Transposable Elements Co-Option in Genome Evolution and Gene Regulation
Abstract
1. Introduction
2. The Biology of Transposable Elements
2.1. TE Classification and Distribution in the Human Genome
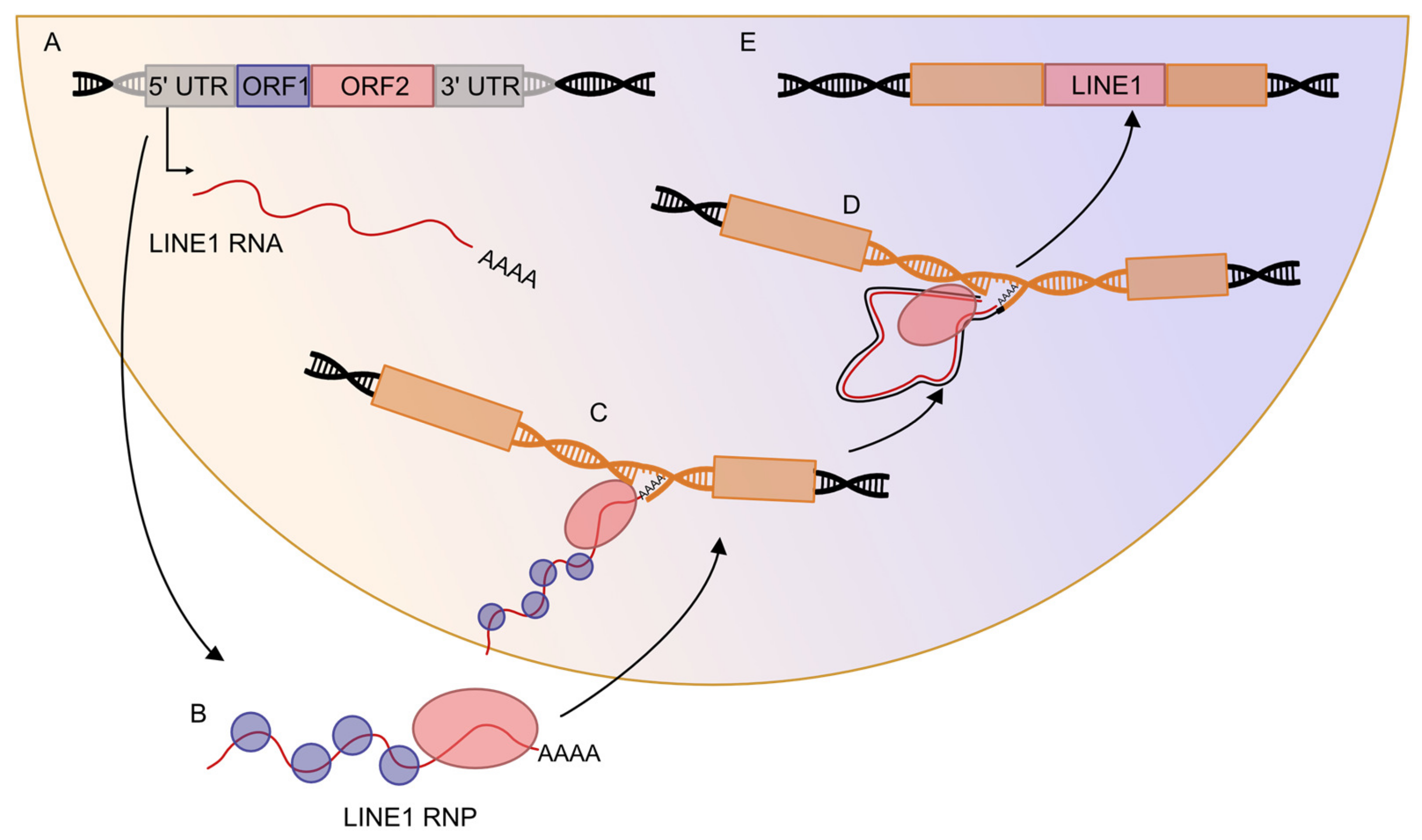
2.2. Retrotransposons Can Mobilize in the Genome
3. How TEs Promote Genome Evolution beyond Retrotransposition
3.1. Horizontal Transposon Transfer Is an Ancestral Mechanism of DNA Motion
3.2. Mobile Elements Induce DNA Structural Rearrangements
4. Transposable Elements Orchestrate Cell Identity and Genome Plasticity
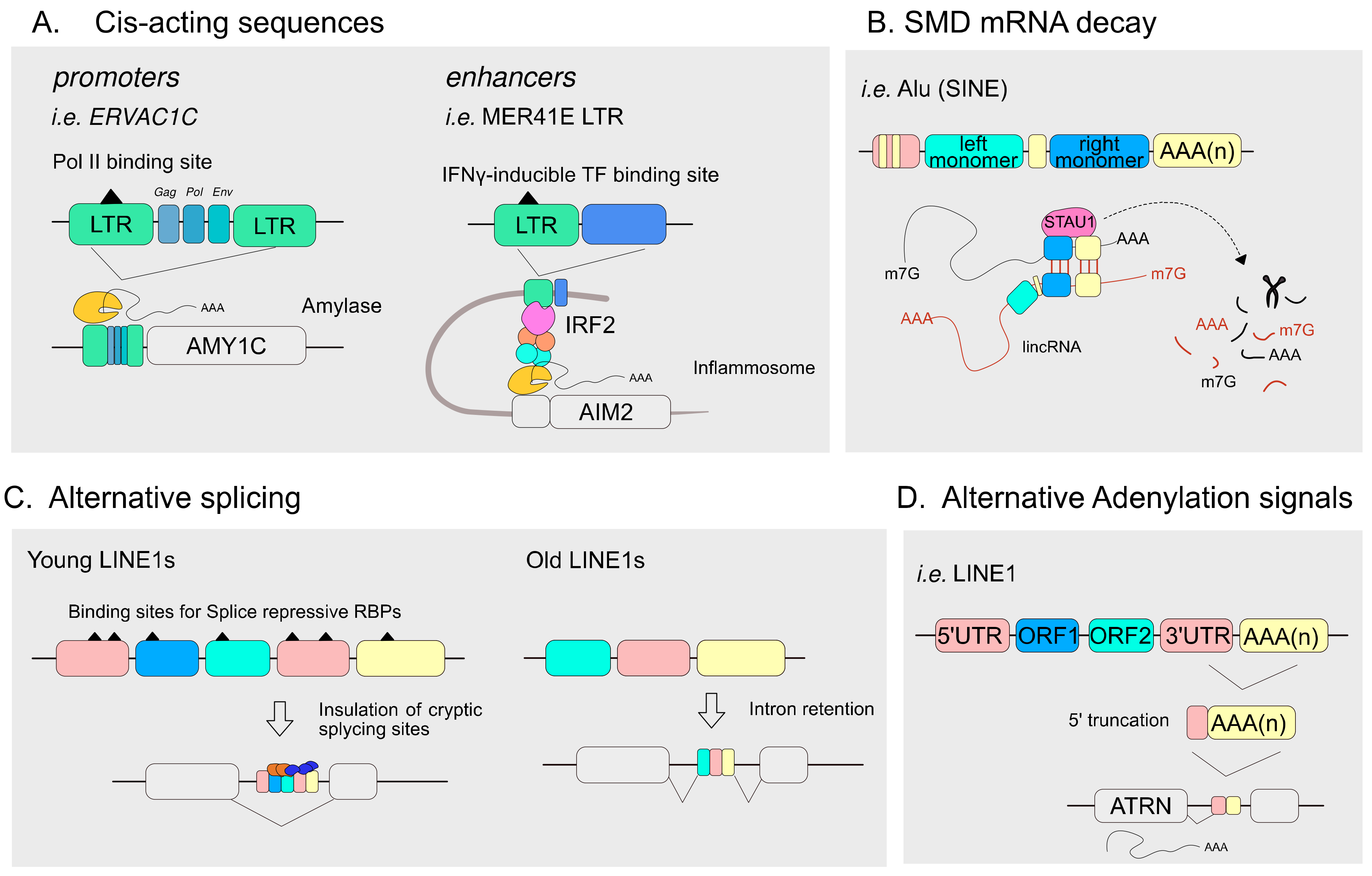
4.1. TEs Articulate the Transcriptional Landscape of Host Genomes
4.2. TEs Guide the Three-Dimensional Genome Organization
5. Linking Deregulation of TE Expression and Activity to Diseases
5.1. Host Genomes Evolved Sophisticated Strategies to Govern TE Expression and Activity
5.2. Deregulated TE Activity and Expression Drives Pathological States
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mc, C.B. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 1951, 16, 13–47. [Google Scholar]
- Mc, C.B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar]
- McClintock, B. Intranuclear systems controlling gene action and mutation. Brookhaven Symp. Biol. 1956, 8, 58–74. [Google Scholar]
- McClintock, B. Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 1956, 21, 197–216. [Google Scholar] [CrossRef]
- Feschotte, C.; Pritham, E.J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Panaud, O. Horizontal transfers of transposable elements in eukaryotes. The flying genes. Comptes Rendus Biol. 2016, 339, 296–299. [Google Scholar] [CrossRef]
- Fueyo, R.; Judd, J.; Feschotte, C.; Wysocka, J. Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell Biol. 2022, 23, 481–497. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The complete sequence of a human genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef]
- Munoz-Lopez, M.; Garcia-Perez, J.L. DNA transposons. Nature and applications in genomics. Curr. Genom. 2010, 11, 115–128. [Google Scholar] [CrossRef]
- Whitcomb, J.M.; Hughes, S.H. Retroviral reverse transcription and integration. Progress and problems. Annu. Rev. Cell Biol. 1992, 8, 275–306. [Google Scholar] [CrossRef]
- Luan, D.D.; Korman, M.H.; Jakubczak, J.L.; Eickbush, T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site. A mechanism for non-LTR retrotransposition. Cell 1993, 72, 595–605. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvak, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008, 9, 411–412. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Piegu, B.; Bire, S.; Arensburger, P.; Bigot, Y. A survey of transposable element classification systems--a call for a fundamental update to meet the challenge of their diversity and complexity. Mol. Phylogenet. Evol. 2015, 86, 90–109. [Google Scholar] [CrossRef]
- Pisano, M.P.; Grandi, N.; Tramontano, E. Human Endogenous Retroviruses (HERVs) and Mammalian Apparent LTRs Retrotransposons (MaLRs) Are Dynamically Modulated in Different Stages of Immunity. Biology 2021, 10, 405. [Google Scholar] [CrossRef]
- Gifford, R.J.; Blomberg, J.; Coffin, J.M.; Fan, H.; Heidmann, T.; Mayer, J.; Stoye, J.; Tristem, M.; Johnson, W.E. Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology 2018, 15, 59. [Google Scholar] [CrossRef]
- Jansz, N.; Faulkner, G.J. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef] [PubMed]
- Marasca, F.; Gasparotto, E.; Polimeni, B.; Vadala, R.; Ranzani, V.; Bodega, B. The Sophisticated Transcriptional Response Governed by Transposable Elements in Human Health and Disease. Int. J. Mol. Sci. 2020, 21, 3201. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.R.; Garcia-Perez, J.L.; Badge, R.M.; Moran, J.V. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum. Genet. 2011, 12, 187–215. [Google Scholar] [CrossRef] [PubMed]
- Bodega, B.; Orlando, V. Repetitive elements dynamics in cell identity programming, maintenance and disease. Curr. Opin. Cell Biol. 2014, 31, 67–73. [Google Scholar] [CrossRef]
- Ade, C.; Roy-Engel, A.M.; Deininger, P.L. Alu elements. An intrinsic source of human genome instability. Curr. Opin. Virol. 2013, 3, 639–645. [Google Scholar] [CrossRef]
- Deininger, P. Alu elements. Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef]
