MEK Is a Potential Indirect Target in Subtypes of Head and Neck Cancers
Abstract
1. Introduction
2. Results
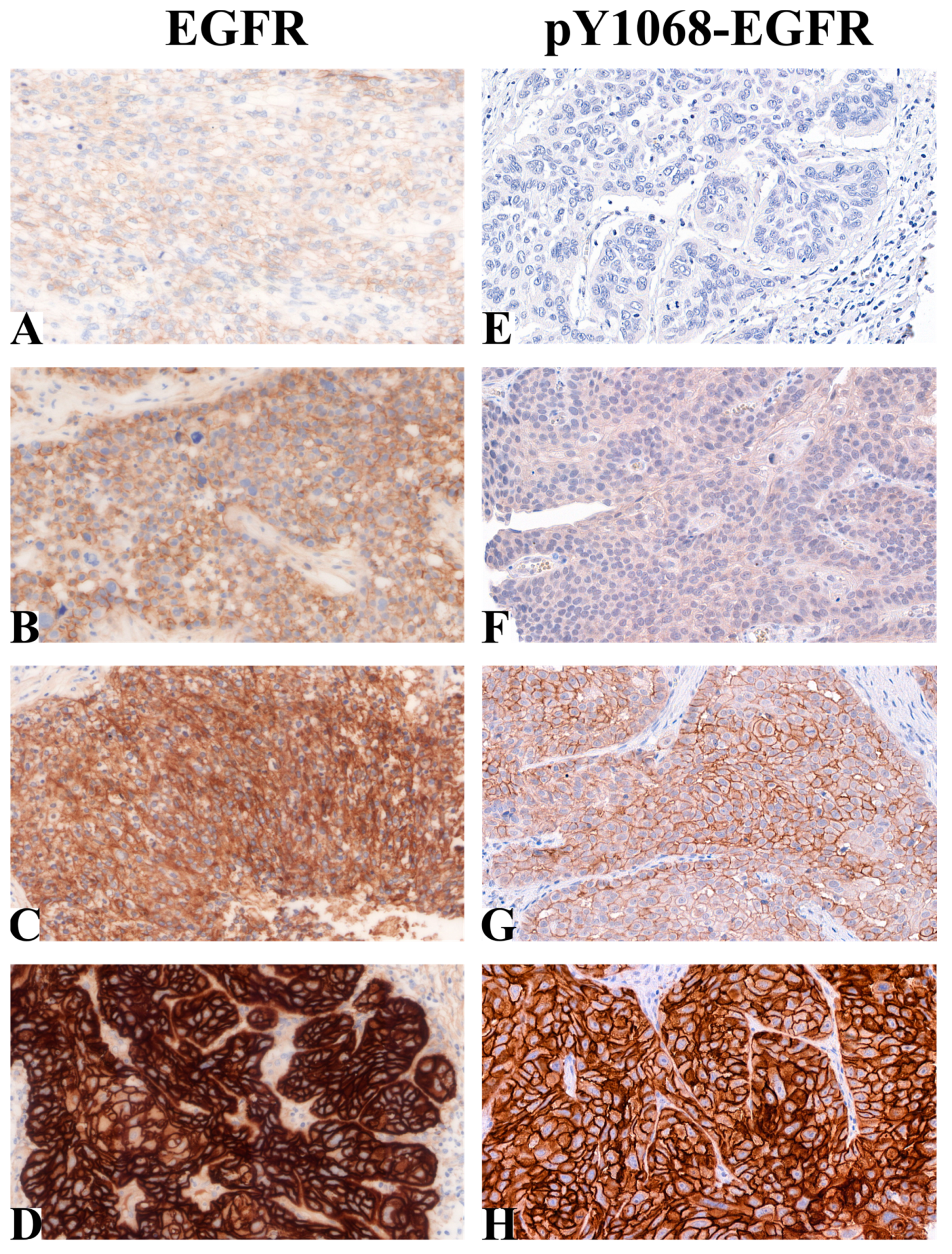
2.1. EGFR and pY1068-EGFR Protein Levels in Patient-Derived HNSCC Samples
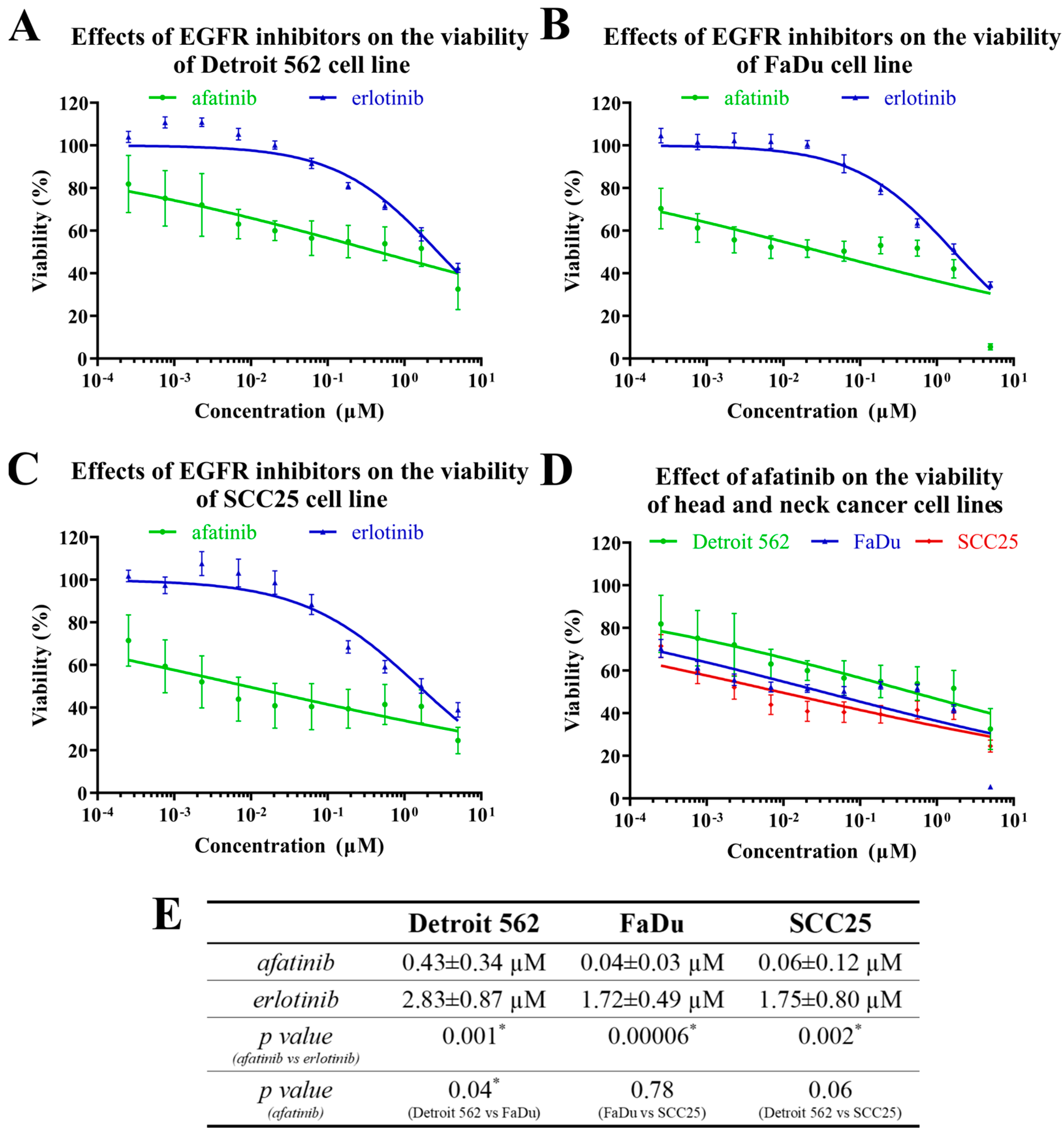
2.2. The Effects of EGFR Inhibitors on Head-and-Neck-Cancer-Cell Viability
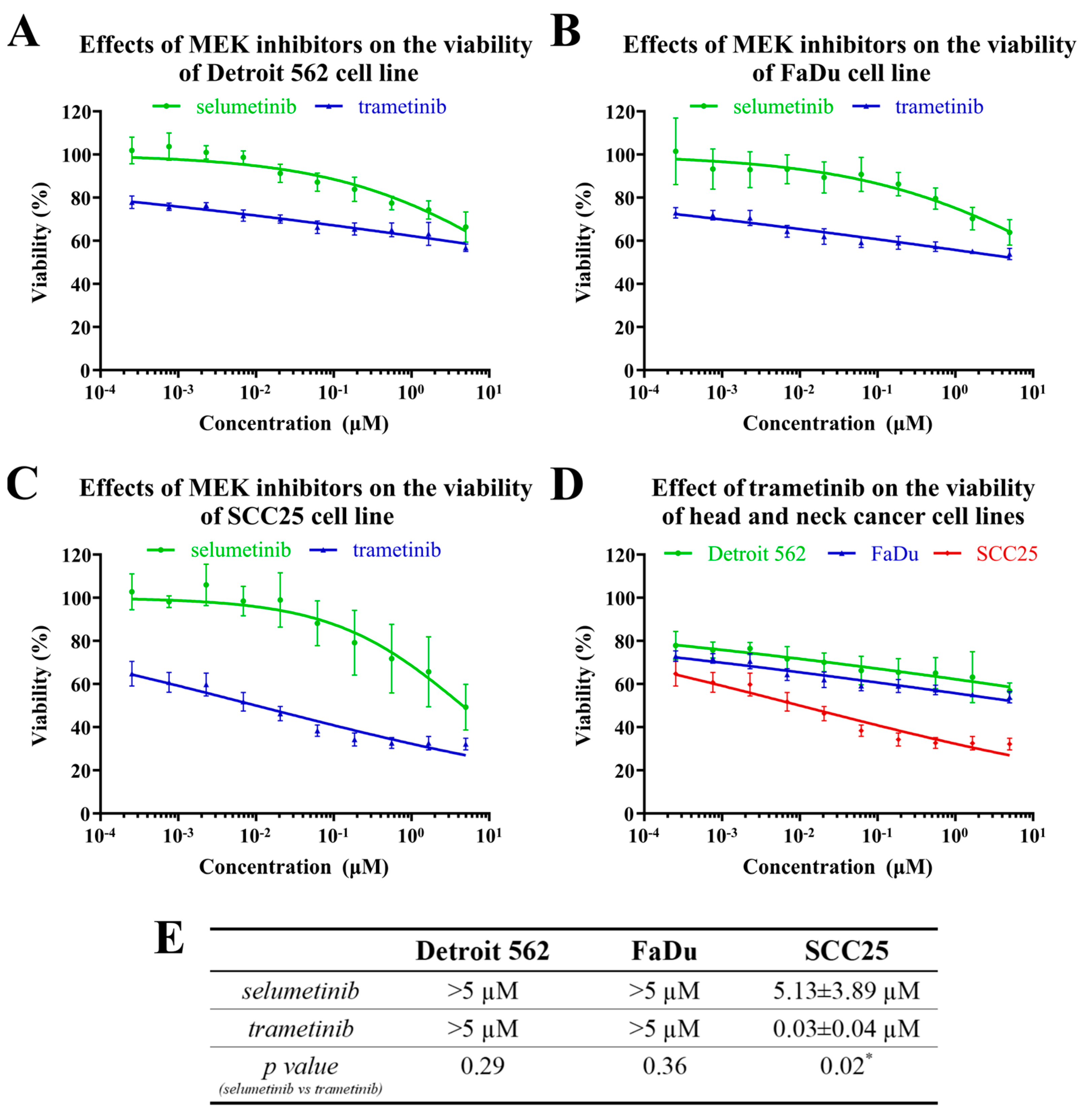
2.3. The Effects of MEK Inhibitors on the Viability of Head-and-Neck-Cancer-Cell Lines
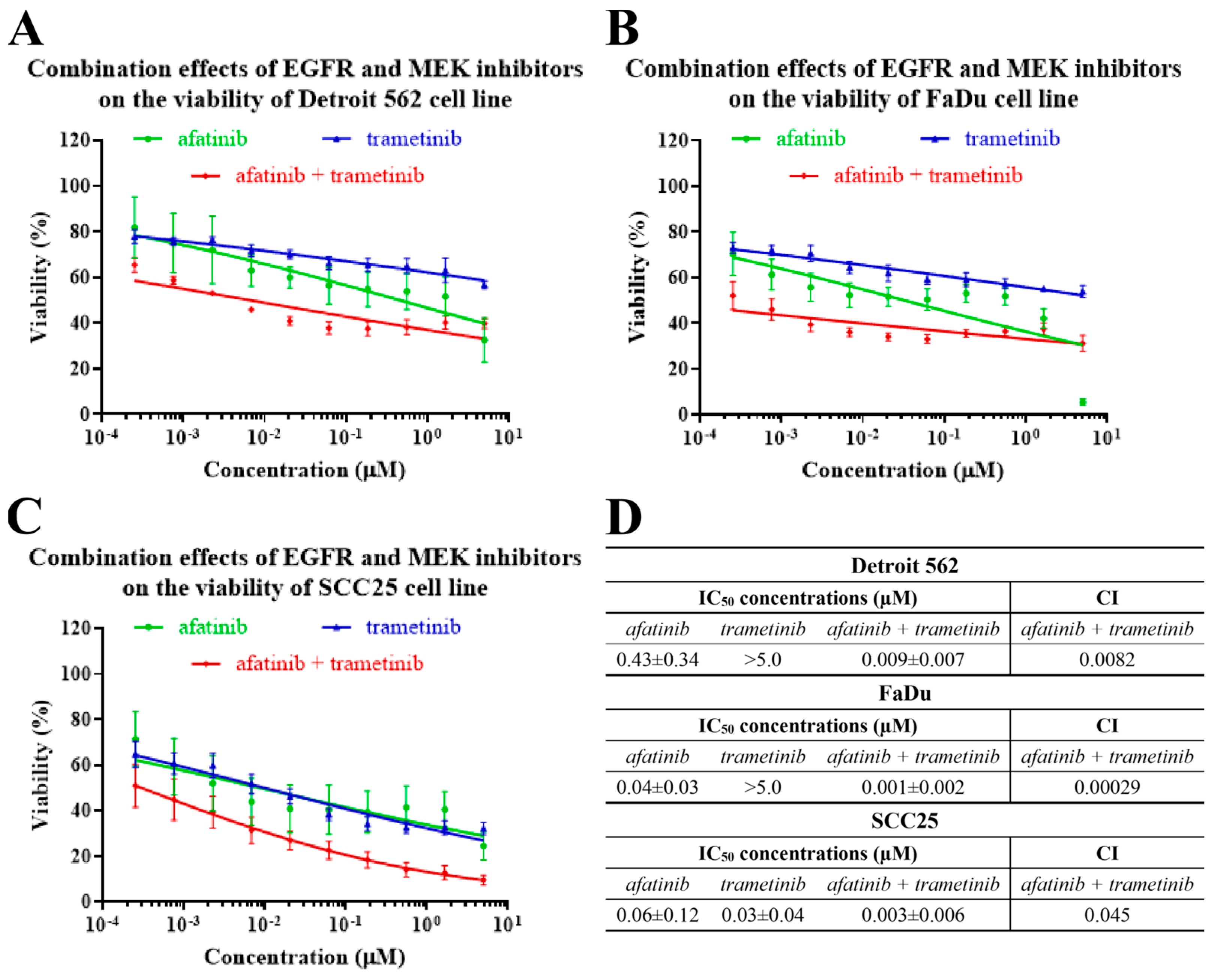
2.4. The Effects of EGFR- and MEK-Inhibitor Combinations on the Viability of Head-and-Neck-Cancer-Cell Lines
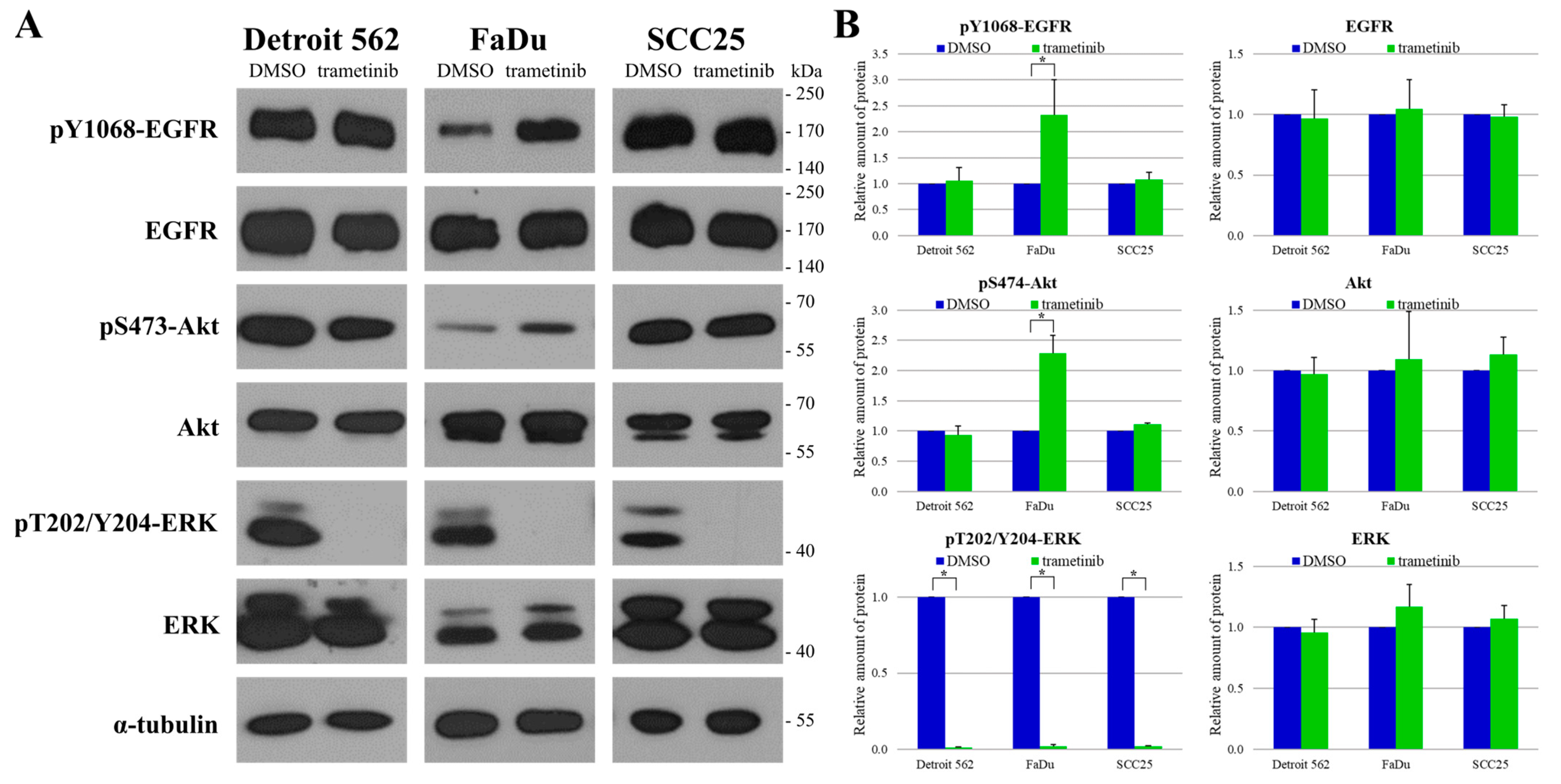
2.5. Protein Expression and Phosphorylation Analysis of Head and Neck Cancer Cell Lines
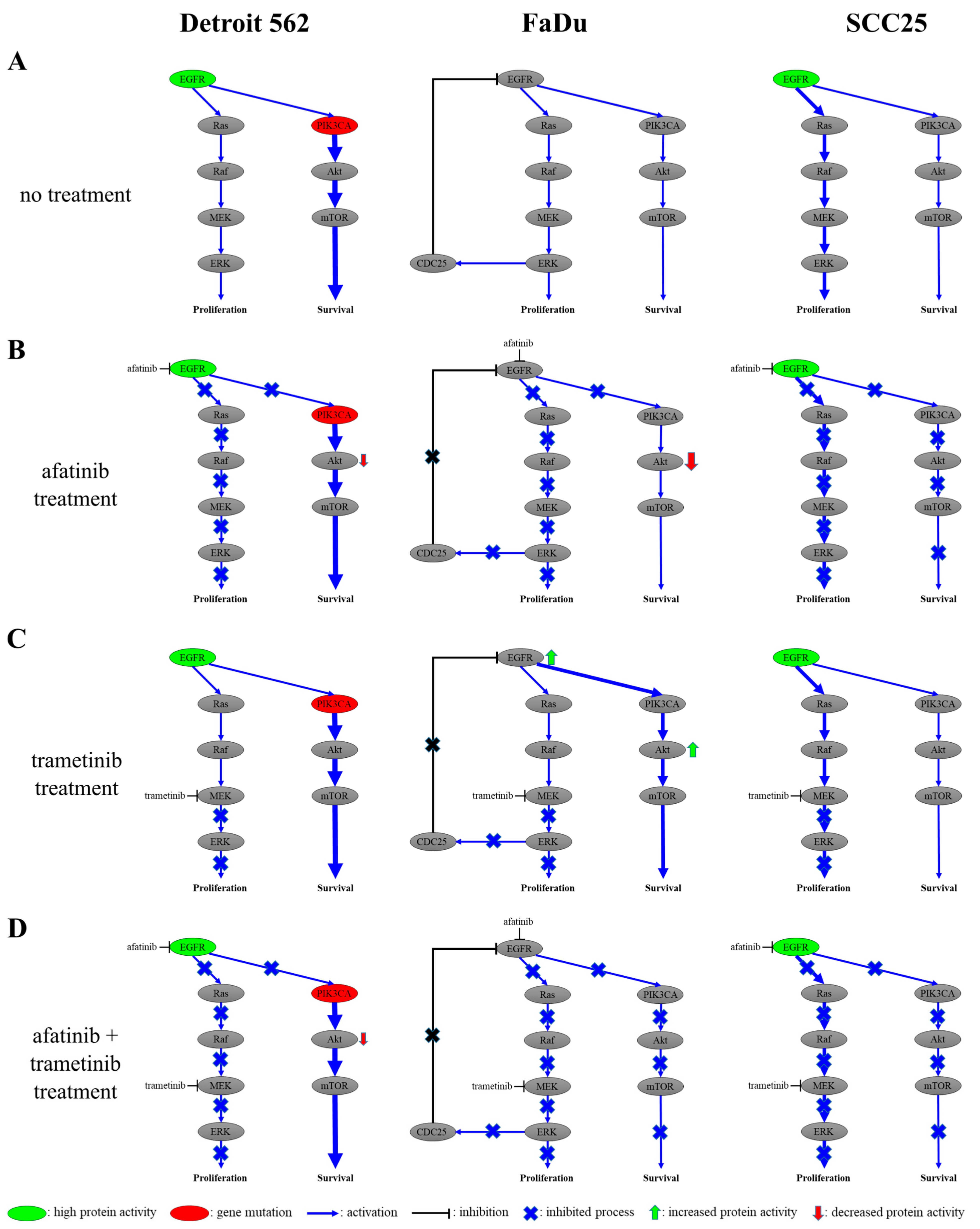
2.6. The Responses of Head-and-Neck-Cancer-Cell Lines to Trametinib Treatment
3. Discussion
4. Materials and Methods
4.1. Cell Culturing and Inhibitors
4.2. Cell Viability Assay and Drug Synergism
4.3. Western-Blot Analysis
4.4. Patients
4.5. Tissue Microarray (TMA) and Immunohistochemistry
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, M.E.; Chiocca, S. Human papillomavirus as a driver of head and neck cancers. Br. J. Cancer 2020, 122, 306–314. [Google Scholar] [CrossRef]
- Wise-Draper, T.M.; Bahig, H.; Karivedu, V.; Burtness, B. Current Therapy for Metastatic Head and Neck Cancer: Evidence, Opportunities, and Challenges. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Kordbacheh, F.; Farah, C.S. Current and Emerging Molecular Therapies for Head and Neck Squamous Cell Carcinoma. Cancers 2021, 13, 5471. [Google Scholar] [CrossRef]
