Non-Coding RNAs Regulating Mitochondrial Functions and the Oxidative Stress Response as Putative Targets against Age-Related Macular Degeneration (AMD)
Abstract
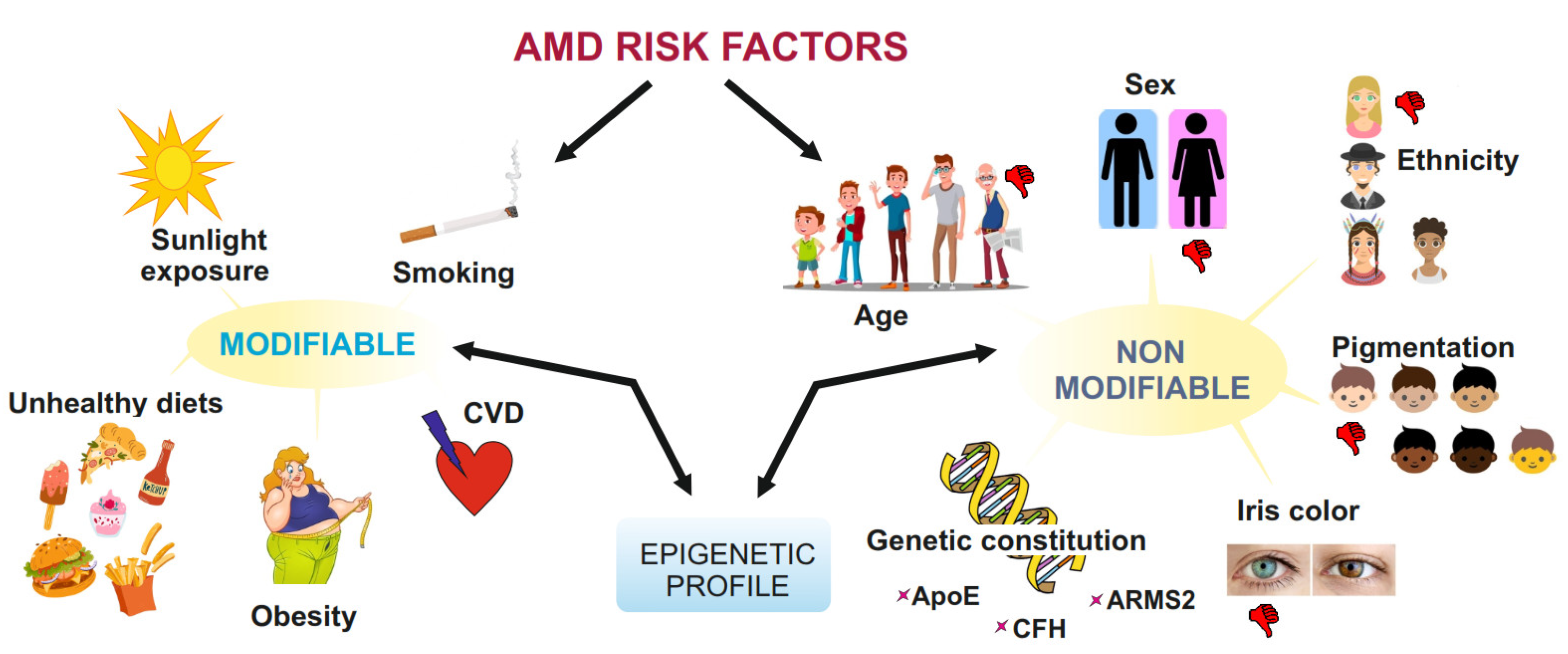
1. Introduction
2. AMD—General
3. Oxidative Stress and Mitochondrial Function in AMD
4. Non-Coding RNAs
4.1. Overview
4.2. Micro-RNAs
4.3. Long Non-Coding RNAs
4.4. Circular Non-Coding RNAs
5. Non-Coding RNAs in Mitochondrial Regulation
5.1. MiRNAs as Bioenergy Regulators in the Mitochondria
| miRNAs | Targets | Effect | Model | References |
|---|---|---|---|---|
| 1 ↑ | MINOS, GPD2, LRPPRC | Mitochondrial damage ↑, mitophagy ↑ | Human breast cancer and melanoma cells | [72] |
| 7 ↑ | KEAP1 | Antioxidant response ↑ | Human neuroblastoma cells | [73] |
| 9 ↑ | a PGC-1α ↑ | Mitochondrial function ↑ | Human kidney cells | [74] |
| 15b, 16, 95 ↑ | Arl2 | ATP production ↓ | Rat cardiomyocytes | [75] |
| 17, 18a, 19a/b, 20a, 92 ↑ | MFN1 | Mitochondrial fusion ↓ | Human osteosarcoma cells | [76] |
| 19b-3p, 221-3p, 222-3p ↑ | PPARGC1A | Mitochondrial function ↓ | Human atherosclerotic vessel | [77] |
| 23a ↑ | GLS1 MnSOD | Glutamine metabolism ↓ Mitochondrial function ↓ | Human RPE cells Mouse cardiomyocytes | [78,79] |
| 23a-3p ↑ | PPARGC1A | Mitochondrial function and fatty acid metabolism ↓ | Mouse liver | [80] |
| 24-3p ↑ | KEAP1 | Antioxidant response ↑ | Mouse cardiomyocytes | [81] |
| 26a/b ↑ | COX5a | OXPHOS ↓ | Rat myoblasts, rat | [65] |
| 27a ↑ | NFE2L2 | Antioxidant response ↓ | Human and rat kidney cells | [82] |
| 27a/b ↑ | PINK1 | Oxidative stress ↑ | Human cervical cancer and neuroblastoma cells | [83] |
| 29a/b, 124 ↑ | MCT1 | Pyruvate circulation ↓ | Human and mouse pancreatic cells | [84,85] |
| 33a/b ↑ | CROT | Fatty acid oxidation ↓ | Monkey liver cells | [86] |
| 34a ↑ | NFE2L2 PINK1 | Antioxidant response ↓ Mitophagy ↓ | Neuroblastoma cells Human kidney cells, mouse | [87,88] |
| 34b/c ↓ | a Parkin ↓, DJ-1 ↓ | Mitochondrial function ↓ | Parkinson’s disease human tissue | [89] |
| 98 ↓ | Hey2 (Notch signaling) | Oxidative stress ↑, mitochondrial function ↓, apoptosis ↓, and viability ↓ | Alzheimer’s disease mouse model | [90] |
| 101 ↑ | PRDM16 | Mitochondrial function ↓, apoptosis ↑ | Human astrocytoma cells, in silico | [91] |
| 130-3p ↑ | PPARGC1A | Mitochondrial function ↓, TFAM ↓ | Human placental cells | [92] |
| 142, 144, 153 ↑ | NFE2L2 | Antioxidant response ↓ | Human neuroblastoma cells | [93] |
| 181a ↑ | PARKIN | Mitophagy ↓ | Human neuroblastoma cells | [94] |
| 181a/b ↑ | NRF1, COX11, COQ10B, PRDX3 | Mitochondrial biogenesis and function ↓ | Mouse retinal neurons | [95] |
| 181c ↑ | COX1 | OXPHOS ↓ | Rat myocytes | [66,67] |
| 204 ↑ | PPARGC1A | Mitochondrial copy number ↓, citrate cycle function ↓, autophagy ↓ | Mouse myoblast cells | [96] |
| 210 ↑ | COX10 | OXPHOS ↓ | Human primary fibroblasts | [69] |
| 210 ↑ | Ephrin-A3 | Tubulogenesis and chemotaxis ↑ | Human umbilical vein and osteosarcoma cells | [97] |
| 338 ↑ | COX4, ATP5G1 | OXPHOS ↓ | Primary rat neuronal cells | [98,99] |
| 494 ↑ | PARK7 | Antioxidant response ↓ | Mouse adipocyte and neuroblastoma cells | [100] |
| 762 ↑ | ND2 | OXPHOS ↓ | Mouse cardiomyocytes | [101] |
5.2. MiRNAs Affecting Additional Mitochondrial Functions Other Than Energy Supply
5.3. Effects of Selected lncRNAs on Mitochondria
| LncRNA | Target/Mediator | Effects | Model | References |
|---|---|---|---|---|
| Cyt b ↑ | mtDNA (?) | Mitochondrial gene expression regulation (?) | Human cervical cancer cells | [111,112] |
| FENDRR ↑ | PPARGC1A/miR-18-5p | Mitochondrial disorder ↓ | Human coronary cells | [117] |
| GAS5 ↑ | Sirt1/miR-579-3p | Mitochondrial disorder ↓, antioxidant response ↑ | Renal injury mouse | [118] |
| LINC00842 ↑ | Acetylated PGC-α | OXPHOS ↓, fatty acid synthesis ↑ | Human adeno-carcinoma cells | [116] |
| MALAT1 ↑ | NFE2L2 | Antioxidant response ↓ | Mouse | [119] |
| MALAT1 ↑ | SMAD 2/3 pathway | EMT ↑ | Human RPE cells | [120] |
| MEG3 ↑ | Drp1 | Mitochondrial fission ↑ | Diabetic mouse model | [113] |
| MEG3 ↑ | MMP-2 | Fibrosis ↑ | Mouse cardiac fibroblasts | [121] |
| MEG3 ↑ | Sirt1/miR-204 | Oxidative stress↓, inflammation ↓ | Muller cells of mouse retina | [122] |
| MEG3 ↑ | NFE2L2/miR-93 | Apoptosis and inflammation ↓ | Human RPE cells | [123] |
| MEG3 ↑ | Pax6/miR-7 | RPE differentiation ↑ | Human RPE cells | [114] |
| ND5 and ND6 ↑ | mtDNA (?) | Mitochondrial gene expression regulation (?) | Human cervical carcinoma cells | [111,112] |
| NRAL ↑ | NFE2L2/miR-340 | Antioxidant response ↑ | Human liver carcinoma cells | [124] |
| PWRN2 ↑ | Not known | Cell death ↑, mitochondrial damage ↑ | Human RPE cells | [125] |
| TUG1 ↑ | PPARGC1A | Mitochondrial function ↑ | Diabetic mouse model | [126] |
| TUG1 ↑ | NFE2L2 | Antioxidant response ↑ | Glaucoma mouse model, mouse retinal ganglion cells | [127] |
| UCA1 ↑ | NFE2L2/miR-495 | Antioxidant response ↑, apoptosis ↓ | Rat epilepsy model | [128] |
5.4. Mitochondrial Actions of Circular Selected Non-Coding RNAs
6. Non-Coding RNAs in Antioxidant Response Pathway
6.1. Overview
6.2. MiRNAs
6.3. LncRNAs
6.4. CircRNAs
7. Non-Coding RNAs as Therapeutic Targets
7.1. General Aspects
7.2. MiRNAs
7.3. LncRNAs
7.4. Circular ncRNAs
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| AKT | protein kinase B |
| AMD | age-related macular degeneration |
| ASO | antisense oligonucleotide |
| circRNA | circular RNA |
| COQ10 | coenzyme Q10; ubiquinone |
| COX | cytochrome c oxidase subunit |
| CREB | cAMP response element binding protein |
| CROT | carnitine-O-acetyltransferase |
| dAMD | dry AMD |