- Hara, T.; Hirai, Y.; Baicharoen, S.; Hayakawa, T.; Hirai, H.; Koga, A. A novel composite retrotransposon derived from or generated independently of the SVA (SINE/VNTR/Alu) transposon has undergone proliferation in gibbon genomes. Genes Genet. Syst. 2012, 87, 181–190. [Google Scholar] [CrossRef]
- Ostertag, E.M.; Goodier, J.L.; Zhang, Y.; Kazazian, H.H., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003, 73, 1444–1451. [Google Scholar] [CrossRef]
- Wang, H.; Xing, J.; Grover, D.; Hedges, D.J.; Han, K.; Walker, J.A.; Batzer, M.A. SVA elements. A hominid-specific retroposon family. J. Mol. Biol. 2005, 354, 994–1007. [Google Scholar] [CrossRef]
- Moran, J.V.; Holmes, S.E.; Naas, T.P.; DeBerardinis, R.J.; Boeke, J.D.; Kazazian, H.H., Jr. High frequency retrotransposition in cultured mammalian cells. Cell 1996, 87, 917–927. [Google Scholar] [CrossRef]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef]
- Martin, S.L. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010, 7, 706–711. [Google Scholar] [CrossRef]
- Feng, Q.; Moran, J.V.; Kazazian, H.H., Jr.; Boeke, J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996, 87, 905–916. [Google Scholar] [CrossRef]
- Mathias, S.L.; Scott, A.F.; Kazazian, H.H., Jr.; Boeke, J.D.; Gabriel, A. Reverse transcriptase encoded by a human transposable element. Science 1991, 254, 1808–1810. [Google Scholar] [CrossRef]
- Richardson, S.R.; Doucet, A.J.; Kopera, H.C.; Moldovan, J.B.; Garcia-Perez, J.L.; Moran, J.V. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiol. Spectr. 2015, 3, MDNA3-0061-2014. [Google Scholar] [CrossRef]
- Wei, W.; Gilbert, N.; Ooi, S.L.; Lawler, J.F.; Ostertag, E.M.; Kazazian, H.H.; Boeke, J.D.; Moran, J.V. Human L1 retrotransposition. Cis preference versus trans complementation. Mol. Cell Biol. 2001, 21, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Mita, P.; Wudzinska, A.; Sun, X.; Andrade, J.; Nayak, S.; Kahler, D.J.; Badri, S.; LaCava, J.; Ueberheide, B.; Yun, C.Y.; et al. LINE-1 protein localization and functional dynamics during the cell cycle. Elife 2018, 7, e30058. [Google Scholar] [CrossRef]
- Jurka, J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. USA 1997, 94, 1872–1877. [Google Scholar] [CrossRef]
- Khadgi, B.B.; Govindaraju, A.; Christensen, S.M. Completion of LINE integration involves an open ‘4-way’ branched DNA intermediate. Nucleic. Acids Res. 2019, 47, 8708–8719. [Google Scholar] [CrossRef]
- Bannert, N.; Kurth, R. Retroelements and the human genome. New perspectives on an old relation. Proc. Natl. Acad. Sci. USA 2004, 101 (Suppl. S2), 14572–14579. [Google Scholar] [CrossRef]
- Shin, W.; Lee, J.; Son, S.Y.; Ahn, K.; Kim, H.S.; Han, K. Human-specific HERV-K insertion causes genomic variations in the human genome. PLoS ONE 2013, 8, e60605. [Google Scholar] [CrossRef]
- Kovalskaya, E.; Buzdin, A.; Gogvadze, E.; Vinogradova, T.; Sverdlov, E. Functional human endogenous retroviral LTR transcription start sites are located between the R and U5 regions. Virology 2006, 346, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.; Wilhelm, F.X. Reverse transcription of retroviruses and LTR retrotransposons. Cell Mol. Life Sci. 2001, 58, 1246–1262. [Google Scholar] [CrossRef] [PubMed]
- Haig, D. Transposable elements. Self-seekers of the germline, team-players of the soma. Bioessays 2016, 38, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, G.; Singh, M.; Ghanbarian, A.T.; Rasko, T.; Szvetnik, A.; Cai, H.; Besser, D.; Prigione, A.; Fuchs, N.V.; et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 2014, 516, 405–409. [Google Scholar] [CrossRef]
- Gerdes, P.; Richardson, S.R.; Mager, D.L.; Faulkner, G.J. Transposable elements in the mammalian embryo. Pioneers surviving through stealth and service. Genome Biol. 2016, 17, 100. [Google Scholar] [CrossRef]
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Yeo, G.W.; Mu, Y.; Lovci, M.T.; Morell, M.; O’Shea, K.S.; Moran, J.V.; Gage, F.H. L1 retrotransposition in human neural progenitor cells. Nature 2009, 460, 1127–1131. [Google Scholar] [CrossRef]
- Baillie, J.K.; Barnett, M.W.; Upton, K.R.; Gerhardt, D.J.; Richmond, T.A.; De Sapio, F.; Brennan, P.M.; Rizzu, P.; Smith, S.; Fell, M.; et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 2011, 479, 534–537. [Google Scholar] [CrossRef]
- Ravel-Godreuil, C.; Znaidi, R.; Bonnifet, T.; Joshi, R.L.; Fuchs, J. Transposable elements as new players in neurodegenerative diseases. FEBS Lett. 2021, 595, 2733–2755. [Google Scholar] [CrossRef]
