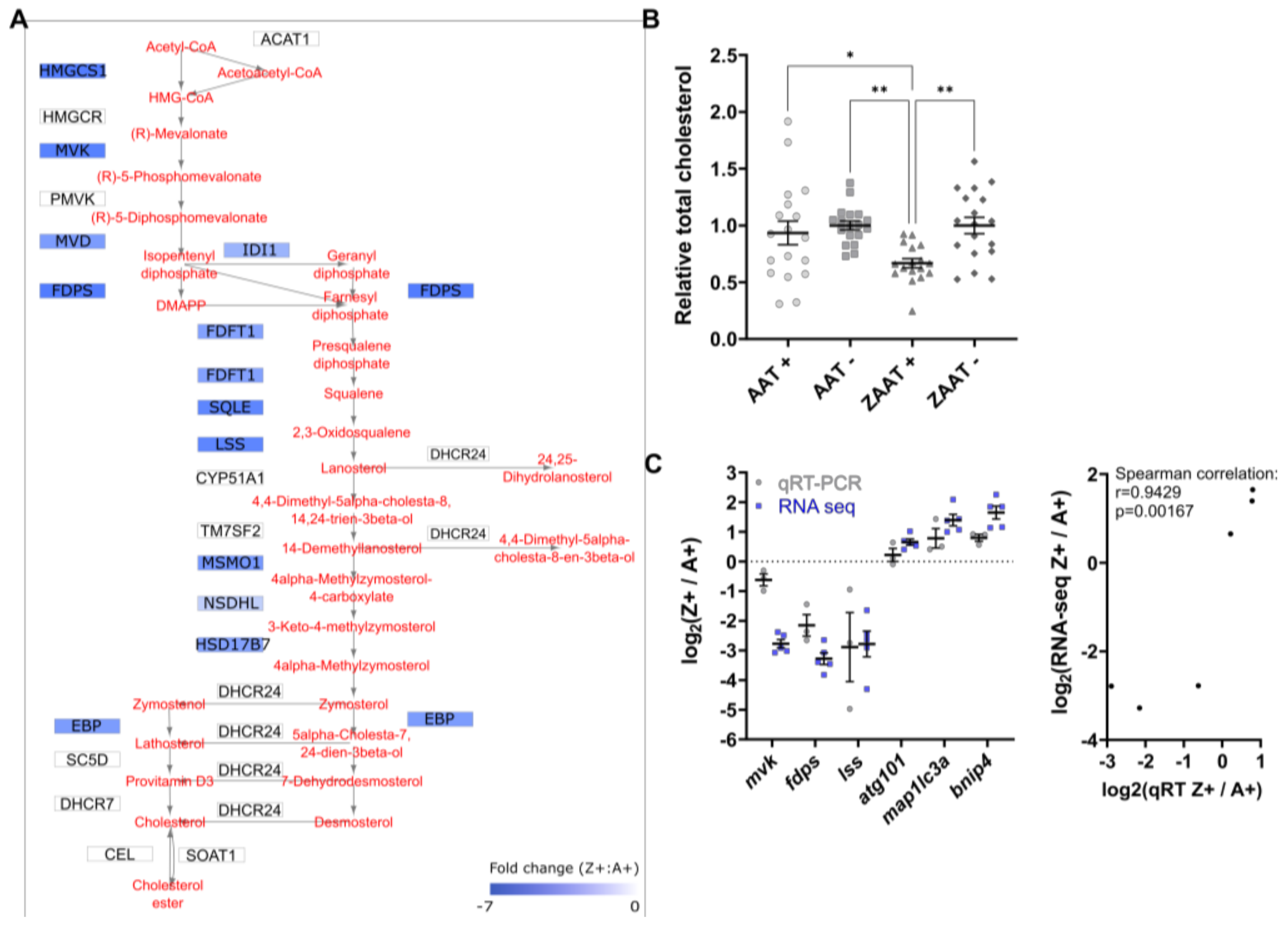
2.1. Expression of ZAAT in Zebrafish Liver Results in Suppression of Cholesterol Biosynthesis
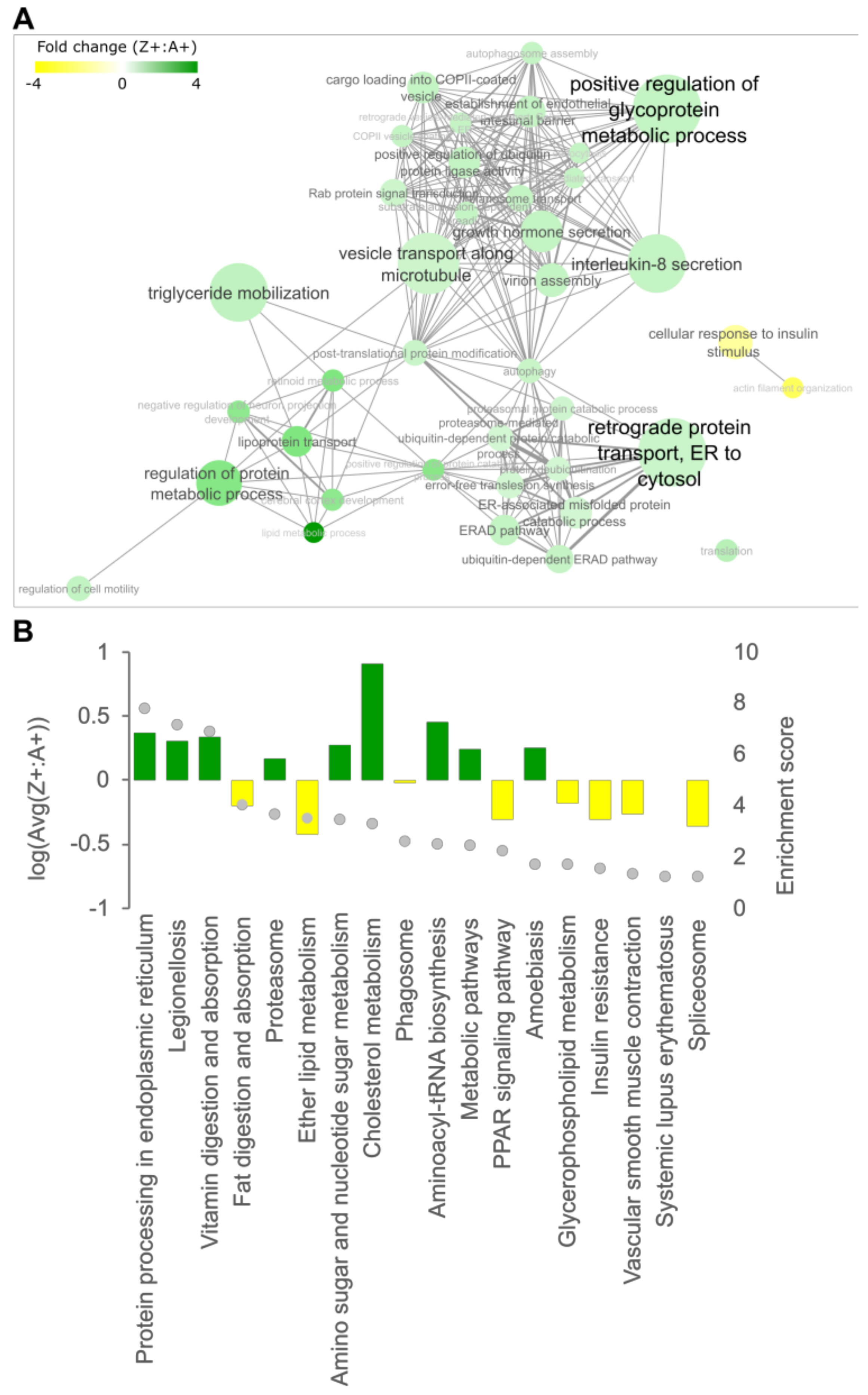
2.2. Cholesterol Biosynthesis Suppression Is a ZAAT-Specific Response in Both Zebrafish and Mammalian Models
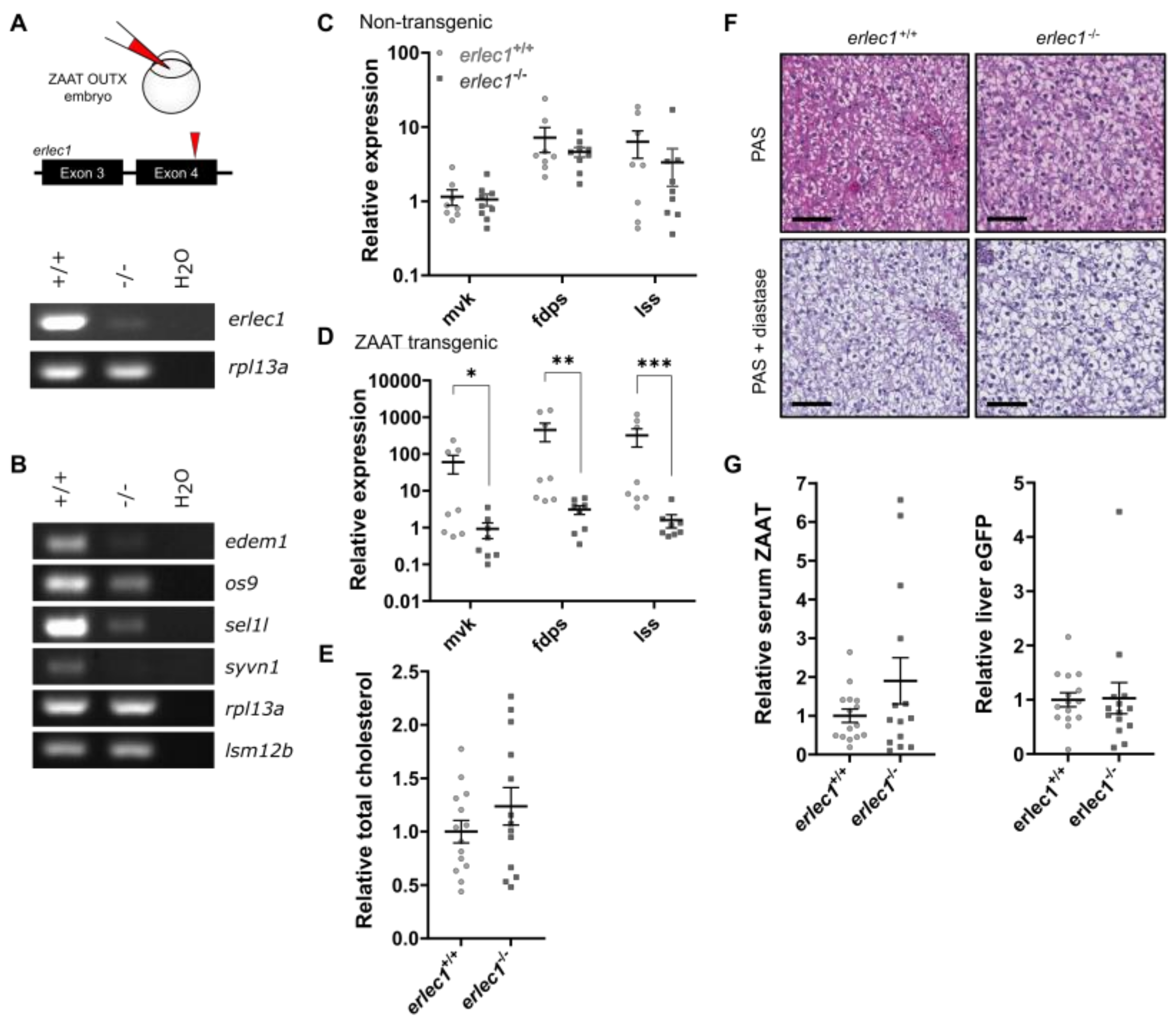
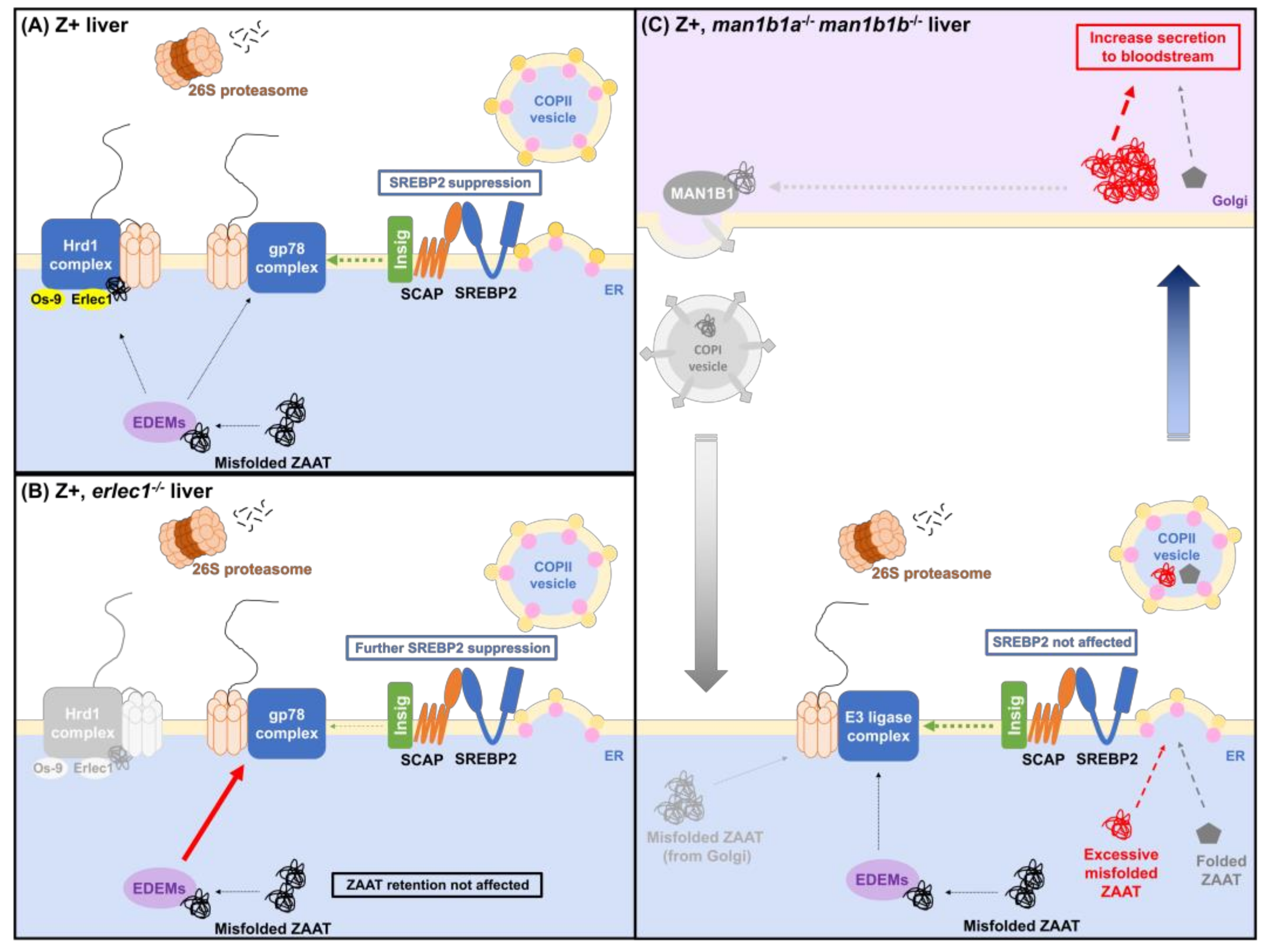
2.3. Compromising the HRD1 E3 Ligase Complex Further Suppresses Cholesterol Biosynthesis but Does Not Influence ZAAT Accumulation or Secretion
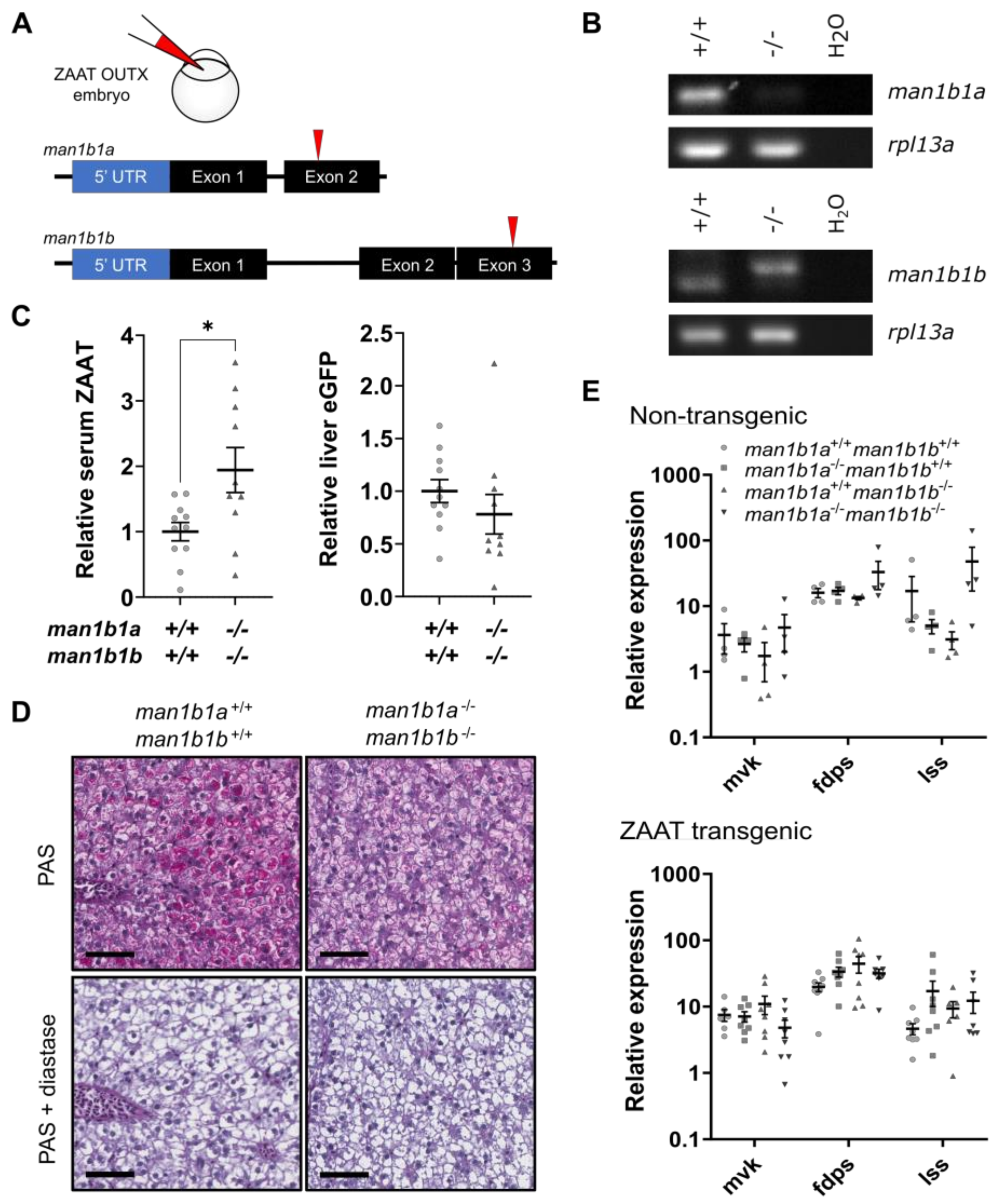
2.4. Deletion of Man1b1 Mannosidases Increases ZAAT Secretion without Inducing Intracellular Accumulation
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Cell Culturing and Transfection
4.3. Reverse Transcriptase PCR
4.4. Real-Time Quantitative PCR
4.5. Zebrafish Maintenance
4.6. Mutant Zebrafish Lines
4.7. Liquid Chromatography-Mass Spectrometry (LC-MS/MS)
4.8. RNA-Sequencing
4.9. Differential Gene/Protein Analysis
4.10. Antibodies
4.11. Protein Lysates and Immunoblotting
4.12. Total Cholesterol Assay
4.13. Periodic Acid-Schiff (PAS) Diastase (D) Staining
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beatty, K.; Bieth, J.; Travis, J. Kinetics of association of serine proteinases with native and oxidized alpha-1-proteinase inhibitor and alpha-1-antichymotrypsin. J. Biol. Chem. 1980, 255, 3931–3934. [Google Scholar] [CrossRef] [PubMed]
- Cazzola, M.; Stolz, D.; Rogliani, P.; Matera, M.G. alpha(1)-Antitrypsin deficiency and chronic respiratory disorders. Eur. Respir. Rev. 2020, 29, 190073. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.; Oliveira, M.J.; Guimaraes, M.; Lima, R.; Gomes, S.; Seixas, S. Alpha-1-antitrypsin (SERPINA1) mutation spectrum: Three novel variants and haplotype characterization of rare deficiency alleles identified in Portugal. Respir. Med. 2016, 116, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Stoller, J.K.; Snider, G.L.; Brantly, M.L.; Fallat, R.J.; Stockley, R.A.; Turino, G.M.; Konietzko, N.; Dirksen, A.; Eden, E.; Luisetti, M.; et al. American Thoracic Society/European Respiratory Society statement: Standards for the diagnosis and management of individuals with alpha-1 antitrypsin deficiency. Am. J. Respir. Crit. Care Med. 2003, 168, 818–900. [Google Scholar] [CrossRef]
- Laffranchi, M.; Berardelli, R.; Ronzoni, R.; Lomas, D.A.; Fra, A. Heteropolymerization of alpha-1-antitrypsin mutants in cell models mimicking heterozygosity. Hum. Mol. Genet. 2018, 27, 1785–1793. [Google Scholar] [CrossRef]