| EMT | epithelial-to-endosomal transition |
| ETC | electron transport chain |
| KEAP1 | Kelch-like ECH-associated protein 1 |
| LNA | locked nucleic acid |
| lncRNA | long non-coding RNA |
| miRNA | microRNA |
| MnSOD | manganese superoxide dismutase |
| mtDNA | mitochondrial DNA |
| ncRNA | non-coding RNA |
| ND | (mitochondrial) NADH dehydrogenase subunit |
| NFE2L2 | nuclear factor erythroid 2-related factor 2; NFE2 like bZIP transcription factor 2 |
| NOX | NADPH oxidase |
| NRF | nuclear respiratory factor |
| nt | nucleotide |
| OXPHOS | oxidative phosphorylation |
| PD | Parkinson’s disease |
| PGC-1α | peroxisome proliferator-activated receptor gamma coactivator-1 alpha |
| PINK1 | PTEN-induced putative kinase 1 |
| PPARGC1A | gene coding PGC-1α |
| ROS | reactive oxygen species |
| RPE | retinal pigment epithelium |
| SIRT1 | silent information regulator factor 2-related enzyme |
| TFAM | mitochondrial transcription factor |
| TGF | transforming growth factor |
| UTR | untranslated region |
| VEGF | vascular endothelial growth factor |
| wAMD | wet AMD |
References
- Cenini, G.; Lloret, A.; Cascella, R. Oxidative stress in neurodegenerative diseases: From a mitochondrial point of view. Oxid. Med. Cell Longev. 2019, 2019, 2105607. [Google Scholar] [CrossRef] [PubMed]
- Kaarniranta, K.; Uusitalo, H.; Blasiak, J.; Felszeghy, S.; Kannan, R.; Kauppinen, A.; Salminen, A.; Sinha, D.; Ferrington, D. Mechanisms of mitochondrial dysfunction and their impact on age-related macular degeneration. Prog. Retin Eye Res. 2020, 7, 100858. [Google Scholar] [CrossRef] [PubMed]
- Blasiak, J.; Hyttinen, J.M.T.; Szczepanska, J.; Pawlowska, E.; Kaarniranta, K. Potential of long non-Coding RNAs in age-related macular degeneration. Int. J. Mol. Sci. 2021, 22, 9178. [Google Scholar] [CrossRef] [PubMed]
- Hyttinen, J.M.T.; Blasiak, J.; Felszeghy, S.; Kaarniranta, K. MicroRNAs in the regulation of autophagy and their possible use in age-related macular degeneration therapy. Ageing Res. Rev. 2021, 67, 101260. [Google Scholar] [CrossRef]
- Konovalova, J.; Gerasymchuk, D.; Parkkinen, I.; Chmielarz, P.; Domanskyi, A. Interplay between microRNAs and oxidative stress in neurodegenerative diseases. Int. J. Mol. Sci. 2019, 20, 6055. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Wan, Y.; Zhao, Y.; Wen, Q.; Tang, X.; Shen, J.; Wu, X.; Li, M.; Li, X.; et al. Circular RNAs in the regulation of oxidative stress. Front. Pharmacol. 2021, 12, 697903. [Google Scholar] [CrossRef]
- Fleckenstein, M.; Keenan, T.D.L.; Guymer, R.H.; Chakravarthy, U.; Schmitz-Valckenberg, S.; Klaver, C.C.; Wong, W.T.; Chew, E.Y. Age-related macular degeneration. Nat. Rev. Dis. Primers 2021, 7, 31. [Google Scholar] [CrossRef]
- Jun, S.; Datta, S.; Wang, L.; Pegany, R.; Cano, M.; Handa, J.T. The impact of lipids, lipid oxidation, and inflammation on AMD, and the potential role of miRNAs on lipid metabolism in the RPE. Exp. Eye Res. 2019, 181, 346–355. [Google Scholar] [CrossRef]
- Schulz, N.M.; Bhardwaj, S.; Barclay, C.; Gaspar, L.; Schwartz, J. burden of dry age-related macular degeneration: A targeted literature review. Clin. Ther. 2021, 43, 1792–1818. [Google Scholar] [CrossRef]
- Kaarniranta, K.; Blasiak, J.; Liton, P.; Boulton, M.; Klionsky, D.J.; Sinha, D. Autophagy in age-related macular degeneration. Autophagy 2022, in press. [Google Scholar] [CrossRef]
- Double, K.L.; Dedov, V.N.; Fedorow, H.; Kettle, E.; Halliday, G.M.; Garner, B.; Brunk, U.T. The comparative biology of neuromelanin and lipofuscin in the human brain. Cell Mol. Life Sci. 2008, 65, 1669–1682. [Google Scholar] [CrossRef] [PubMed]
- Ambati, J.; Fowler, B.J. Mechanisms of age-related macular degeneration. Neuron 2012, 75, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Gil-Martínez, M.; Santos-Ramos, P.; Fernández-Rodríguez, M.; Abraldes, M.J.; Rodríguez-Cid, M.J.; Santiago-Varela, M.; Fernández-Ferreiro, A.; Gómez-Ulla, F. Pharmacological advances in the treatment of age-related macular degeneration. Curr. Med. Chem. 2020, 27, 583–598. [Google Scholar] [CrossRef] [PubMed]
- Ohno-Matsui, K. Parallel findings in age-related macular degeneration and Alzheimer’s disease. Prog. Retin Eye Res 2011, 30, 217–238. [Google Scholar] [CrossRef] [PubMed]
- Spaide, R.F.; Jaffe, G.J.; Sarraf, D.; Freund, K.B.; Sadda, S.R.; Staurenghi, G.; Waheed, N.K.; Chakravarthy, U.; Rosenfeld, P.J.; Holz, F.G.; et al. Consensus nomenclature for reporting neovascular age-related macular degeneration data: Consensus on Neovascular Age-related Macular Degeneration Nomenclature Study Group. Ophthalmology 2020, 127, 616–636. [Google Scholar] [CrossRef]
- Campochiaro, P. Ocular neovascularization. J. Mol. Med. 2013, 91, 311–321. [Google Scholar] [CrossRef]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders—A step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 863, 1066–1077. [Google Scholar] [CrossRef]
- Dunn, J.D.; Alvarez, L.A.; Zhang, X.; Soldati, T. Reactive oxygen species and mitochondria: A nexus of cellular homeostasis. Redox. Biol. 2015, 6, 472–485. [Google Scholar] [CrossRef]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Fisher, C.R.; Kowluru, R.A. Mitochondrial defects drive degenerative retinal diseases. Trends Mol. Med. 2020, 26, 105–118. [Google Scholar] [CrossRef]
- Bilbao-Malavé, V.; González-Zamora, J.; de la Puente, M.; Recalde, S.; Fernandez-Robredo, P.; Hernandez, M.; Layana, A.G.; Saenz de Viteri, M. Mitochondrial dysfunction and endoplasmic reticulum stress in age related macular degeneration, role in pathophysiology, and possible new therapeutic strategies. Antioxidants 2021, 10, 1170. [Google Scholar] [CrossRef] [PubMed]