- Kiss, F.; Pohoczky, K.; Gorbe, A.; Dembrovszky, F.; Kiss, S.; Hegyi, P.; Szako, L.; Toth, L.; Ezer, E.S.; Szalai, E.; et al. Addition of epidermal growth factor receptor inhibitors to standard chemotherapy increases survival of advanced head and neck squamous cell carcinoma patients: A systematic review and meta-analysis. Oral Dis. 2022. ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.; Siwak, D.R.; Chai, R.; LaValle, C.; Seethala, R.R.; Wang, L.; Cieply, K.; Sherer, C.; Joy, C.; Mills, G.B.; et al. Tumor epidermal growth factor receptor and EGFR PY1068 are independent prognostic indicators for head and neck squamous cell carcinoma. Clin. Cancer Res. 2012, 18, 2278–2289. [Google Scholar] [CrossRef]
- Bossi, P.; Resteghini, C.; Paielli, N.; Licitra, L.; Pilotti, S.; Perrone, F. Prognostic and predictive value of EGFR in head and neck squamous cell carcinoma. Oncotarget 2016, 7, 74362–74379. [Google Scholar] [CrossRef]
- Fasano, M.; Della Corte, C.M.; Viscardi, G.; Di Liello, R.; Paragliola, F.; Sparano, F.; Iacovino, M.L.; Castrichino, A.; Doria, F.; Sica, A.; et al. Head and neck cancer: The role of anti-EGFR agents in the era of immunotherapy. Ther. Adv. Med. Oncol. 2021, 13, 1758835920949418. [Google Scholar] [CrossRef]
- Kriegs, M.; Clauditz, T.S.; Hoffer, K.; Bartels, J.; Buhs, S.; Gerull, H.; Zech, H.B.; Bussmann, L.; Struve, N.; Rieckmann, T.; et al. Analyzing expression and phosphorylation of the EGF receptor in HNSCC. Sci. Rep. 2019, 9, 13564. [Google Scholar] [CrossRef]