- Hancks, D.C.; Kazazian, H.H., Jr. Roles for retrotransposon insertions in human disease. Mob. DNA 2016, 7, 9. [Google Scholar] [CrossRef]
- Sturm, A.; Ivics, Z.; Vellai, T. The mechanism of ageing. Primary role of transposable elements in genome disintegration. Cell Mol. Life Sci. 2015, 72, 1839–1847. [Google Scholar] [CrossRef]
- Jonsson, M.E.; Garza, R.; Johansson, P.A.; Jakobsson, J. Transposable Elements. A Common Feature of Neurodevelopmental and Neurodegenerative Disorders. Trends Genet. 2020, 36, 610–623. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Wang, H.; Belancio, V.P.; Cordaux, R.; Deininger, P.L.; Batzer, M.A. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc. Natl. Acad. Sci. USA 2006, 103, 17608–17613. [Google Scholar] [CrossRef]
- Gilbert, C.; Feschotte, C. Horizontal acquisition of transposable elements and viral sequences. Patterns and consequences. Curr. Opin. Genet. Dev. 2018, 49, 15–24. [Google Scholar] [CrossRef]
- Wallau, G.L.; Ortiz, M.F.; Loreto, E.L. Horizontal transposon transfer in eukarya. Detection, bias, and perspectives. Genome Biol. Evol. 2012, 4, 689–699. [Google Scholar] [CrossRef]
- Wallau, G.L.; Vieira, C.; Loreto, E.L.S. Genetic exchange in eukaryotes through horizontal transfer. Connected by the mobilome. Mob. DNA 2018, 9, 6. [Google Scholar] [CrossRef]
- Silva, J.C.; Kidwell, M.G. Horizontal transfer and selection in the evolution of P elements. Mol. Biol. Evol. 2000, 17, 1542–1557. [Google Scholar] [CrossRef]
- Ludwig, A.; Valente, V.L.; Loreto, E.L. Multiple invasions of Errantivirus in the genus Drosophila. Insect Mol. Biol. 2008, 17, 113–124. [Google Scholar] [CrossRef]
- Dotto, B.R.; Carvalho, E.L.; da Silva, A.F.; Dezordi, F.Z.; Pinto, P.M.; Campos, T.L.; Rezende, A.M.; Wallau, G.D.L. HTT-DB. New features and updates. Database 2018, 2018, bax102. [Google Scholar] [CrossRef]
- Dotto, B.R.; Carvalho, E.L.; Silva, A.F.; Duarte Silva, L.F.; Pinto, P.M.; Ortiz, M.F.; Wallau, G.L. HTT-DB. Horizontally transferred transposable elements database. Bioinformatics 2015, 31, 2915–2917. [Google Scholar] [CrossRef]
- Wallau, G.L.; Capy, P.; Loreto, E.; Le Rouzic, A.; Hua-Van, A. VHICA, a New Method to Discriminate between Vertical and Horizontal Transposon Transfer. Application to the Mariner Family within Drosophila. Mol. Biol. Evol. 2016, 33, 1094–1109. [Google Scholar] [CrossRef] [PubMed]
- Piskurek, O.; Okada, N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc. Natl. Acad. Sci. USA 2007, 104, 12046–12051. [Google Scholar] [CrossRef] [PubMed]
- Arnold, B.J.; Huang, I.T.; Hanage, W.P. Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 2022, 20, 206–218. [Google Scholar] [CrossRef]
- Gilbert, C.; Chateigner, A.; Ernenwein, L.; Barbe, V.; Bezier, A.; Herniou, E.A.; Cordaux, R. Population genomics supports baculoviruses as vectors of horizontal transfer of insect transposons. Nat. Commun. 2014, 5, 3348. [Google Scholar] [CrossRef]
- Daniels, S.B.; Peterson, K.R.; Strausbaugh, L.D.; Kidwell, M.G.; Chovnick, A. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 1990, 124, 339–355. [Google Scholar] [CrossRef]
- Gilbert, C.; Cordaux, R. Viruses as vectors of horizontal transfer of genetic material in eukaryotes. Curr. Opin. Virol. 2017, 25, 16–22. [Google Scholar] [CrossRef]
- Ivancevic, A.M.; Kortschak, R.D.; Bertozzi, T.; Adelson, D.L. Horizontal transfer of BovB and L1 retrotransposons in eukaryotes. Genome Biol. 2018, 19, 85. [Google Scholar] [CrossRef]
- Kordis, D.; Gubensek, F. Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc. Natl. Acad. Sci. USA 1998, 95, 10704–10709. [Google Scholar] [CrossRef]
- Lin, X.; Faridi, N.; Casola, C. An Ancient Transkingdom Horizontal Transfer of Penelope-Like Retroelements from Arthropods to Conifers. Genome Biol. Evol. 2016, 8, 1252–1266. [Google Scholar]
- Gao, D.; Chu, Y.; Xia, H.; Xu, C.; Heyduk, K.; Abernathy, B.; Ozias-Akins, P.; Leebens-Mack, J.H.; Jackson, S.A. Horizontal Transfer of Non-LTR Retrotransposons from Arthropods to Flowering Plants. Mol. Biol. Evol. 2018, 35, 354–364. [Google Scholar] [CrossRef]
- Langley, C.H.; Montgomery, E.; Hudson, R.; Kaplan, N.; Charlesworth, B. On the role of unequal exchange in the containment of transposable element copy number. Genet. Res. 1988, 52, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Cho, C.S.; Han, K.; Lee, J. Structural Variation of Alu Element and Human Disease. Genom. Inf. 2016, 14, 70–77. [Google Scholar] [CrossRef]