- Hyttinen, J.M.T.; Viiri, J.; Kaarniranta, K.; Błasiak, J. Mitochondrial quality control in AMD: Does mitophagy play a pivotal role? Cell Mol. Life Sci. 2018, 75, 2991–3008. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Cano, M.; Satyanarayana, G.; Liu, T.; Wang, L.; Wang, J.; Cheng, J.; Itoh, K.; Sharma, A.; Bhutto, I.; et al. Mitophagy initiates retrograde mitochondrial-nuclear signaling to guide retinal pigment cell heterogeneity. Autophagy 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Jannig, P.R.; Dumesic, P.A.; Spiegelman, B.M.; Ruas, J.L. SnapShot: Regulation and biology of PGC-1α. Cell 2022, 185, 1444. [Google Scholar] [CrossRef] [PubMed]
- Hyttinen, J.; Blasiak, J.; Tavi, P.; Kaarniranta, K. Therapeutic potential of PGC-1α in age-related macular degeneration (AMD)—The involvement of mitochondrial quality control, autophagy, and antioxidant response. Expert. Opin. Ther. Targets 2021, 25, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.A. New insights into PGC-1 coactivators: Redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 2014, 282, 647–672. [Google Scholar] [CrossRef]
- Felszeghy, S.; Viiri, J.; Paterno, J.J.; Hyttinen, J.M.T.; Koskela, A.; Chen, M.; Leinonen, H.; Tanila, H.; Kivinen, N.; Koistinen, A.; et al. Loss of NRF-2 and PGC-1α genes leads to retinal pigment epithelium damage resembling dry age-related macular degeneration. Redox. Biol. 2019, 20, 1–12. [Google Scholar] [CrossRef]
- Blasiak, J.; Koskela, A.; Pawlowska, E.; Liukkonen, M.; Ruuth, J.; Toropainen, E.; Hyttinen, J.M.T.; Viiri, J.; Eriksson, J.E.; Xu, H.; et al. Epithelial-mesenchymal transition and senescence in the retinal pigment epithelium of NFE2L2/PGC-1α double knock-out mice. Int. J. Mol. Sci. 2021, 22, 1684. [Google Scholar] [CrossRef]
- Rosales, M.A.B.; Shum, D.Y.; Iacovelli, J.; Saint-Geniez, M. Loss of PGC-1α in RPE induces mesenchymal transition and promotes retinal degeneration. Life Sci. Alliance 2019, 2, e201800212. [Google Scholar] [CrossRef]
- Satish, S.; Philipose, H.; Rosales, M.A.B.; Saint-Geniez, M. Pharmaceutical induction of PGC-1α promotes retinal pigment epithelial cell metabolism and protects against oxidative damage. Oxid. Med. Cell Longev. 2018, 2018, 9248640. [Google Scholar] [CrossRef]
- Shu, D.Y.; Butcher, E.R.; Saint-Geniez, M. Suppression of PGC-1α drives metabolic dysfunction in TGFβ2-induced EMT of retinal pigment epithelial cells. Int. J. Mol. Sci. 2021, 22, 4701. [Google Scholar] [CrossRef]
- Liukkonen, M.P.K.; Paterno, J.J.; Kivinen, N.; Siintamo, L.; Koskela, A.K.J.; Kaarniranta, K. Epithelial-mesenchymal transition-related serum markers ET-1.; IL-8 and TGF-β2 are elevated in a Finnish wet age-related macular degeneration cohort. Acta Ophthalmol. 2022, 100, e1153–e1162. [Google Scholar] [CrossRef] [PubMed]
- Merry, T.L.; Ristow, M. Nuclear factor erythroid-derived 2-like 2 (NFE2L2, Nrf2) mediates exercise-induced mitochondrial biogenesis and the anti-oxidant response in mice. J. Physiol. 2016, 594, 5195–5207. [Google Scholar] [CrossRef]
- Hyttinen, J.M.T.; Kannan, R.; Felszeghy, S.; Niittykoski, M.; Salminen, A.; Kaarniranta, K. The regulation of NFE2L2 (NRF2) signalling and epithelial-to-mesenchymal transition in age-related macular degeneration pathology. Int. J. Mol. Sci. 2019, 20, 5800. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Chen, Y.; Wang, J.; Sternberg, P.; Freeman, M.L.; Grossniklaus, H.E.; Cai, J. Age-related retinopathy in NRF2-deficient mice. PLoS ONE 2011, 6, e19456. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Zhang, Z.; Krause, H.M. Long noncoding RNAs and repetitive elements: Junk or intimate evolutionary partners? Trends Genet. 2019, 35, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan microRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Zogg, H.; Singh, R.; Ro, S. Current advances in RNA therapeutics for human diseases. Int. J. Mol. Sci. 2022, 23, 2736. [Google Scholar] [CrossRef]
- Zhang, C.; Owen, L.A.; Lillvis, J.H.; Zhang, S.X.; Kim, I.K.; DeAngelis, M.M. AMD genomics: Non-coding RNAs as biomarkers and therapeutic targets. J. Clin. Med. 2022, 11, 1484. [Google Scholar] [CrossRef] [PubMed]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Wright, C.B.; Uehara, H.; Kim, Y.; Yasuma, T.; Yasuma, R.; Hirahara, S.; Makin, R.D.; Apicella, I.; Pereira, F.; Nagasaka, Y.; et al. Chronic Dicer1 deficiency promotes atrophic and neovascular outer retinal pathologies in mice. Proc. Natl. Acad. Sci. USA 2020, 117, 2579–2587. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H.; Dridi, S.; Tarallo, V.; Gelfand, B.D.; Fowler, B.J.; Cho, W.G.; Kleinman, M.E.; Ponicsan, S.L.; Hauswirth, W.W.; Chiodo, V.A.; et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 2011, 471, 325–330. [Google Scholar] [CrossRef]
- Fukuda, S.; Narendran, S.; Varshney, A.; Nagasaka, Y.; Wang, S.B.; Ambati, K.; Apicella, I.; Pereira, F.; Fowler, B.J.; Yasuma, T.; et al. Alu complementary DNA is enriched in atrophic macular degeneration and triggers retinal pigmented epithelium toxicity via cytosolic innate immunity. Sci. Adv. 2021, 7, eabj3658. [Google Scholar] [CrossRef]
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2018, 47, D135–D139. [Google Scholar] [CrossRef]
- Borkiewicz, L.; Kalafut, J.; Dudziak, K.; Przybyszewska-Podstawka, A.; Telejko, I. Decoding lncRNAs. Cancers 2021, 13, 2643. [Google Scholar] [CrossRef]