- Zhao, Y.; Adjei, A.A. The clinical development of MEK inhibitors. Nat. Rev. Clin. Oncol. 2014, 11, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Hauschild, A.; Santinami, M.; Atkinson, V.; Mandala, M.; Chiarion-Sileni, V.; Larkin, J.; Nyakas, M.; Dutriaux, C.; Haydon, A.; et al. Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N. Engl. J. Med. 2017, 377, 1813–1823. [Google Scholar] [CrossRef] [PubMed]
- Stockley, T.L.; Oza, A.M.; Berman, H.K.; Leighl, N.B.; Knox, J.J.; Shepherd, F.A.; Chen, E.X.; Krzyzanowska, M.K.; Dhani, N.; Joshua, A.M.; et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: The Princess Margaret IMPACT/COMPACT trial. Genome Med. 2016, 8, 109. [Google Scholar] [CrossRef] [PubMed]
- Prahallad, A.; Sun, C.; Huang, S.; Di Nicolantonio, F.; Salazar, R.; Zecchin, D.; Beijersbergen, R.L.; Bardelli, A.; Bernards, R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012, 483, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wawrose, J.S.; Gooding, W.E.; Garraway, L.A.; Lui, V.W.; Peyser, N.D.; Grandis, J.R. Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: A rational approach to preclinical model selection. Mol. Cancer Res. 2014, 12, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Bareschino, M.A.; Schettino, C.; Troiani, T.; Martinelli, E.; Morgillo, F.; Ciardiello, F. Erlotinib in cancer treatment. Ann. Oncol. 2007, 18, vi35–vi41. [Google Scholar] [CrossRef]
- Maarof, N.N.N.; Alsalahi, A.; Abdulmalek, E.; Fakurazi, S.; Tejo, B.A.; Abdul Rahman, M.B. Efficacy of Afatinib in the Treatment of Patients with Non-Small Cell Lung Cancer and Head and Neck Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. Cancers 2021, 13, 688. [Google Scholar] [CrossRef]