- Lee, J.; Han, K.; Meyer, T.J.; Kim, H.S.; Batzer, M.A. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLoS ONE 2008, 3, e4047. [Google Scholar] [CrossRef]
- Pascarella, G.; Hon, C.C.; Hashimoto, K.; Busch, A.; Luginbuhl, J.; Parr, C.; Hin Yip, W.; Abe, K.; Kratz, A.; Bonetti, A.; et al. Recombination of repeat elements generates somatic complexity in human genomes. Cell 2022, 185, 3025–3040.e3026. [Google Scholar] [CrossRef]
- Erwin, J.A.; Paquola, A.C.; Singer, T.; Gallina, I.; Novotny, M.; Quayle, C.; Bedrosian, T.A.; Alves, F.I.; Butcher, C.R.; Herdy, J.R.; et al. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat. Neurosci. 2016, 19, 1583–1591. [Google Scholar] [CrossRef]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Bejerano, G.; Lowe, C.B.; Ahituv, N.; King, B.; Siepel, A.; Salama, S.R.; Rubin, E.M.; Kent, W.J.; Haussler, D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 2006, 441, 87–90. [Google Scholar] [CrossRef]
- Lanciano, S.; Cristofari, G. Measuring and interpreting transposable element expression. Nat. Rev. Genet. 2020, 21, 721–736. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements. From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Jacques, P.E.; Jeyakani, J.; Bourque, G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 2013, 9, e1003504. [Google Scholar] [CrossRef] [PubMed]
- Novati, A.; Roman, V.; Cetin, T.; Hagewoud, R.; den Boer, J.A.; Luiten, P.G.; Meerlo, P. Chronically restricted sleep leads to depression-like changes in neurotransmitter receptor sensitivity and neuroendocrine stress reactivity in rats. Sleep 2008, 31, 1579–1585. [Google Scholar] [CrossRef]
- Pontis, J.; Pulver, C.; Playfoot, C.J.; Planet, E.; Grun, D.; Offner, S.; Duc, J.; Manfrin, A.; Lutolf, M.P.; Trono, D. Primate-specific transposable elements shape transcriptional networks during human development. Nat. Commun. 2022, 13, 7178. [Google Scholar] [CrossRef]
- Babarinde, I.A.; Ma, G.; Li, Y.; Deng, B.; Luo, Z.; Liu, H.; Abdul, M.M.; Ward, C.; Chen, M.; Fu, X.; et al. Transposable element sequence fragments incorporated into coding and noncoding transcripts modulate the transcriptome of human pluripotent stem cells. Nucleic. Acids Res. 2021, 49, 9132–9153. [Google Scholar] [CrossRef]
- Brosius, J.; Gould, S.J. On “genomenclature”. A comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc. Natl. Acad. Sci. USA 1992, 89, 10706–10710. [Google Scholar] [CrossRef]
- Batut, P.; Dobin, A.; Plessy, C.; Carninci, P.; Gingeras, T.R. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013, 23, 169–180. [Google Scholar] [CrossRef]
- Roman, A.C.; Benitez, D.A.; Carvajal-Gonzalez, J.M.; Fernandez-Salguero, P.M. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 1632–1637. [Google Scholar] [CrossRef]
- Sundaram, V.; Cheng, Y.; Ma, Z.; Li, D.; Xing, X.; Edge, P.; Snyder, M.P.; Wang, T. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014, 24, 1963–1976. [Google Scholar] [CrossRef]
- Attig, J.; Agostini, F.; Gooding, C.; Chakrabarti, A.M.; Singh, A.; Haberman, N.; Zagalak, J.A.; Emmett, W.; Smith, C.W.J.; Luscombe, N.M.; et al. Heteromeric RNP Assembly at LINEs Controls Lineage-Specific RNA Processing. Cell 2018, 174, 1067–1081.e1017. [Google Scholar] [CrossRef]
- Tang, W.; Gunn, T.M.; McLaughlin, D.F.; Barsh, G.S.; Schlossman, S.F.; Duke-Cohan, J.S. Secreted and membrane attractin result from alternative splicing of the human ATRN gene. Proc. Natl. Acad. Sci. USA 2000, 97, 6025–6030. [Google Scholar] [CrossRef]
- Chen, J.; Sun, M.; Hurst, L.D.; Carmichael, G.G.; Rowley, J.D. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 2005, 21, 326–329. [Google Scholar] [CrossRef] [PubMed]
- Mager, D.L.; Stoye, J.P. Mammalian Endogenous Retroviruses. Microbiol. Spectr. 2015, 3, MDNA3-0009-2014. [Google Scholar] [CrossRef]
- Ostertag, E.M.; Kazazian, H.H., Jr. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001, 35, 501–538. [Google Scholar] [CrossRef] [PubMed]
- Speek, M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell Biol. 2001, 21, 1973–1985. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Hou, Y.; Maeng, J.H.; Shah, N.M.; Chen, Y.; Lawson, H.A.; Yang, H.; Yue, F.; Wang, T. Epigenomic analysis reveals prevalent contribution of transposable elements to cis-regulatory elements, tissue-specific expression, and alternative promoters in zebrafish. Genome Res. 2022, 32, 1424–1436. [Google Scholar] [CrossRef]