- Fritah, S.; Niclou, S.P.; Azuaje, F. Databases for lncRNAs: A comparative evaluation of emerging tools. RNA 2014, 20, 1655–1665. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Gloss, B.S.; Dinger, M.E. The specificity of long noncoding RNA expression. Biochim. Biophys. Acta 2016, 1859, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.J.; Xu, G.; Chen, L.L. Mechanisms of long noncoding RNA nuclear retention. Trends Biochem. Sci. 2020, 45, 947–950. [Google Scholar] [CrossRef] [PubMed]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Bach, D.H.; Lee, S.K.; Sood, A.K. Circular RNAs in cancer. Mol. Ther. Nucleic Acids 2019, 16, 118–129. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Tang, Q.; Hann, S.S. Biological roles and mechanisms of circular RNA in human cancers. Onco. Targets Ther. 2020, 13, 2067–2092. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Meganck, R.M.; Liu, J.; Hale, A.E.; Simon, K.E.; Fanous, M.M.; Vincent, H.A.; Wilusz, J.E.; Moorman, N.J.; Marzluff, W.F.; Asokan, A. highly efficient backsplicing and translation of synthetic circRNAs. Mol. Ther. Nucleic Acids 2021, 23, 821–834. [Google Scholar] [CrossRef]
- Talhouarne, G.J.S.; Gall, J.G. Lariat intronic RNAs in the cytoplasm of vertebrate cells. Proc. Natl. Acad. Sci. USA 2018, 115, E7970–E7977. [Google Scholar] [CrossRef]
- Jiao, S.; Wu, S.; Huang, S.; Liu, M.; Gao, B. Advances in the identification of circular RNAs and research into circRNAs in human diseases. Front. Genet. 2021, 12, 665233. [Google Scholar] [CrossRef] [PubMed]
- Nisar, S.; Bhat, A.A.; Singh, M.; Karedath, T.; Rizwan, A.; Hashem, S.; Bagga, P.; Reddy, R.; Jamal, F.; Uddin, S.; et al. Insights into the role of circRNAs: Biogenesis, characterization, functional, and clinical impact in human malignancies. Front. Cell Dev. Biol 2021, 9, 617281. [Google Scholar] [CrossRef] [PubMed]
- Prats, A.C.; David, F.; Diallo, L.H.; Roussel, E.; Tatin, F.; Garmy-Susini, B.; Lacazette, E. Circular RNA, the key for translation. Int. J. Mol. Sci. 2020, 21, 8591. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.E.; Kim, S.W.; Jeong, S.; Moon, H.; Choi, W.S.; Lim, S.; Lee, S.; Hwang, K.C.; Choi, J.W. MicroRNA-26a/b-5p promotes myocardial infarction-induced cell death by downregulating cytochrome c oxidase 5a. Exp. Mol. Med. 2021, 53, 1332–1343. [Google Scholar] [CrossRef]
- Das, S.; Ferlito, M.; Kent, O.A.; Fox-Talbot, K.; Wang, R.; Liu, D.; Raghavachari, N.; Yang, Y.; Wheelan, S.J.; Murphy, E.; et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012, 110, 1596–1603. [Google Scholar] [CrossRef]
- Das, S.; Bedja, D.; Campbell, N.; Dunkerly, B.; Chenna, V.; Maitra, A.; Steenbergen, C. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS ONE 2014, 9, e96820. [Google Scholar] [CrossRef]
- Jung, K.A.; Lee, S.; Kwak, M.K. NFE2L2/NRF2 activity is linked to mitochondria and AMP-activated protein kinase signaling in cancers through miR-181c/mitochondria-encoded cytochrome c oxidase regulation. Antioxid. Redox. Signal. 2017, 27, 945–961. [Google Scholar] [CrossRef]
- Colleoni, F.; Padmanabhan, N.; Yung, H.W.; Watson, E.D.; Cetin, I.; Tissot van Patot, M.C.; Burton, G.J.; Murray, A.J. Suppression of mitochondrial electron transport chain function in the hypoxic human placenta: A role for miRNA-210 and protein synthesis inhibition. PLoS ONE 2013, 8, e55194. [Google Scholar] [CrossRef] [PubMed]
- Qiao, A.; Khechaduri, A.; Kannan Mutharasan, R.; Wu, R.; Nagpal, V.; Ardehali, H. MicroRNA-210 decreases heme levels by targeting ferrochelatase in cardiomyocytes. J. Am. Heart Assoc. 2013, 2, e000121. [Google Scholar] [CrossRef]
- Liu, C.H.; Wang, Z.; Sun, Y.; SanGiovanni, J.P.; Chen, J. Retinal expression of small non-coding RNAs in a murine model of proliferative retinopathy. Sci. Rep. 2016, 6, 33947. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, C.; Zhang, X. Mitochondrial damage mediated by miR-1 overexpression in cancer stem cells. Mol. Ther. Nucleic Acids 2019, 18, 938–953. [Google Scholar] [CrossRef]
- Kabaria, S.; Choi, D.C.; Chaudhuri, A.D.; Jain, M.R.; Li, H.; Junn, E. MicroRNA-7 activates Nrf2 pathway by targeting Keap1. Free Radic. Biol. Med. 2015, 89, 548–556. [Google Scholar] [CrossRef]
- Fierro-Fernández, M.; Miguel, V.; Márquez-Expósito, L.; Nuevo-Tapioles, C.; Herrero, J.I.; Blanco-Ruiz, E.; Tituaña, J.; Castillo, C.; Cannata, P.; Monsalve, M.; et al. MiR-9-5p protects from kidney fibrosis by metabolic reprogramming. FASEB J. 2020, 34, 410–431. [Google Scholar] [CrossRef] [PubMed]
- Nishi, H.; Ono, K.; Iwanaga, Y.; Horie, T.; Nagao, K.; Takemura, G.; Kinoshita, M.; Kuwabara, Y.; Mori, R.T.; Hasegawa, K.; et al. MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J. Biol. Chem. 2010, 285, 4920–4930. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, F.S.; Wu, Z.Y.; Lin, J.L.; Lan, W.B.; Lin, J.H. MicroRNA-19b targets Mfn1 to inhibit Mfn1-induced apoptosis in osteosarcoma cells. Neoplasma 2014, 61, 265–273. [Google Scholar] [CrossRef]
- Xue, Y.; Wei, Z.; Ding, H.; Wang, Q.; Zhou, Z.; Zheng, S.; Zhang, Y.; Hou, D.; Liu, Y.; Zen, K.; et al. MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1α in the progression of atherosclerosis. Atherosclerosis 2021, 241, 671–681. [Google Scholar] [CrossRef]
- Li, D.D.; Zhong, B.W.; Zhang, H.X.; Zhou, H.Y.; Luo, J.; Liu, Y.; Xu, G.C.; Luan, C.S.; Fang, J. Inhibition of the oxidative stress-induced miR-23a protects the human retinal pigment epithelium (RPE) cells from apoptosis through the upregulation of glutaminase and glutamine uptake. Mol. Biol. Rep. 2016, 43, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Long, B.; Gan, Y.; Zhang, R.C.; Zhang, Y.H. miR-23a regulates cardiomyocyte apoptosis by targeting manganese superoxide dismutase. Mol. Cell 2017, 40, 542–549. [Google Scholar] [CrossRef]