- Sullivan, I.; Planchard, D. Next-Generation EGFR Tyrosine Kinase Inhibitors for Treating EGFR-Mutant Lung Cancer beyond First Line. Front. Med. 2016, 3, 76. [Google Scholar] [CrossRef]
- Truong, D.H.; Le, V.K.H.; Pham, T.T.; Dao, A.H.; Pham, T.P.D.; Tran, T.H. Delivery of erlotinib for enhanced cancer treatment: An update review on particulate systems. J. Drug Deliv. Sci. Technol. 2020, 55, 101348. [Google Scholar] [CrossRef]
- Cheng, Y.; Tian, H. Current Development Status of MEK Inhibitors. Molecules 2017, 22, 1551. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur. J. Cancer 2001, 37 (Suppl. 4), 3–8. [Google Scholar] [CrossRef]
- Wang, F.; Wang, S.; Wang, Z.; Duan, J.; An, T.; Zhao, J.; Bai, H.; Wang, J. Phosphorylated EGFR expression may predict outcome of EGFR-TKIs therapy for the advanced NSCLC patients with wild-type EGFR. J. Exp. Clin. Cancer Res. CR 2012, 31, 65. [Google Scholar] [CrossRef] [PubMed]
- Brauswetter, D.; Gurbi, B.; Varga, A.; Varkondi, E.; Schwab, R.; Banhegyi, G.; Fabian, O.; Keri, G.; Valyi-Nagy, I.; Petak, I. Molecular subtype specific efficacy of MEK inhibitors in pancreatic cancers. PLoS ONE 2017, 12, e0185687. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Windon, M.J.; D’Souza, G.; Rettig, E.M.; Westra, W.H.; van Zante, A.; Wang, S.J.; Ryan, W.R.; Mydlarz, W.K.; Ha, P.K.; Miles, B.A.; et al. Increasing prevalence of human papillomavirus-positive oropharyngeal cancers among older adults. Cancer 2018, 124, 2993–2999. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.Y.; Lam, J.W.; Chan, K.C. Clinical utility of circulating Epstein-Barr virus DNA analysis for the management of nasopharyngeal carcinoma. Chin. Clin. Oncol. 2016, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Rehmani, H.S.; Issaeva, N. EGFR in head and neck squamous cell carcinoma: Exploring possibilities of novel drug combinations. Ann. Transl. Med. 2020, 8, 813. [Google Scholar] [CrossRef]