- Curiel, D.T.; Holmes, M.D.; Okayama, H.; Brantly, M.L.; Vogelmeier, C.; Travis, W.D.; Stier, L.E.; Perks, W.H.; Crystal, R.G. Molecular basis of the liver and lung disease associated with the alpha 1-antitrypsin deficiency allele Mmalton. J. Biol. Chem. 1989, 264, 13938–13945. [Google Scholar] [CrossRef]
- Roberts, E.A.; Cox, D.W.; Medline, A.; Wanless, I.R. Occurrence of alpha-1-antitrypsin deficiency in 155 patients with alcoholic liver disease. Am. J. Clin. Pathol. 1984, 82, 424–427. [Google Scholar] [CrossRef]
- Callea, F.; Giovannoni, I.; Francalanci, P.; Boldrini, R.; Faa, G.; Medicina, D.; Nobili, V.; Desmet, V.J.; Ishak, K.; Seyama, K.; et al. Mineralization of alpha-1-antitrypsin inclusion bodies in Mmalton alpha-1-antitrypsin deficiency. Orphanet. J. Rare Dis. 2018, 13, 79. [Google Scholar] [CrossRef]
- Janciauskiene, S.; Eriksson, S.; Callea, F.; Mallya, M.; Zhou, A.; Seyama, K.; Hata, S.; Lomas, D.A. Differential detection of PAS-positive inclusions formed by the Z, Siiyama, and Mmalton variants of alpha1-antitrypsin. Hepatology 2004, 40, 1203–1210. [Google Scholar] [CrossRef]
- Elliott, P.R.; Stein, P.E.; Bilton, D.; Carrell, R.W.; Lomas, D.A. Structural explanation for the deficiency of S alpha 1-antitrypsin. Nat. Struct. Biol. 1996, 3, 910–911. [Google Scholar] [CrossRef]
- Miranda, E.; Perez, J.; Ekeowa, U.I.; Hadzic, N.; Kalsheker, N.; Gooptu, B.; Portmann, B.; Belorgey, D.; Hill, M.; Chambers, S.; et al. A novel monoclonal antibody to characterize pathogenic polymers in liver disease associated with alpha1-antitrypsin deficiency. Hepatology 2010, 52, 1078–1088. [Google Scholar] [CrossRef]
- Annunziata, A.; Ferrarotti, I.; Coppola, A.; Lanza, M.; Imitazione, P.; Spinelli, S.; Micco, P.D.; Fiorentino, G. Alpha-1 Antitrypsin Screening in a Selected Cohort of Patients Affected by Chronic Pulmonary Diseases in Naples, Italy. J. Clin. Med. 2021, 10, 1546. [Google Scholar] [CrossRef]
- Hofker, M.H.; Nukiwa, T.; van Paassen, H.M.; Nelen, M.; Kramps, J.A.; Klasen, E.C.; Frants, R.R.; Crystal, R.G. A Pro—Leu substitution in codon 369 of the alpha-1-antitrypsin deficiency variant PI MHeerlen. Hum. Genet. 1989, 81, 264–268. [Google Scholar] [CrossRef]
- Donato, L.J.; Jenkins, S.M.; Smith, C.; Katzmann, J.A.; Snyder, M.R. Reference and interpretive ranges for alpha(1)-antitrypsin quantitation by phenotype in adult and pediatric populations. Am. J. Clin. Pathol. 2012, 138, 398–405. [Google Scholar] [CrossRef]
- Clark, V.C.; Marek, G.; Liu, C.; Collinsworth, A.; Shuster, J.; Kurtz, T.; Nolte, J.; Brantly, M. Clinical and histologic features of adults with alpha-1 antitrypsin deficiency in a non-cirrhotic cohort. J. Hepatol. 2018, 69, 1357–1364. [Google Scholar] [CrossRef]