- Wang, L.; Kong, L.; Xu, S.; Wang, X.; Huang, K.; Wang, S.; Wu, J.; Wang, C.; Sun, H.; Liu, K.; et al. Isoliquiritigenin-mediated miR-23a-3p inhibition activates PGC-1α to alleviate alcoholic liver injury. Phytomedicine 2022, 96, 153845. [Google Scholar] [CrossRef]
- Xiao, X.; Lu, Z.; Lin, V.; May, A.; Shaw, D.H.; Wang, Z.; Che, B.; Tran, K.; Du, H.; Shaw, P.X. MicroRNA miR-24-3p reduces apoptosis and regulates Keap1-Nrf2 pathway in mouse cardiomyocytes responding to ischemia/reperfusion injury. Oxid. Med. Cell Longev. 2018, 2018, 7042105. [Google Scholar] [CrossRef]
- Song, J.; Zhang, H.; Sun, Y.; Guo, R.; Zhong, D.; Xu, R.; Song, M. Omentin-1 protects renal function of mice with type 2 diabetic nephropathy via regulating miR-27a-Nrf2/Keap1 axis. Biomed. Pharmacother. 2018, 107, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Fiesel, F.C.; Belmonte, K.C.; Hudec, R.; Wang, W.X.; Kim, C.; Nelson, P.T.; Springer, W.; Kim, J. miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol. Neurodegener. 2016, 11, 55. [Google Scholar] [CrossRef]
- Pullen, T.J.; da Silva Xavier, G.; Kelsey, G.; Rutter, G.A. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol. Cell Biol. 2011, 31, 3182–3194. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.H.; Liang, H.; Lu, S.N.; Wang, H.; Su, Z.L.; Zhang, L.; Ma, J.Q.; Guo, M.; Tai, S.; Yu, S. miR-124 suppresses pancreatic ductal adenocarcinoma growth by regulating monocarboxylate transporter 1-mediated cancer lactate metabolism. Cell. Physiol. Biochem. 2018, 50, 924–935. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Esau, C.C.; Hussain, F.N.; McDaniel, A.L.; Marshall, S.M.; van Gils, J.M.; Ray, T.D.; Sheedy, F.J.; Goedeke, L.; Liu, X.; et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 2011, 478, 404–407. [Google Scholar] [CrossRef]
- Tai, Y.; Pu, M.; Yuan, L.; Guo, H.; Qiao, J.; Lu, H.; Wang, G.; Chen, J.; Qi, X.; Tao, Z.; et al. miR-34a-5p regulates PINK1-mediated mitophagy via multiple modes. Life Sci. 2021, 276, 119415. [Google Scholar] [CrossRef]
- Ba, Q.; Cui, C.; Wen, L.; Feng, S.; Zhou, J.; Yang, K. Schisandrin B shows neuroprotective effect in 6-OHDA-induced Parkinson’s disease via inhibiting the negative modulation of miR-34a on Nrf2 pathway. Biomed. Pharmacother. 2015, 75, 165–172. [Google Scholar] [CrossRef]
- Miñones-Moyano, E.; Porta, S.; Escaramís, G.; Rabionet, R.; Iraola, S.; Kagerbauer, B.; Espinosa-Parrilla, Y.; Ferrer, I.; Estivill, X.; Martí, E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011, 20, 3067–3078. [Google Scholar] [CrossRef]
- Chen, F.; Zhao, Y.; Chen, H. MicroRNA-98 reduces amyloid β-protein production and improves oxidative stress and mitochondrial dysfunction through the Notch signaling pathway via HEY2 in Alzheimer’s disease mice. Int. J. Mol. Med. 2019, 43, 91–102. [Google Scholar] [CrossRef]
- Lei, Q.; Liu, X.; Fu, H.; Sun, Y.; Wang, L.; Xu, G.; Wang, W.; Yu, Z.; Liu, C.; Li, P.; et al. miR-101 reverses hypomethylation of the PRDM16 promoter to disrupt mitochondrial function in astrocytoma cells. Oncotarget 2015, 7, 5007–5022. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Teague, A.M.; Tryggestad, J.B.; Chernausek, S.D. Role of microRNA-130b in placental PGC-1α/TFAM mitochondrial biogenesis pathway. Biochem. Biophys. Res. Commun. 2017, 487, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, M.; Patel, D.; Vedpathak, D.; Rathinam, M.; Henderson, G.; Mahimainathan, L. Identification of novel microRNAs in post-transcriptional control of Nrf2 expression and redox homeostasis in neuronal, SH-SY5Y cells. PLoS ONE 2012, 7, e51111. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Liu, L.; Lao, Y.; Liao, W.; Liao, M.; Luo, X.; Wu, J.; Xie, W.; Zhang, Y.; Xu, N. MicroRNA-181a suppresses parkin-mediated mitophagy and sensitizes neuroblastoma cells to mitochondrial uncoupler-induced apoptosis. Oncotarget 2016, 7, 42274–42287. [Google Scholar] [CrossRef] [PubMed]
- Indrieri, A.; Carrella, S.; Romano, A.; Spaziano, A.; Marrocco, E.; Fernandez-Vizarra, E.; Barbato, S.; Pizzo, M.; Ezhova, Y.; Golia, F.M.; et al. miR-181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol. Med. 2019, 11, e8734. [Google Scholar] [CrossRef] [PubMed]
- Houzelle, A.; Dahlmans, D.; Nascimento, E.B.M.; Schaart, G.; Jörgensen, J.A.; Moonen-Kornips, E.; Kersten, S.; Wang, X.; Hoeks, J. MicroRNA-204-5p modulates mitochondrial biogenesis in C2C12 myotubes and associates with oxidative capacity in humans. J. Cell Physiol. 2020, 235, 9851–9863. [Google Scholar] [CrossRef] [PubMed]
- Fasanaro, P.; D’Alessandra, Y.; Di Stefano, V.; Melchionna, R.; Romani, S.; Pompilio, G.; Capogrossi, M.C.; Martelli, F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 2008, 283, 15878–15883. [Google Scholar] [CrossRef] [PubMed]
- Aschrafi, A.; Schwechter, A.D.; Mameza, M.G.; Natera-Naranjo, O.; Gioio, A.E.; Kaplan, B.B. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J. Neurosci. 2008, 28, 12581–12590. [Google Scholar] [CrossRef]
- Aschrafi, A.; Kar, A.N.; Natera-Naranjo, O.; MacGibeny, M.A.; Gioio, A.E.; Kaplan, B.B. MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell Mol. Life Sci. 2012, 69, 4017–4027. [Google Scholar] [CrossRef]
- Xiong, R.; Wang, Z.; Zhao, Z.; Li, H.; Chen, W.; Zhang, B.; Wang, L.; Wu, L.; Li, W.; Ding, J.; et al. MicroRNA-494 reduces DJ-1 expression and exacerbates neurodegeneration. Neurobiol. Aging 2014, 35, 705–714. [Google Scholar] [CrossRef]