- Ngan, H.L.; Law, C.H.; Choi, Y.C.Y.; Chan, J.Y.; Lui, V.W.Y. Precision drugging of the MAPK pathway in head and neck cancer. NPJ Genom. Med. 2022, 7, 20. [Google Scholar] [CrossRef]
- Mercer, K.E.; Pritchard, C.A. Raf proteins and cancer: B-Raf is identified as a mutational target. Biochim. Biophys. Acta 2003, 1653, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Langhanki, L.; Sommerer, F.; Markwarth, A.; Wittekind, C.; Tannapfel, A. Mutations of the BRAF gene in squamous cell carcinoma of the head and neck. Oncogene 2003, 22, 4757–4759. [Google Scholar] [CrossRef]
- Bruckman, K.C.; Schonleben, F.; Qiu, W.; Woo, V.L.; Su, G.H. Mutational analyses of the BRAF, KRAS, and PIK3CA genes in oral squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2010, 110, 632–637. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, T.G.; De Carvalho, A.C.; Maia, D.C.; Ogawa, J.K.; Carvalho, A.L.; Vettore, A.L. Search for mutations in signaling pathways in head and neck squamous cell carcinoma. Oncol. Rep. 2013, 30, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Zhu, A.; Gu, X. Mitogen-activated protein kinase inhibition-induced modulation of epidermal growth factor receptor signaling in human head and neck squamous cell carcinoma. Head Neck 2021, 43, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Neuzillet, C.; Tijeras-Raballand, A.; de Mestier, L.; Cros, J.; Faivre, S.; Raymond, E. MEK in cancer and