- Schneider, C.V.; Hamesch, K.; Gross, A.; Mandorfer, M.; Moeller, L.S.; Pereira, V.; Pons, M.; Kuca, P.; Reichert, M.C.; Benini, F.; et al. Liver Phenotypes of European Adults Heterozygous or Homozygous for Pi *Z Variant of AAT (Pi *MZ vs Pi *ZZ genotype) and Noncarriers. Gastroenterology 2020, 159, 534–548. [Google Scholar] [CrossRef]
- Sveger, T. Liver disease in alpha1-antitrypsin deficiency detected by screening of 200,000 infants. N. Engl. J. Med. 1976, 294, 1316–1321. [Google Scholar] [CrossRef]
- Sveger, T.; Eriksson, S. The liver in adolescents with alpha 1-antitrypsin deficiency. Hepatology 1995, 22, 514–517. [Google Scholar] [CrossRef]
- Mostafavi, B.; Diaz, S.; Tanash, H.A.; Piitulainen, E. Liver function in alpha-1-antitrypsin deficient individuals at 37 to 40 years of age. Medicine 2017, 96, e6180. [Google Scholar] [CrossRef]
- Cox, D.W.; Smyth, S. Risk for liver disease in adults with alpha 1-antitrypsin deficiency. Am. J. Med. 1983, 74, 221–227. [Google Scholar] [CrossRef]
- Teckman, J.H.; Burrows, J.; Hidvegi, T.; Schmidt, B.; Hale, P.D.; Perlmutter, D.H. The proteasome participates in degradation of mutant alpha 1-antitrypsin Z in the endoplasmic reticulum of hepatoma-derived hepatocytes. J. Biol. Chem. 2001, 276, 44865–44872. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Teckman, J.H.; Omura, S.; Perlmutter, D.H. Degradation of a mutant secretory protein, alpha1-antitrypsin Z, in the endoplasmic reticulum requires proteasome activity. J. Biol. Chem. 1996, 271, 22791–22795. [Google Scholar] [CrossRef] [PubMed]
- Teckman, J.H.; Gilmore, R.; Perlmutter, D.H. Role of ubiquitin in proteasomal degradation of mutant alpha(1)-antitrypsin Z in the endoplasmic reticulum. Am. J. Physiol. Gastrointest Liver Physiol. 2000, 278, G39–G48. [Google Scholar] [CrossRef]
- Shen, Y.; Ballar, P.; Fang, S. Ubiquitin ligase gp78 increases solubility and facilitates degradation of the Z variant of alpha-1-antitrypsin. Biochem. Biophys. Res. Commun. 2006, 349, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Q.; Shen, Y.; Sun, A.; Zhu, X.; Fang, S.; Shen, Y. The ubiquitin ligase Hrd1 promotes degradation of the Z variant alpha 1-antitrypsin and increases its solubility. Mol. Cell. Biochem. 2011, 346, 137–145. [Google Scholar] [CrossRef]
- Kamimoto, T.; Shoji, S.; Hidvegi, T.; Mizushima, N.; Umebayashi, K.; Perlmutter, D.H.; Yoshimori, T. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J. Biol. Chem. 2006, 281, 4467–4476. [Google Scholar] [CrossRef]