- Faulkner, G.J.; Kimura, Y.; Daub, C.O.; Wani, S.; Plessy, C.; Irvine, K.M.; Schroder, K.; Cloonan, N.; Steptoe, A.L.; Lassmann, T.; et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009, 41, 563–571. [Google Scholar] [CrossRef]
- Ting, C.N.; Rosenberg, M.P.; Snow, C.M.; Samuelson, L.C.; Meisler, M.H. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 1992, 6, 1457–1465. [Google Scholar] [CrossRef]
- Emera, D.; Wagner, G.P. Transformation of a transposon into a derived prolactin promoter with function during human pregnancy. Proc. Natl. Acad. Sci. USA 2012, 109, 11246–11251. [Google Scholar] [CrossRef]
- Jang, H.S.; Shah, N.M.; Du, A.Y.; Dailey, Z.Z.; Pehrsson, E.C.; Godoy, P.M.; Zhang, D.; Li, D.; Xing, X.; Kim, S.; et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet. 2019, 51, 611–617. [Google Scholar] [CrossRef]
- Babaian, A.; Romanish, M.T.; Gagnier, L.; Kuo, L.Y.; Karimi, M.M.; Steidl, C.; Mager, D.L. Onco-exaptation of an endogenous retroviral LTR drives IRF5 expression in Hodgkin lymphoma. Oncogene 2016, 35, 2542–2546. [Google Scholar] [CrossRef]
- Senft, A.D.; Macfarlan, T.S. Transposable elements shape the evolution of mammalian development. Nat. Rev. Genet. 2021, 22, 691–711. [Google Scholar] [CrossRef]
- Nishihara, H. Retrotransposons spread potential cis-regulatory elements during mammary gland evolution. Nucleic Acids Res. 2019, 47, 11551–11562. [Google Scholar] [CrossRef] [PubMed]
- Notwell, J.H.; Chung, T.; Heavner, W.; Bejerano, G. A family of transposable elements co-opted into developmental enhancers in the mouse neocortex. Nat. Commun. 2015, 6, 6644. [Google Scholar] [CrossRef]
- Sasaki, T.; Nishihara, H.; Hirakawa, M.; Fujimura, K.; Tanaka, M.; Kokubo, N.; Kimura-Yoshida, C.; Matsuo, I.; Sumiyama, K.; Saitou, N.; et al. Possible involvement of SINEs in mammalian-specific brain formation. Proc. Natl. Acad. Sci. USA 2008, 105, 4220–4225. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef]
- Judd, J.; Sanderson, H.; Feschotte, C. Evolution of mouse circadian enhancers from transposable elements. Genome Biol. 2021, 22, 193. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Xia, B.; Zhang, W.; Wudzinska, A.; Huang, E.; Brosh, R.; Pour, M.; Miller, A.; Jeremy, S.; Matthew, T.M.; Sang, Y.; et al. The genetic basis of tail-loss evolution in humans and apes. BioRxiv 2021. [Google Scholar] [CrossRef]
- Pinson, M.E.; Court, F.; Masson, A.; Renaud, Y.; Fantini, A.; Bacoeur-Ouzillou, O.; Barriere, M.; Pereira, B.; Guichet, P.O.; Chautard, E.; et al. L1 chimeric transcripts are expressed in healthy brain and their deregulation in glioma follows that of their host locus. Hum. Mol. Genet. 2022, 31, 2606–2622. [Google Scholar] [CrossRef]
- Percharde, M.; Lin, C.J.; Yin, Y.; Guan, J.; Peixoto, G.A.; Bulut-Karslioglu, A.; Biechele, S.; Huang, B.; Shen, X.; Ramalho-Santos, M. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell 2018, 174, 391–405.e319. [Google Scholar] [CrossRef]
- Marasca, F.; Sinha, S.; Vadala, R.; Polimeni, B.; Ranzani, V.; Paraboschi, E.M.; Burattin, F.V.; Ghilotti, M.; Crosti, M.; Negri, M.L.; et al. LINE1 are spliced in non-canonical transcript variants to regulate T cell quiescence and exhaustion. Nat. Genet. 2022, 54, 180–193. [Google Scholar] [CrossRef]
- Schmidt, D.; Schwalie, P.C.; Wilson, M.D.; Ballester, B.; Goncalves, A.; Kutter, C.; Brown, G.D.; Marshall, A.; Flicek, P.; Odom, D.T. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 2012, 148, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Lunyak, V.V.; Prefontaine, G.G.; Nunez, E.; Cramer, T.; Ju, B.G.; Ohgi, K.A.; Hutt, K.; Roy, R.; Garcia-Diaz, A.; Zhu, X.; et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 2007, 317, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Bourque, G.; Leong, B.; Vega, V.B.; Chen, X.; Lee, Y.L.; Srinivasan, K.G.; Chew, J.L.; Ruan, Y.; Wei, C.L.; Ng, H.H.; et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008, 18, 1752–1762. [Google Scholar] [CrossRef]
- Kaaij, L.J.T.; Mohn, F.; van der Weide, R.H.; de Wit, E.; Buhler, M. The ChAHP Complex Counteracts Chromatin Looping at CTCF Sites that Emerged from SINE Expansions in Mouse. Cell 2019, 178, 1437–1451.e1414. [Google Scholar] [CrossRef]