cancer therapy. Pharmacol. Ther. 2014, 141, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Chappell, W.H.; Steelman, L.S.; Long, J.M.; Kempf, R.C.; Abrams, S.L.; Franklin, R.A.; Bäsecke, J.; Stivala, F.; Donia, M.; Fagone, P.; et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: Rationale and importance to inhibiting these pathways in human health. Oncotarget 2011, 2, 135–164. [Google Scholar] [CrossRef] [PubMed]
- Britten, C.D. PI3K and MEK inhibitor combinations: Examining the evidence in selected tumor types. Cancer Chemother. Pharmacol. 2013, 71, 1395–1409. [Google Scholar] [CrossRef]
- Avraham, R.; Yarden, Y. Feedback regulation of EGFR signalling: Decision making by early and delayed loops. Nat. Reviews. Mol. Cell Biol. 2011, 12, 104–117. [Google Scholar] [CrossRef]
- Wang, R.; He, G.; Nelman-Gonzalez, M.; Ashorn, C.L.; Gallick, G.E.; Stukenberg, P.T.; Kirschner, M.W.; Kuang, J. Regulation of Cdc25C by ERK-MAP kinases during the G2/M transition. Cell 2007, 128, 1119–1132. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, M.; Lazo, J.S.; Carr, B.I. Identification of epidermal growth factor receptor as a target of Cdc25A protein phosphatase. J. Biol. Chem. 2002, 277, 19470–19475. [Google Scholar] [CrossRef]
- Ligasova, A.; Vydrzalova, M.; Burianova, R.; Bruckova, L.; Vecerova, R.; Janostakova, A.; Koberna, K. A New Sensitive Method for the Detection of Mycoplasmas Using Fluorescence Microscopy. Cells 2019, 8, 1510. [Google Scholar] [CrossRef]