- Feng, L.; Zhang, J.; Zhu, N.; Ding, Q.; Zhang, X.; Yu, J.; Qiang, W.; Zhang, Z.; Ma, Y.; Huang, D.; et al. Ubiquitin ligase SYVN1/HRD1 facilitates degradation of the SERPINA1 Z variant/alpha-1-antitrypsin Z variant via SQSTM1/p62-dependent selective autophagy. Autophagy 2017, 13, 686–702. [Google Scholar] [CrossRef]
- Kroeger, H.; Miranda, E.; MacLeod, I.; Perez, J.; Crowther, D.C.; Marciniak, S.J.; Lomas, D.A. Endoplasmic reticulum-associated degradation (ERAD) and autophagy cooperate to degrade polymerogenic mutant serpins. J. Biol. Chem. 2009, 284, 22793–22802. [Google Scholar] [CrossRef]
- Fregno, I.; Fasana, E.; Bergmann, T.J.; Raimondi, A.; Loi, M.; Solda, T.; Galli, C.; D’Antuono, R.; Morone, D.; Danieli, A.; et al. ER-to-lysosome-associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J. 2018, 37, e99259. [Google Scholar] [CrossRef]
- Yip, E.; Giousoh, A.; Fung, C.; Wilding, B.; Prakash, M.D.; Williams, C.; Verkade, H.; Bryson-Richardson, R.J.; Bird, P.I. A transgenic zebrafish model of hepatocyte function in human Z alpha1-antitrypsin deficiency. Biol. Chem. 2019, 400, 1603–1616. [Google Scholar] [CrossRef]
- Marcus, N.Y.; Perlmutter, D.H. Glucosidase and mannosidase inhibitors mediate increased secretion of mutant alpha1 antitrypsin Z. J. Biol. Chem. 2000, 275, 1987–1992. [Google Scholar] [CrossRef]
- Cabral, C.M.; Liu, Y.; Moremen, K.W.; Sifers, R.N. Organizational diversity among distinct glycoprotein endoplasmic reticulum-associated degradation programs. Mol. Biol. Cell 2002, 13, 2639–2650. [Google Scholar] [CrossRef]
- Kaushal, S.; Annamali, M.; Blomenkamp, K.; Rudnick, D.; Halloran, D.; Brunt, E.M.; Teckman, J.H. Rapamycin reduces intrahepatic alpha-1-antitrypsin mutant Z protein polymers and liver injury in a mouse model. Exp. Biol. Med. (Maywood) 2010, 235, 700–709. [Google Scholar] [CrossRef]
- Hidvegi, T.; Ewing, M.; Hale, P.; Dippold, C.; Beckett, C.; Kemp, C.; Maurice, N.; Mukherjee, A.; Goldbach, C.; Watkins, S.; et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science 2010, 329, 229–232. [Google Scholar] [CrossRef]
- Teckman, J.H.; Perlmutter, D.H. Retention of mutant alpha(1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am. J. Physiol. Gastrointest Liver Physiol. 2000, 279, G961–G974. [Google Scholar] [CrossRef]
- Hidvegi, T.; Schmidt, B.Z.; Hale, P.; Perlmutter, D.H. Accumulation of mutant alpha1-antitrypsin Z in the endoplasmic reticulum activates caspases-4 and -12, NFkappaB, and BAP31 but not the unfolded protein response. J. Biol. Chem. 2005, 280, 39002–39015. [Google Scholar] [CrossRef]
- Ordonez, A.; Snapp, E.L.; Tan, L.; Miranda, E.; Marciniak, S.J.; Lomas, D.A. Endoplasmic reticulum polymers impair luminal protein mobility and sensitize to cellular stress in alpha1-antitrypsin deficiency. Hepatology 2013, 57, 2049–2060. [Google Scholar] [CrossRef]