- Yan, K.; An, T.; Zhai, M.; Huang, Y.; Wang, Q.; Wang, Y.; Zhang, R.; Wang, T.; Liu, J.; Zhang, Y.; et al. Mitochondrial miR-762 regulates apoptosis and myocardial infarction by impairing ND2. Cell Death Dis. 2019, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Bisbach, C.M.; Hass, D.T.; Thomas, E.D.; Cherry, T.J.; Hurley, J.B. Monocarboxylate transporter 1 (MCT1) mediates succinate export in the retina. Invest. Ophthalmol. Vis. Sci. 2022, 63, 1. [Google Scholar] [CrossRef] [PubMed]
- Sridevi Gurubaran, I.; Viiri, J.; Koskela, A.; Hyttinen, J.M.T.; Paterno, J.J.; Kis, G.; Antal, M.; Urtti, A.; Kauppinen, A.; Felszeghy, S.; et al. Mitophagy in the retinal pigment epithelium of dry age-related macular degeneration investigated in the NFE2L2/PGC-1α mouse model. Int. J. Mol. Sci. 2020, 21, 1976. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.Y.; Datta, S.; Bandeira, E.; Cano, M.; Mallick, E.; Rai, U.; Powell, B.; Tian, J.; Witwer, K.W.; Handa, J.T.; et al. Release of extracellular vesicle miR-494-3p by ARPE-19 cells with impaired mitochondria. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129598. [Google Scholar] [CrossRef] [PubMed]
- Lemecha, M.; Morino, K.; Imamura, T.; Iwasaki, H.; Ohashi, N.; Ida, S.; Sato, D.; Sekine, O.; Ugi, S.; Maegawa, H. MiR-494-3p regulates mitochondrial biogenesis and thermogenesis through PGC1-α signalling in beige adipocytes. Sci. Rep. 2018, 8, 15096. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.J.; Krebs, M.P.; Mao, H.; Jones, K.; Conners, M.; Lewin, A.S. Pathological consequences of long-term mitochondrial oxidative stress in the mouse retinal pigment epithelium. Exp. Eye Res. 2012, 101, 60–71. [Google Scholar] [CrossRef]
- Kang, H.M.; Ahn, S.H.; Choi, P.; Ko, Y.A.; Han, S.H.; Chinga, F.; Park, A.S.; Tao, J.; Sharma, K.; Pullman, J.; et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015, 21, 37–46. [Google Scholar] [CrossRef]
- Feher, J.; Kovacs, B.; Kovacs, I.; Schveoller, M.; Papale, A.; Balacco Gabrieli, C. Improvement of visual functions and fundus alterations in early age-related macular degeneration treated with a combination of acetyl-L-carnitine, n-3 fatty acids, and coenzyme Q10. Ophthalmologica 2005, 219, 154–166. [Google Scholar] [CrossRef]
- Chiang, Y.W.; Su, C.H.; Sun, H.Y.; Chen, S.P.; Chen, C.J.; Chen, W.Y.; Chang, C.C.; Chen, C.M.; Kuan, Y.H. Bisphenol A induced apoptosis via oxidative stress generation involved Nrf2/HO-1 pathway and mitochondrial dependent pathways in human retinal pigment epithelium (ARPE-19) cells. Environ. Toxicol. 2022, 37, 131–141. [Google Scholar] [CrossRef]
- Kutty, R.K.; Samuel, W.; Jaworski, C.; Duncan, T.; Nagineni, C.N.; Raghavachari, N.; Wiggert, B.; Redmond, T.M. MicroRNA expression in human retinal pigment epithelial (ARPE-19) cells: Increased expression of microRNA-9 by N-(4-hydroxyphenyl)retinamide. Mol. Vis. 2010, 16, 1475–1486. [Google Scholar] [PubMed]
- Liu, X.; Shan, G. Mitochondria encoded non-coding RNAs in cell physiology. Front. Cell Dev. Biol. 2021, 9, 713729. [Google Scholar] [CrossRef] [PubMed]
- Rackham, O.; Shearwood, A.M.; Mercer, T.R.; Davies, S.M.; Mattick, J.S.; Filipovska, A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Wen, R.; Liu, S.; Chen, X.; Song, S.; Li, X.; Su, Z.; Wang, C. Increased long noncoding RNA maternally expressed gene 3 contributes to podocyte injury induced by high glucose through regulation of mitochondrial fission. Cell Death Dis. 2020, 11, 814. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.J.; Zhang, F.F.; Xiao, Q.; Xu, J.; Zhu, L.J. lncRNA MEG3.; Acting as a ceRNA.; modulates RPE differentiation through the miR-7-5p/Pax6 axis. Biochem. Genet. 2021, 59, 1617–1630. [Google Scholar] [CrossRef] [PubMed]
- Bäumer, N.; Marquardt, T.; Stoykova, A.; Spieler, D.; Treichel, D.; Ashery-Padan, R.; Gruss, P. Retinal pigmented epithelium determination requires the redundant activities of Pax2 and Pax6. Development 2003, 130, 2903–2915. [Google Scholar] [CrossRef]
- Huang, X.; Pan, L.; Zuo, Z.; Li, M.; Zeng, L.; Li, R.; Ye, Y.; Zhang, J.; Wu, G.; Bai, R.; et al. LINC00842 inactivates transcription co-regulator PGC-1α to promote pancreatic cancer malignancy through metabolic remodelling. Nat. Commun. 2021, 12, 3830. [Google Scholar] [CrossRef]
- Wang, G.; Yang, Y.; Ma, H.; Shi, L.; Jia, W.; Hao, X.; Liu, W. LncRNA FENDRR inhibits ox-LDL induced mitochondrial energy metabolism disorder in aortic endothelial cells via miR-18a-5p/PGC-1α signaling pathway. Front. Endocrinol. 2021, 12, 622665. [Google Scholar] [CrossRef]
- Ling, H.; Li, Q.; Duan, Z.P.; Wang, Y.J.; Hu, B.Q.; Dai, X.G. LncRNA GAS5 inhibits miR-579-3p to activate SIRT1/PGC-1α/Nrf2 signaling pathway to reduce cell pyroptosis in sepsis-associated renal injury. Am. J. Physiol. Cell Physiol. 2021, 321, C117–C133. [Google Scholar] [CrossRef]
- Chen, J.; Ke, S.; Zhong, L.; Wu, J.; Tseng, A.; Morpurgo, B.; Golovko, A.; Wang, G.; Cai, J.J.; Ma, X.; et al. Long noncoding RNA MALAT1 regulates generation of reactive oxygen species and the insulin responses in male mice. Biochem. Pharmacol. 2018, 152, 94–103. [Google Scholar] [CrossRef]
- Yang, S.; Yao, H.; Li, M.; Li, H.; Wang, F. Long Non-Coding RNA MALAT1 mediates transforming growth factor beta1-induced epithelial-mesenchymal transition of retinal pigment epithelial cells. PLoS ONE 2016, 11, e0152687. [Google Scholar] [CrossRef]
- Piccoli, M.T.; Gupta, S.K.; Viereck, J.; Foinquinos, A.; Samolovac, S.; Kramer, F.L.; Garg, A.; Remke, J.; Zimmer, K.; Batkai, S.; et al. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ. Res. 2017, 121, 575–583. [Google Scholar] [CrossRef]