- Chou, T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Calder, J.; Nagelberg, A.; Luu, J.; Lu, D.; Lockwood, W.W. Resistance to BET inhibitors in lung adenocarcinoma is mediated by casein kinase phosphorylation of BRD4. Oncogenesis 2021, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Gurbi, B.; Brauswetter, D.; Varga, A.; Gyulavari, P.; Penzes, K.; Muranyi, J.; Zambo, V.; Birtalan, E.; Krenacs, T.; Becker, D.L.; et al. The Potential Impact of Connexin 43 Expression on Bcl-2 Protein Level and Taxane Sensitivity in Head and Neck Cancers-In Vitro Studies. Cancers 2019, 11, 1848. [Google Scholar] [CrossRef] [PubMed]
- Szentkuti, G.; Danos, K.; Brauswetter, D.; Kiszner, G.; Krenacs, T.; Csako, L.; Repassy, G.; Tamas, L. Correlations between prognosis and regional biomarker profiles in head and neck squamous cell carcinomas. Pathol. Oncol. Res. 2015, 21, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Kim, S.H.; Park, S.Y.; Yoo, J.; Kim, S.K.; Kim, H.K. Identification of EGFR Mutations by Immunohistochemistry with EGFR Mutation-Specific Antibodies in Biopsy and Resection Specimens from Pulmonary Adenocarcinoma. Cancer Res. Treat. 2015, 47, 653–660. [Google Scholar] [CrossRef]
- Wen, Y.H.; Brogi, E.; Hasanovic, A.; Ladanyi, M.; Soslow, R.A.; Chitale, D.; Shia, J.; Moreira, A.L. Immunohistochemical staining with EGFR mutation-specific antibodies: High specificity as a diagnostic marker for lung adenocarcinoma. Mod. Pathol. 2013, 26, 1197–1203. [Google Scholar] [CrossRef]


| Variable | No. of Patients |
|---|---|
| Total no. of patients | 97 |
| Sex | |
| Male | 79 |
| Female | 18 |
| Age (year) | |
| Mean | 61 (43–81) |
| Localization | |
| Oropharynx | 36 |
| Hypopharynx | 35 |
| Supraglottis | 24 |
| Glottis | 2 |
| TNM 1 T parameter | |
| 1 | 14 |
| 2 | 29 |
| 3 | 27 |