- Lawless, M.W.; Greene, C.M.; Mulgrew, A.; Taggart, C.C.; O’Neill, S.J.; McElvaney, N.G. Activation of endoplasmic reticulum-specific stress responses associated with the conformational disease Z alpha 1-antitrypsin deficiency. J. Immunol. 2004, 172, 5722–5726. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Tsyganov, K.; Perry, A.J.; Archer, S.K.; Powell, D. RNAsik: A Pipeline for complete and reproducible RNA-seq analysis that runs anywhere with speed and ease. J. Open Source Softw. 2018, 3, 583. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cobanoglu, M.C.; Li, J.; Hidvegi, T.; Hale, P.; Ewing, M.; Chu, A.S.; Gong, Z.; Muzumdar, R.; Pak, S.C.; et al. An analog of glibenclamide selectively enhances autophagic degradation of misfolded alpha1-antitrypsin Z. PLoS ONE 2019, 14, e0209748. [Google Scholar] [CrossRef]
- Choi, S.M.; Kim, Y.; Shim, J.S.; Park, J.T.; Wang, R.H.; Leach, S.D.; Liu, J.O.; Deng, C.; Ye, Z.; Jang, Y.Y. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology 2013, 57, 2458–2468. [Google Scholar] [CrossRef]
- Karatas, E.; Raymond, A.A.; Leon, C.; Dupuy, J.W.; Di-Tommaso, S.; Senant, N.; Collardeau-Frachon, S.; Ruiz, M.; Lachaux, A.; Saltel, F.; et al. Hepatocyte proteomes reveal the role of protein disulfide isomerase 4 in alpha 1-antitrypsin deficiency. JHEP Rep. 2021, 3, 100297. [Google Scholar] [CrossRef] [PubMed]
- Daemen, S.; Kutmon, M.; Evelo, C.T. A pathway approach to investigate the function and regulation of SREBPs. Genes. Nutr. 2013, 8, 289–300. [Google Scholar] [CrossRef]
- Horton, J.D.; Shimomura, I.; Brown, M.S.; Hammer, R.E.; Goldstein, J.L.; Shimano, H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Investig. 1998, 101, 2331–2339. [Google Scholar] [CrossRef]
- Lee, J.N.; Song, B.; DeBose-Boyd, R.A.; Ye, J. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J. Biol. Chem. 2006, 281, 39308–39315. [Google Scholar] [CrossRef]
- Amano, T.; Yamasaki, S.; Yagishita, N.; Tsuchimochi, K.; Shin, H.; Kawahara, K.; Aratani, S.; Fujita, H.; Zhang, L.; Ikeda, R.; et al. Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev. 2003, 17, 2436–2449. [Google Scholar] [CrossRef]
- Francisco, A.B.; Singh, R.; Li, S.; Vani, A.K.; Yang, L.; Munroe, R.J.; Diaferia, G.; Cardano, M.; Biunno, I.; Qi, L.; et al. Deficiency of suppressor enhancer Lin12 1 like (SEL1L) in mice leads to systemic endoplasmic reticulum stress and embryonic lethality. J. Biol. Chem. 2010, 285, 13694–13703. [Google Scholar] [CrossRef]