- Tu, Y.; Song, E.; Wang, Z.; Ji, N.; Zhu, L.; Wang, K.; Sun, H.; Zhang, Y.; Zhu, Q.; Liu, X.; et al. Melatonin attenuates oxidative stress and inflammation of Müller cells in diabetic retinopathy via activating the Sirt1 pathway. Biomed. Pharmacother. 2021, 137, 111274. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Jin, H.; Li, L.; Hu, Y.X.; Xiao, F. Long noncoding RNA MEG3 inhibits apoptosis of retinal pigment epithelium cells induced by high glucose via the miR-93/Nrf2 Axis. Am. J. Pathol. 2020, 190, 1813–1822. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.L.; Cai, W.P.; Lei, X.; Shi, K.Q.; Lin, X.Y.; Shi, L. NRAL mediates cisplatin resistance in hepatocellular carcinoma via miR-340-5p/Nrf2 axis. J. Cell Commun. Signal. 2019, 13, 99–112. [Google Scholar] [CrossRef]
- Yu, X.; Luo, Y.; Chen, G.; Liu, H.; Tian, N.; Zen, X.; Huang, Y. Long non-coding RNA PWRN2 regulates cytotoxicity in an in vitro model of age-related macular degeneration. Biochem. Biophys. Res. Commun. 2021, 535, 39–46. [Google Scholar] [CrossRef]
- Long, J.; Badal, S.S.; Ye, Z.; Wang, Y.; Ayanga, B.A.; Galvan, D.L.; Green, N.H.; Chang, B.H.; Overbeek, P.A.; Danesh, F.R. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Invest. 2016, 126, 4205–4218. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Li, J.; Zhu, G.; Wang, Y.; Zheng, G.; Kan, Q. Chlorogenic acid relieved oxidative stress injury in retinal ganglion cells through IncRNA-TUG1/Nrf2. Cell Cycle 2019, 18, 1549–1559. [Google Scholar] [CrossRef]
- Geng, J.F.; Liu, X.; Zhao, H.B.; Fan, W.F.; Geng, J.J.; Liu, X.Z. LncRNA UCA1 inhibits epilepsy and seizure-induced brain injury by regulating miR-495/Nrf2-ARE signal pathway. Int. J. Biochem. Cell Biol. 2018, 99, 133–139. [Google Scholar] [CrossRef]
- Fernández-Silva, P.; Enriquez, J.A.; Montoya, J. Replication and transcription of mammalian mitochondrial DNA. Exp. Physiol. 2003, 88, 41–56. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, W.; Zhong, L.; Li, H.; Bai, L.; Chen, X.; Lin, Y.; Zheng, D. circ-AKT3 aggravates renal ischaemia-reperfusion injury via regulating miR-144-5p/Wnt/β-catenin pathway and oxidative stress. J. Cell Mol. Med. 2020, 26, 1766–1775. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Y.; Sun, R.; Sun, Y.; Liu, D.; Lin, M.; Chen, Z.; Zhou, J.; Lv, L.; Tian, X.; et al. circ-CBFB upregulates p66Shc to perturb mitochondrial dynamics in APAP-induced liver injury. Cell Death Dis. 2020, 11, 953. [Google Scholar] [CrossRef]
- Liu, Z.; He, Q.; Liu, Y.; Zhang, Y.; Cui, M.; Peng, H.; Wang, Y.; Chen, S.; Li, D.; Chen, L.; et al. Hsa_circ_0005915 promotes N,N-dimethylformamide-induced oxidative stress in HL-7702 cells through NRF2/ARE axis. Toxicology 2021, 458, 152838. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ren, F.; Sun, D.; Liu, J.; Liu, B.; He, Y.; Pang, S.; Shi, B.; Zhou, F.; Yao, L.; et al. CircKEAP1 suppresses the progression of lung adenocarcinoma via the miR-141-3p/KEAP1/NRF2 axis. Front. Oncol. 2021, 11, 672586. [Google Scholar] [CrossRef]
- Wu, Z.; Sun, H.; Wang, C.; Liu, W.; Liu, M.; Zhu, Y.; Xu, W.; Jin, H.; Li, J. Mitochondrial genome-derived circRNA mc-COX2 functions as an oncogene in chronic lymphocytic leukemia. Mol. Ther. Nucleic Acids 2020, 20, 801–811. [Google Scholar] [CrossRef]
- Li, M.; Ding, W.; Tariq, M.A.; Chang, W.; Zhang, X.; Xu, W.; Hou, L.; Wang, Y.; Wang, J. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics 2018, 8, 5855–5869. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Cao, X.; Xue, L.; Xia, L.; Xu, Y. CircPRKCI-miR-545/589-E2F7 axis dysregulation mediates hydrogen peroxide-induced neuronal cell injury. Biochem. Biophys. Res. Commun. 2019, 514, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Henderson, N.C.; Mackinnon, A.C.; Farnworth, S.L.; Kipari, T.; Haslett, C.; Iredale, J.P.; Liu, F.T.; Hughes, J.; Sethi, T. Galectin-3 expression and secretion links macrophages to the promotion of renal fibrosis. Am. J. Pathol. 2008, 172, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhou, X.; Luo, G.; Zhang, J.; Yang, M.; Song, C. CircRNA RERE promotes the oxidative stress-induced apoptosis and autophagy of nucleus pulposus cells through the miR-299-5p/Galectin-3 axis. J. Health Eng. 2021, 2021, 2771712. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, J.; Deng, H.; Ma, R.; Liao, J.Y.; Liang, H.; Hu, J.; Li, J.; Guo, Z.; Cai, J.; et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 2020, 183, 76–93.e22. [Google Scholar] [CrossRef]
- Hanan, M.; Simchovitz, A.; Yayon, N.; Vaknine, S.; Cohen-Fultheim, R.; Karmon, M.; Madrer, N.; Rohrlich, T.M.; Maman, M.; Bennett, E.R.; et al. A Parkinson’s disease circRNAs resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol. Med. 2020, 12, e11942. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, L.; Li, Y.J.; Mei, R.; Yu, H.L.; Gong, Y.; Du, M.Y.; Wang, F. MicroRNA-128 protects dopamine neurons from apoptosis and upregulates the expression of excitatory amino acid Transporter 4 in Parkinson’s disease by binding to AXIN1. Cell Physiol. Biochem. 2018, 51, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, Z.; Cao, K.; Shan, W.; Liu, J.; Wen, Q.; Wang, R. Circ-SPECC1 modulates TGFβ2 and autophagy under oxidative stress by sponging miR-33a to promote hepatocellular carcinoma tumorigenesis. Cancer Med. 2020, 9, 5999–6008. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, B.; Hao, C.; Huang, X.; Li, X.; Huang, Y.; Luo, Z. Omentin-a novel adipokine in respiratory diseases. Int. J. Mol. Sci. 2017, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Kaarniranta, K.; Paananen, J.; Nevalainen, T.; Sorri, I.; Seitsonen, S.; Immonen, I.; Salminen, A.; Pulkkinen, L.; Uusitupa, M. Adiponectin receptor 1 gene (ADIPOR1) variant is associated with advanced age-related macular degeneration in Finnish population. Neurosci. Lett. 2012, 513, 233–237. [Google Scholar] [CrossRef]