- Hall, L.L.; Carone, D.M.; Gomez, A.V.; Kolpa, H.J.; Byron, M.; Mehta, N.; Fackelmayer, F.O.; Lawrence, J.B. Stable C0T-1 Repeat RNA Is Abundant and Is Associated with Euchromatic Interphase Chromosomes. Cell 2014, 156, 907–919. [Google Scholar] [CrossRef]
- Jachowicz, J.W.; Bing, X.; Pontabry, J.; Boskovic, A.; Rando, O.J.; Torres-Padilla, M.E. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 2017, 49, 1502–1510. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, T.; Preissl, S.; Amaral, M.L.; Grinstein, J.D.; Farah, E.N.; Destici, E.; Qiu, Y.; Hu, R.; Lee, A.Y.; et al. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat. Genet. 2019, 51, 1380–1388. [Google Scholar] [CrossRef]
- Lu, J.Y.; Shao, W.; Chang, L.; Yin, Y.; Li, T.; Zhang, H.; Hong, Y.; Percharde, M.; Guo, L.; Wu, Z.; et al. Genomic Repeats Categorize Genes with Distinct Functions for Orchestrated Regulation. Cell Rep. 2020, 30, 3296–3311.e3295. [Google Scholar] [CrossRef]
- Henikoff, S.; Matzke, M.A. Exploring and explaining epigenetic effects. Trends Genet. 1997, 13, 293–295. [Google Scholar] [CrossRef]
- Huda, A.; Jordan, I.K. Epigenetic regulation of Mammalian genomes by transposable elements. Ann. N. Y. Acad. Sci. 2009, 1178, 276–284. [Google Scholar] [CrossRef]
- Haws, S.A.; Simandi, Z.; Barnett, R.J.; Phillips-Cremins, J.E. 3D genome, on repeat. Higher-order folding principles of the heterochromatinized repetitive genome. Cell 2022, 185, 2690–2707. [Google Scholar] [CrossRef] [PubMed]
- Frank, L.; Rippe, K. Repetitive RNAs as Regulators of Chromatin-Associated Subcompartment Formation by Phase Separation. J. Mol. Biol. 2020, 432, 4270–4286. [Google Scholar] [CrossRef]
- Asimi, V.; Sampath Kumar, A.; Niskanen, H.; Riemenschneider, C.; Hetzel, S.; Naderi, J.; Fasching, N.; Popitsch, N.; Du, M.; Kretzmer, H.; et al. Hijacking of transcriptional condensates by endogenous retroviruses. Nat. Genet. 2022, 54, 1238–1247. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H., Jr.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370. [Google Scholar] [CrossRef]
- Gasior, S.L.; Wakeman, T.P.; Xu, B.; Deininger, P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006, 357, 1383–1393. [Google Scholar] [CrossRef]
- Bourgeois, Y.; Boissinot, S. On the Population Dynamics of Junk. A Review on the Population Genomics of Transposable Elements. Genes 2019, 10, 419. [Google Scholar] [CrossRef]
- Kato, Y.; Kaneda, M.; Hata, K.; Kumaki, K.; Hisano, M.; Kohara, Y.; Okano, M.; Li, E.; Nozaki, M.; Sasaki, H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 2007, 16, 2272–2280. [Google Scholar] [CrossRef]
- Liu, S.; Brind’Amour, J.; Karimi, M.M.; Shirane, K.; Bogutz, A.; Lefebvre, L.; Sasaki, H.; Shinkai, Y.; Lorincz, M.C. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2014, 28, 2041–2055. [Google Scholar] [CrossRef]
- Bulut-Karslioglu, A.; De La Rosa-Velázquez, I.A.; Ramirez, F.; Barenboim, M.; Onishi-Seebacher, M.; Arand, J.; Galán, C.; Winter, G.E.; Engist, B.; Gerle, B.; et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell 2014, 55, 277–290. [Google Scholar] [CrossRef]
- Turelli, P.; Castro-Diaz, N.; Marzetta, F.; Kapopoulou, A.; Raclot, C.; Duc, J.; Tieng, V.; Quenneville, S.; Trono, D. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014, 24, 1260–1270. [Google Scholar] [CrossRef]
- Jacobs, F.M.; Greenberg, D.; Nguyen, N.; Haeussler, M.; Ewing, A.D.; Katzman, S.; Paten, B.; Salama, S.R.; Haussler, D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 2014, 516, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Sachidanandam, R.; Bourc’his, D.; Schaefer, C.; Pezic, D.; Toth, K.F.; Bestor, T.; Hannon, G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 2008, 31, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Totoki, Y.; Toyoda, A.; Ikawa, M.; Asada, N.; Kojima, K.; Yamaguchi, Y.; Ijiri, T.W.; et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008, 22, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs. The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Muotri, A.R.; Chu, V.T.; Marchetto, M.C.; Deng, W.; Moran, J.V.; Gage, F.H. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 2005, 435, 903–910. [Google Scholar] [CrossRef]