- Eura, Y.; Yanamoto, H.; Arai, Y.; Okuda, T.; Miyata, T.; Kokame, K. Derlin-1 deficiency is embryonic lethal, Derlin-3 deficiency appears normal, and Herp deficiency is intolerant to glucose load and ischemia in mice. PLoS ONE 2012, 7, e34298. [Google Scholar] [CrossRef]
- Christianson, J.C.; Shaler, T.A.; Tyler, R.E.; Kopito, R.R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008, 10, 272–282. [Google Scholar] [CrossRef]
- Hosokawa, N.; Wada, I.; Nagasawa, K.; Moriyama, T.; Okawa, K.; Nagata, K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 2008, 283, 20914–20924. [Google Scholar] [CrossRef]
- Hosokawa, N.; Kamiya, Y.; Kamiya, D.; Kato, K.; Nagata, K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J. Biol. Chem. 2009, 284, 17061–17068. [Google Scholar] [CrossRef]
- Yamaguchi, D.; Hu, D.; Matsumoto, N.; Yamamoto, K. Human XTP3-B binds to alpha1-antitrypsin variant null(Hong Kong) via the C-terminal MRH domain in a glycan-dependent manner. Glycobiology 2010, 20, 348–355. [Google Scholar] [CrossRef]
- van der Goot, A.T.; Pearce, M.M.P.; Leto, D.E.; Shaler, T.A.; Kopito, R.R. Redundant and Antagonistic Roles of XTP3B and OS9 in Decoding Glycan and Non-glycan Degrons in ER-Associated Degradation. Mol. Cell 2018, 70, 516–530. [Google Scholar] [CrossRef]
- Joo, J.H.; Wang, B.; Frankel, E.; Ge, L.; Xu, L.; Iyengar, R.; Li-Harms, X.; Wright, C.; Shaw, T.I.; Lindsten, T.; et al. The Noncanonical Role of ULK/ATG1 in ER-to-Golgi Trafficking Is Essential for Cellular Homeostasis. Mol. Cell 2016, 62, 491–506. [Google Scholar] [CrossRef] [PubMed]
- Rajak, S.; Iannucci, L.F.; Zhou, J.; Anjum, B.; George, N.; Singh, B.K.; Ghosh, S.; Yen, P.M.; Sinha, R.A. Loss of ULK1 Attenuates Cholesterogenic Gene Expression in Mammalian Hepatic Cells. Front. Cell Dev. Biol. 2020, 8, 523550. [Google Scholar] [CrossRef]
- Li, T.Y.; Sun, Y.; Liang, Y.; Liu, Q.; Shi, Y.; Zhang, C.S.; Zhang, C.; Song, L.; Zhang, P.; Zhang, X.; et al. ULK1/2 Constitute a Bifurcate Node Controlling Glucose Metabolic Fluxes in Addition to Autophagy. Mol. Cell. 2016, 62, 359–370. [Google Scholar] [CrossRef]
- Hidvegi, T.; Mirnics, K.; Hale, P.; Ewing, M.; Beckett, C.; Perlmutter, D.H. Regulator of G Signaling 16 is a marker for the distinct endoplasmic reticulum stress state associated with aggregated mutant alpha1-antitrypsin Z in the classical form of alpha1-antitrypsin deficiency. J. Biol. Chem. 2007, 282, 27769–27780. [Google Scholar] [CrossRef]
- Khodayari, N.; Wang, R.L.; Oshins, R.; Lu, Y.; Millett, M.; Aranyos, A.M.; Mostofizadeh, S.; Scindia, Y.; Flagg, T.O.; Brantly, M. The Mechanism of Mitochondrial Injury in Alpha-1 Antitrypsin Deficiency Mediated Liver Disease. Int. J. Mol. Sci. 2021, 22, 13255. [Google Scholar] [CrossRef]