- Arun, G.; Aggarwal, D.; Spector, D.L. MALAT1 long non-coding RNA: Functional implications. Noncoding RNA 2020, 6, 22. [Google Scholar] [CrossRef]
- Dang, R.; Yang, M.; Cui, C.; Wang, C.; Zhang, W.; Geng, C.; Han, W.; Jiang, P. Activation of angiotensin-converting enzyme 2/angiotensin (1-7)/mas receptor axis triggers autophagy and suppresses microglia proinflammatory polarization via forkhead box class O1 signaling. Aging Cell 2021, 20, e13480. [Google Scholar] [CrossRef]
- Verzella, D.; Pescatore, A.; Capece, D.; Vecchiotti, D.; Ursini, M.V.; Franzoso, G.; Alesse, E.; Zazzeroni, F. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 2020, 11, 210. [Google Scholar] [CrossRef]
- Shu, D.Y.; Butcher, E.; Saint-Geniez, M. EMT and EndMT: Emerging roles in age-related macular degeneration. Int. J. Mol. Sci. 2020, 21, 4271. [Google Scholar] [CrossRef]
- Lima, J.F.; Cerqueira, L.; Figueiredo, C.; Oliveira, C.; Azevedo, N.F. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018, 15, 338–352. [Google Scholar] [CrossRef]
- Lu, D.; Thum, T. RNA-based diagnostic and therapeutic strategies for cardiovascular disease. Nat. Rev. Cardiol. 2019, 16, 661–674. [Google Scholar] [CrossRef]
- Gemayel, M.C.; Bhatwadekar, A.D.; Ciulla, T. RNA therapeutics for retinal diseases. Expert. Opin. Biol. Ther. 2021, 21, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Long noncoding RNAs as biomarkers in cancer. Dis. Markers 2017, 2017, 7243968. [Google Scholar] [CrossRef] [PubMed]
- Maestro, S.; Weber, N.D.; Zabaleta, N.; Aldabe, R.; Gonzalez-Aseguinolaza, G. Novel vectors and approaches for gene therapy in liver diseases. JHEP Rep. 2021, 3, 100300. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, Y.X.; Jin, X.; Chen, J.; Peng, L.; Wang, D.L.; Li, Y.; Yao, X.Y.; Liao, J.Y.; He, J.H.; et al. Overexpression of lncRNAs with endogenous lengths and functions using a lncRNA delivery system based on transposon. J. Nanobiotechnol. 2021, 19, 303. [Google Scholar] [CrossRef]
- Shi, Y.; Parag, S.; Patel, R.; Lui, A.; Mur, M.; Cai, J.; Patel, N.A. Stabilization of lncRNA GAS5 by a small molecule and its implications in diabetic adipocytes. Cell Chem. Biol. 2019, 26, 319–330.e6. [Google Scholar] [CrossRef]
- Zhang, H.D.; Jiang, L.H.; Sun, D.W.; Hou, J.C.; Ji, Z.L. CircRNA: A novel type of biomarker for cancer. Breast Cancer 2018, 25, 1–7. [Google Scholar] [CrossRef]
- Garikipati, V.N.S.; Verma, S.K.; Cheng, Z.; Liang, D.; Truongcao, M.M.; Cimini, M.; Yue, Y.; Huang, G.; Wang, C.; Benedict, C.; et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2019, 10, 4317. [Google Scholar] [CrossRef] [PubMed]
- Rincón-Riveros, A.; Morales, D.; Rodríguez, J.A.; Villegas, V.E.; López-Kleine, L. Bioinformatic tools for the analysis and prediction of ncRNA interactions. Int. J. Mol. Sci. 2021, 22, 11397. [Google Scholar] [CrossRef]
- Roth, R.; Kim, S.; Kim, J.; Rhee, S. Single-cell and spatial transcriptomics approaches of cardiovascular development and disease. BMB Rep. 2020, 53, 393–399. [Google Scholar] [CrossRef]


| CircRNA | Target/Mediator | Effects | Models | References |
|---|---|---|---|---|
| AKT3 ↑ | β-catenin-Wnt signaling/miR-144 | Apoptosis ↑, EMT ↑ | Rat renal ischemic model | [130] |
| CBFB ↑ | p66Shc/miR-185 | Mitochondrial ROS ↑ | Mouse liver injury model, and mouse liver cells | [131] |
| circ_0005915 ↑ | NFE2L2 pathway | Antioxidant response ↓ | Human liver cells | [132] |
| KEAP1 ↑ | KEAP1/miR-141-3p | Antioxidant response ↓ | Human lung adenocarcinoma samples | [133] |
| mc-COX2 ↓ | ? | ATP production ↓ | Leukemia samples, leukemia cells | [134] |
| mecciND1 and mecciND5 ↑ | Mitochondrial proteins | Protein import ↑, chaperone function ↑ | Human cervical cancer cells | [111] |
| NCX1 ↑ | CDIP1/miR-133-3p | Apoptosis ↑ | Rat myocardial cells, and mouse ischemia model | [135] |
| PRKCI ↓ | E2F7/miR-545 and miR-589 | Neuronal cell injury ↑ | Human neuroblastoma cells | [136] |
| RERE ↑ | Galectin-3/miR-299 | Apoptosis ↑, fibrosis ↑ | Human nucleus pulposus cells | [137,138] |
| SCAR ↑ | ATP5B | ROS production ↓, fibroblast activation ↓ | Human and mouse fibroblasts | [139] |
| SLC8A1 ↑ | AXIN1/miR-128 | Apoptosis ↑ | Human neuroblastoma cells | [140,141] |
| SPECC1 ↓ | TGFβ2/miR-33a | Apoptosis ↑, proliferation ↓, autophagy ↓ | Human hepatocarcinoma cells | [142] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hyttinen, J.M.T.; Blasiak, J.; Kaarniranta, K. Non-Coding RNAs Regulating Mitochondrial Functions and the Oxidative Stress Response as Putative Targets against Age-Related Macular Degeneration (AMD). Int. J. Mol. Sci. 2023, 24, 2636. https://doi.org/10.3390/ijms24032636
Hyttinen JMT, Blasiak J, Kaarniranta K. Non-Coding RNAs Regulating Mitochondrial Functions and the Oxidative Stress Response as Putative Targets against Age-Related Macular Degeneration (AMD). International Journal of Molecular Sciences. 2023; 24(3):2636. https://doi.org/10.3390/ijms24032636
Chicago/Turabian StyleHyttinen, Juha M. T., Janusz Blasiak, and Kai Kaarniranta. 2023. "Non-Coding RNAs Regulating Mitochondrial Functions and the Oxidative Stress Response as Putative Targets against Age-Related Macular Degeneration (AMD)" International Journal of Molecular Sciences 24, no. 3: 2636. https://doi.org/10.3390/ijms24032636
APA StyleHyttinen, J. M. T., Blasiak, J., & Kaarniranta, K. (2023). Non-Coding RNAs Regulating Mitochondrial Functions and the Oxidative Stress Response as Putative Targets against Age-Related Macular Degeneration (AMD). International Journal of Molecular Sciences, 24(3), 2636. https://doi.org/10.3390/ijms24032636