- Kuwabara, T.; Hsieh, J.; Muotri, A.; Yeo, G.; Warashina, M.; Lie, D.C.; Moore, L.; Nakashima, K.; Asashima, M.; Gage, F.H. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 2009, 12, 1097–1105. [Google Scholar] [CrossRef]
- Erwin, J.A.; Marchetto, M.C.; Gage, F.H. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat. Rev. Neurosci. 2014, 15, 497–506. [Google Scholar] [CrossRef]
- Richardson, S.R.; Morell, S.; Faulkner, G.J. L1 retrotransposons and somatic mosaicism in the brain. Annu. Rev. Genet. 2014, 48, 1–27. [Google Scholar] [CrossRef]
- Fontana, C.; Marasca, F.; Provitera, L.; Mancinelli, S.; Pesenti, N.; Sinha, S.; Passera, S.; Abrignani, S.; Mosca, F.; Lodato, S.; et al. Early maternal care restores LINE-1 methylation and enhances neurodevelopment in preterm infants. BMC Med. 2021, 19, 42. [Google Scholar] [CrossRef]
- Payer, L.M.; Burns, K.H. Transposable elements in human genetic disease. Nat. Rev. Genet. 2019, 20, 760–772. [Google Scholar] [CrossRef]
- Kazazian, H.H., Jr.; Wong, C.; Youssoufian, H.; Scott, A.F.; Phillips, D.G.; Antonarakis, S.E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 1988, 332, 164–166. [Google Scholar] [CrossRef]
- Bouchet, C.; Vuillaumier-Barrot, S.; Gonzales, M.; Boukari, S.; Bizec, C.L.; Fallet, C.; Delezoide, A.L.; Moirot, H.; Laquerriere, A.; Encha-Razavi, F.; et al. Detection of an Alu insertion in the POMT1 gene from three French Walker Warburg syndrome families. Mol. Genet. Metab. 2007, 90, 93–96. [Google Scholar] [CrossRef]
- Meischl, C.; Boer, M.; Ahlin, A.; Roos, D. A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur. J. Hum. Genet. 2000, 8, 697–703. [Google Scholar] [CrossRef]
- Miki, Y.; Nishisho, I.; Horii, A.; Miyoshi, Y.; Utsunomiya, J.; Kinzler, K.W.; Vogelstein, B.; Nakamura, Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992, 52, 643–645. [Google Scholar]
- Scott, E.C.; Gardner, E.J.; Masood, A.; Chuang, N.T.; Vertino, P.M.; Devine, S.E. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016, 26, 745–755. [Google Scholar] [CrossRef]
- Helman, E.; Lawrence, M.S.; Stewart, C.; Sougnez, C.; Getz, G.; Meyerson, M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014, 24, 1053–1063. [Google Scholar] [CrossRef]
- Macciardi, F.; Giulia Bacalini, M.; Miramontes, R.; Boattini, A.; Taccioli, C.; Modenini, G.; Malhas, R.; Anderlucci, L.; Gusev, Y.; Gross, T.J.; et al. A retrotransposon storm marks clinical phenoconversion to late-onset Alzheimer’s disease. Geroscience 2022, 44, 1525–1550. [Google Scholar] [CrossRef]
- Blaudin de The, F.X.; Rekaik, H.; Peze-Heidsieck, E.; Massiani-Beaudoin, O.; Joshi, R.L.; Fuchs, J.; Prochiantz, A. Engrailed homeoprotein blocks degeneration in adult dopaminergic neurons through LINE-1 repression. EMBO J. 2018, 37, e97374. [Google Scholar] [CrossRef]
- Li, W.; Jin, Y.; Prazak, L.; Hammell, M.; Dubnau, J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS ONE 2012, 7, e44099. [Google Scholar] [CrossRef]
- Guffanti, G.; Gaudi, S.; Fallon, J.H.; Sobell, J.; Potkin, S.G.; Pato, C.; Macciardi, F. Transposable elements and psychiatric disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2014, 165, 201–216. [Google Scholar] [CrossRef]
- Karlsson, H.; Bachmann, S.; Schroder, J.; McArthur, J.; Torrey, E.F.; Yolken, R.H. Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with schizophrenia. Proc. Natl. Acad. Sci. USA 2001, 98, 4634–4639. [Google Scholar] [CrossRef] [PubMed]
- Yolken, R.H.; Karlsson, H.; Yee, F.; Johnston-Wilson, N.L.; Torrey, E.F. Endogenous retroviruses and schizophrenia. Brain Res. Brain Res. Rev. 2000, 31, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, Q.; Hou, Y.; Jiang, T.; Zong, L.; Wang, Z.; Luo, X.; Liang, W.; Zhao, H.; Ning, Y.; et al. Hypomethylation of LINE-1 elements in schizophrenia and bipolar disorder. J. Psychiatr. Res. 2018, 107, 68–72. [Google Scholar] [CrossRef]
- Doyle, G.A.; Crist, R.C.; Karatas, E.T.; Hammond, M.J.; Ewing, A.D.; Ferraro, T.N.; Hahn, C.G.; Berrettini, W.H. Analysis of LINE-1 Elements in DNA from Postmortem Brains of Individuals with Schizophrenia. Neuropsychopharmacology 2017, 42, 2602–2611. [Google Scholar] [CrossRef] [PubMed]
- Muotri, A.R.; Marchetto, M.C.; Coufal, N.G.; Oefner, R.; Yeo, G.; Nakashima, K.; Gage, F.H. L1 retrotransposition in neurons is modulated by MeCP2. Nature 2010, 468, 443–446. [Google Scholar] [CrossRef]