- Davis, R.L.; Holohan, P.D.; Shrimpton, A.E.; Tatum, A.H.; Daucher, J.; Collins, G.H.; Todd, R.; Bradshaw, C.; Kent, P.; Feiglin, D.; et al. Familial encephalopathy with neuroserpin inclusion bodies. Am. J. Pathol. 1999, 155, 1901–1913. [Google Scholar] [CrossRef] [PubMed]
- Roussel, B.D.; Newton, T.M.; Malzer, E.; Simecek, N.; Haq, I.; Thomas, S.E.; Burr, M.L.; Lehner, P.J.; Crowther, D.C.; Marciniak, S.J.; et al. Sterol metabolism regulates neuroserpin polymer degradation in the absence of the unfolded protein response in the dementia FENIB. Hum. Mol. Genet. 2013, 22, 4616–4626. [Google Scholar] [CrossRef] [PubMed]
- Menzies, S.A.; Volkmar, N.; van den Boomen, D.J.; Timms, R.T.; Dickson, A.S.; Nathan, J.A.; Lehner, P.J. The sterol-responsive RNF145 E3 ubiquitin ligase mediates the degradation of HMG-CoA reductase together with gp78 and Hrd1. eLife 2018, 7, e40009. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Yang, H.; Song, B.L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef]
- Pan, S.; Wang, S.; Utama, B.; Huang, L.; Blok, N.; Estes, M.K.; Moremen, K.W.; Sifers, R.N. Golgi localization of ERManI defines spatial separation of the mammalian glycoprotein quality control system. Mol. Biol. Cell 2011, 22, 2810–2822. [Google Scholar] [CrossRef]
- Pan, S.; Cheng, X.; Sifers, R.N. Golgi-situated endoplasmic reticulum alpha-1, 2-mannosidase contributes to the retrieval of ERAD substrates through a direct interaction with gamma-COP. Mol. Biol. Cell 2013, 24, 1111–1121. [Google Scholar] [CrossRef]
- Iannotti, M.J.; Figard, L.; Sokac, A.M.; Sifers, R.N. A Golgi-localized mannosidase (MAN1B1) plays a non-enzymatic gatekeeper role in protein biosynthetic quality control. J. Biol. Chem. 2014, 289, 11844–11858. [Google Scholar] [CrossRef]
- Sun, A.H.; Collette, J.R.; Sifers, R.N. The cytoplasmic tail of human mannosidase Man1b1 contributes to catalysis-independent quality control of misfolded alpha1-antitrypsin. Proc. Natl. Acad. Sci. USA 2020, 117, 24825–24836. [Google Scholar] [CrossRef]
- Olivari, S.; Cali, T.; Salo, K.E.; Paganetti, P.; Ruddock, L.W.; Molinari, M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 2006, 349, 1278–1284. [Google Scholar] [CrossRef]
- Ninagawa, S.; Okada, T.; Sumitomo, Y.; Horimoto, S.; Sugimoto, T.; Ishikawa, T.; Takeda, S.; Yamamoto, T.; Suzuki, T.; Kamiya, Y.; et al. Forcible destruction of severely misfolded mammalian glycoproteins by the non-glycoprotein ERAD pathway. J. Cell Biol. 2015, 211, 775–784. [Google Scholar] [CrossRef]