| 4a | 18 |
| 4b | 9 |
| TNM 1 N parameter | |
| 0 | 46 |
| 1 | 16 |
| 2a | 6 |
| 2b | 12 |
| 2c | 14 |
| 3 | 3 |
| TNM 1 M parameter | |
| 0 | 92 |
| 1 | 5 |
| TNM 1 stage | |
| 1 | 26 |
| 2 | 66 |
| 3 | 5 |
| Grade | |
| 1 | 6 |
| 2 | 43 |
| 3 | 34 |
| No data | 14 |
| Tobacco use | |
| Never | 10 |
| Previously yes | 28 |
| Currently | 57 |
| No data | 2 |
| Alcohol use | |
| Never | 23 |
| Previously yes | 32 |
| Currently | 42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurbi, B.; Brauswetter, D.; Pénzes, K.; Varga, A.; Krenács, T.; Dános, K.; Birtalan, E.; Tamás, L.; Csala, M. MEK Is a Potential Indirect Target in Subtypes of Head and Neck Cancers. Int. J. Mol. Sci. 2023, 24, 2782. https://doi.org/10.3390/ijms24032782
Gurbi B, Brauswetter D, Pénzes K, Varga A, Krenács T, Dános K, Birtalan E, Tamás L, Csala M. MEK Is a Potential Indirect Target in Subtypes of Head and Neck Cancers. International Journal of Molecular Sciences. 2023; 24(3):2782. https://doi.org/10.3390/ijms24032782
Chicago/Turabian StyleGurbi, Bianka, Diána Brauswetter, Kinga Pénzes, Attila Varga, Tibor Krenács, Kornél Dános, Ede Birtalan, László Tamás, and Miklós Csala. 2023. "MEK Is a Potential Indirect Target in Subtypes of Head and Neck Cancers" International Journal of Molecular Sciences 24, no. 3: 2782. https://doi.org/10.3390/ijms24032782
APA StyleGurbi, B., Brauswetter, D., Pénzes, K., Varga, A., Krenács, T., Dános, K., Birtalan, E., Tamás, L., & Csala, M. (2023). MEK Is a Potential Indirect Target in Subtypes of Head and Neck Cancers. International Journal of Molecular Sciences, 24(3), 2782. https://doi.org/10.3390/ijms24032782