- Zhao, B.; Wu, Q.; Ye, A.Y.; Guo, J.; Zheng, X.; Yang, X.; Yan, L.; Liu, Q.R.; Hyde, T.M.; Wei, L.; et al. Somatic LINE-1 retrotransposition in cortical neurons and non-brain tissues of Rett patients and healthy individuals. PLoS Genet 2019, 15, e1008043. [Google Scholar] [CrossRef]
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Marchetto, M.C.; Muotri, A.R.; Mu, Y.; Carson, C.T.; Macia, A.; Moran, J.V.; Gage, F.H. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. USA 2011, 108, 20382–20387. [Google Scholar] [CrossRef]
- Mak, M.; Samochowiec, J.; Frydecka, D.; Pelka-Wysiecka, J.; Szmida, E.; Karpinski, P.; Sasiadek, M.M.; Piotrowski, P.; Samochowiec, A.; Misiak, B. First-episode schizophrenia is associated with a reduction of HERV-K methylation in peripheral blood. Psychiatry Res. 2019, 271, 459–463. [Google Scholar] [CrossRef]
- Perron, H.; Hamdani, N.; Faucard, R.; Lajnef, M.; Jamain, S.; Daban-Huard, C.; Sarrazin, S.; LeGuen, E.; Houenou, J.; Delavest, M.; et al. Molecular characteristics of Human Endogenous Retrovirus type-W in schizophrenia and bipolar disorder. Transl. Psychiatry 2012, 2, e201. [Google Scholar] [CrossRef]
- Frank, O.; Giehl, M.; Zheng, C.; Hehlmann, R.; Leib-Mosch, C.; Seifarth, W. Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders. J. Virol. 2005, 79, 10890–10901. [Google Scholar] [CrossRef]
- Bundo, M.; Toyoshima, M.; Okada, Y.; Akamatsu, W.; Ueda, J.; Nemoto-Miyauchi, T.; Sunaga, F.; Toritsuka, M.; Ikawa, D.; Kakita, A.; et al. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron 2014, 81, 306–313. [Google Scholar] [CrossRef]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D.; et al. Author Correction. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019, 572, E5. [Google Scholar] [CrossRef]
- Della Valle, F.; Reddy, P.; Yamamoto, M.; Liu, P.; Saera-Vila, A.; Bensaddek, D.; Zhang, H.; Prieto Martinez, J.; Abassi, L.; Celii, M.; et al. LINE-1 RNA causes heterochromatin erosion and is a target for amelioration of senescent phenotypes in progeroid syndromes. Sci. Transl. Med. 2022, 14, eabl6057. [Google Scholar] [CrossRef]
- Gazquez-Gutierrez, A.; Witteveldt, J.S.R.H.; Macias, S. Sensing of transposable elements by the antiviral innate immune system. RNA 2021, 27, 735–752. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2017, 169, 361. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Du, J.; Peng, Y.; Li, P.; Wang, S.; Wang, Y.; Hou, J.; Kang, J.; Zheng, W.; Hua, S.; et al. LINE1 contributes to autoimmunity through both RIG-I- and MDA5-mediated RNA sensing pathways. J. Autoimmun. 2018, 90, 105–115. [Google Scholar] [CrossRef]
- Takaoka, A.; Wang, Z.; Choi, M.K.; Yanai, H.; Negishi, H.; Ban, T.; Lu, Y.; Miyagishi, M.; Kodama, T.; Honda, K.; et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007, 448, 501–505. [Google Scholar] [CrossRef]
- Sohrab, V.; Lopez-Diaz, C.; Di Pietro, A.; Ma, L.J.; Ayhan, D.H. TEfinder. A Bioinformatics Pipeline for Detecting New Transposable Element Insertion Events in Next-Generation Sequencing Data. Genes 2021, 12, 224. [Google Scholar] [CrossRef]
- Polimeni, B.; Marasca, F.; Ranzani, V.; Bodega, B. IRescue. Uncertainty-aware quantification of transposable elements expression at single cell level. BioRxiv 2022. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gasparotto, E.; Burattin, F.V.; Di Gioia, V.; Panepuccia, M.; Ranzani, V.; Marasca, F.; Bodega, B. Transposable Elements Co-Option in Genome Evolution and Gene Regulation. Int. J. Mol. Sci. 2023, 24, 2610. https://doi.org/10.3390/ijms24032610
Gasparotto E, Burattin FV, Di Gioia V, Panepuccia M, Ranzani V, Marasca F, Bodega B. Transposable Elements Co-Option in Genome Evolution and Gene Regulation. International Journal of Molecular Sciences. 2023; 24(3):2610. https://doi.org/10.3390/ijms24032610
Chicago/Turabian StyleGasparotto, Erica, Filippo Vittorio Burattin, Valeria Di Gioia, Michele Panepuccia, Valeria Ranzani, Federica Marasca, and Beatrice Bodega. 2023. "Transposable Elements Co-Option in Genome Evolution and Gene Regulation" International Journal of Molecular Sciences 24, no. 3: 2610. https://doi.org/10.3390/ijms24032610
APA StyleGasparotto, E., Burattin, F. V., Di Gioia, V., Panepuccia, M., Ranzani, V., Marasca, F., & Bodega, B. (2023). Transposable Elements Co-Option in Genome Evolution and Gene Regulation. International Journal of Molecular Sciences, 24(3), 2610. https://doi.org/10.3390/ijms24032610