- Ninagawa, S.; Okada, T.; Sumitomo, Y.; Kamiya, Y.; Kato, K.; Horimoto, S.; Ishikawa, T.; Takeda, S.; Sakuma, T.; Yamamoto, T.; et al. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J. Cell Biol. 2014, 206, 347–356. [Google Scholar] [CrossRef]
- George, G.; Ninagawa, S.; Yagi, H.; Furukawa, J.I.; Hashii, N.; Ishii-Watabe, A.; Deng, Y.; Matsushita, K.; Ishikawa, T.; Mamahit, Y.P.; et al. Purified EDEM3 or EDEM1 alone produces determinant oligosaccharide structures from M8B in mammalian glycoprotein ERAD. eLife 2021, 10, e70357. [Google Scholar] [CrossRef]
- Jang, B.Y.; Ryoo, H.D.; Son, J.; Choi, K.C.; Shin, D.M.; Kang, S.W.; Kang, M.J. Role of Drosophila EDEMs in the degradation of the alpha-1-antitrypsin Z variant. Int. J. Mol. Med. 2015, 35, 870–876. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, L.R.; Lee, J.; Mohammad, N.S.; Aranyos, A.M.; Gould, C.; Khodayari, N.; Oshins, R.A.; Moneypenny, C.G.; Brantly, M.L. The unfolded protein response to PI*Z alpha-1 antitrypsin in human hepatocellular and murine models. Hepatol. Commun. 2022, 6, 2354–2367. [Google Scholar] [CrossRef]
- Attanasio, S.; Ferriero, R.; Gernoux, G.; De Cegli, R.; Carissimo, A.; Nusco, E.; Campione, S.; Teckman, J.; Mueller, C.; Piccolo, P.; et al. CHOP and c-JUN up-regulate the mutant Z alpha1-antitrypsin, exacerbating its aggregation and liver proteotoxicity. J. Biol. Chem. 2020, 295, 13213–13223. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Garcia-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fung, C.; Wilding, B.; Schittenhelm, R.B.; Bryson-Richardson, R.J.; Bird, P.I. Expression of the Z Variant of α1-Antitrypsin Suppresses Hepatic Cholesterol Biosynthesis in Transgenic Zebrafish. Int. J. Mol. Sci. 2023, 24, 2475. https://doi.org/10.3390/ijms24032475
Fung C, Wilding B, Schittenhelm RB, Bryson-Richardson RJ, Bird PI. Expression of the Z Variant of α1-Antitrypsin Suppresses Hepatic Cholesterol Biosynthesis in Transgenic Zebrafish. International Journal of Molecular Sciences. 2023; 24(3):2475. https://doi.org/10.3390/ijms24032475
Chicago/Turabian StyleFung, Connie, Brendan Wilding, Ralf B. Schittenhelm, Robert J. Bryson-Richardson, and Phillip I. Bird. 2023. "Expression of the Z Variant of α1-Antitrypsin Suppresses Hepatic Cholesterol Biosynthesis in Transgenic Zebrafish" International Journal of Molecular Sciences 24, no. 3: 2475. https://doi.org/10.3390/ijms24032475
APA StyleFung, C., Wilding, B., Schittenhelm, R. B., Bryson-Richardson, R. J., & Bird, P. I. (2023). Expression of the Z Variant of α1-Antitrypsin Suppresses Hepatic Cholesterol Biosynthesis in Transgenic Zebrafish. International Journal of Molecular Sciences, 24(3), 2475. https://doi.org/10.3390/ijms24032475








