Unravelling How Single-Stranded DNA Binding Protein Coordinates DNA Metabolism Using Single-Molecule Approaches
Abstract
1. Introduction
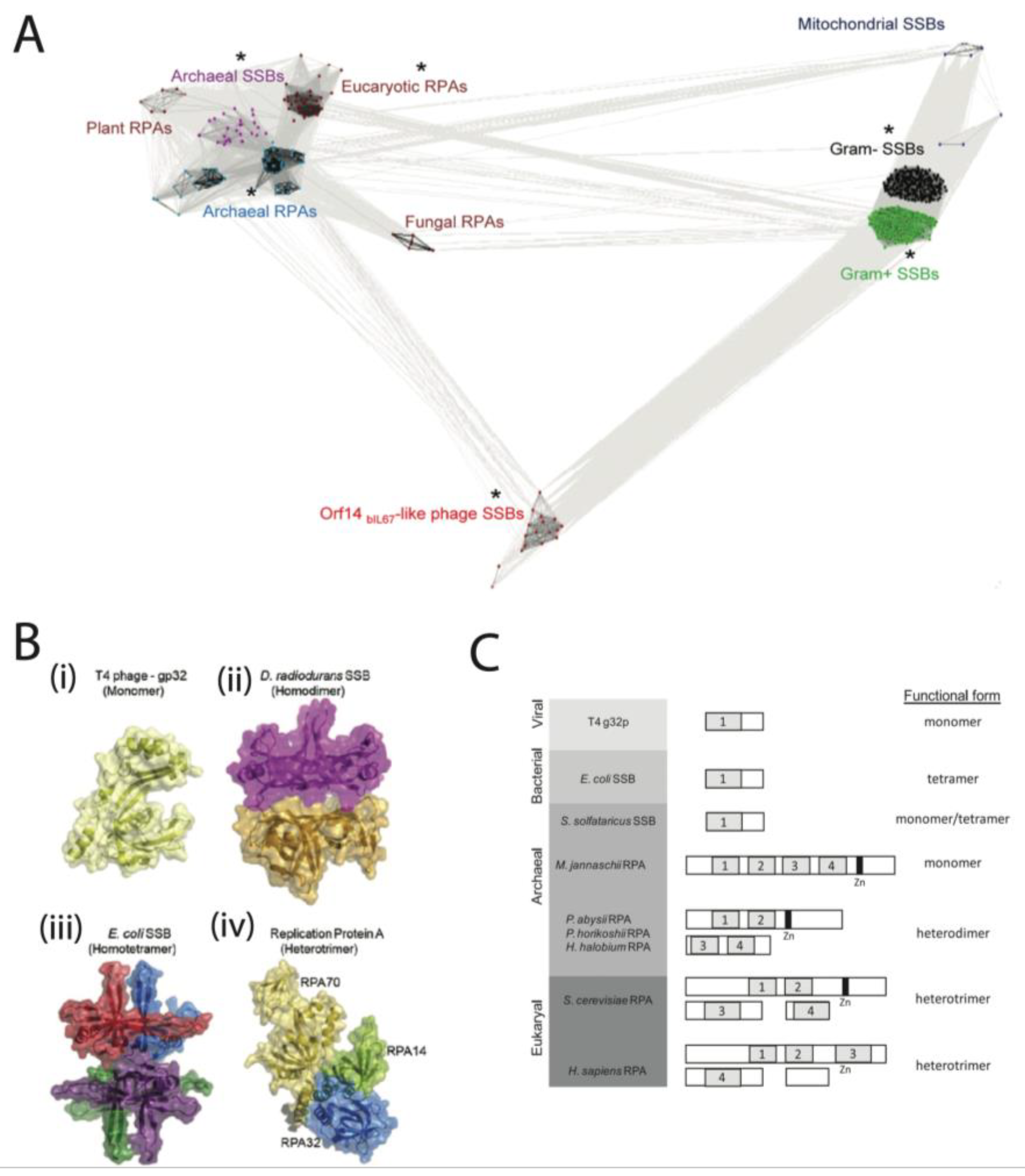
2. Classification of SSB
2.1. Properties of SSB
2.2. Classification of SSB
3. Single-Molecule Toolbox to Study SSB
3.1. Single-Molecule Force Studies of SSB–ssDNA Interactions
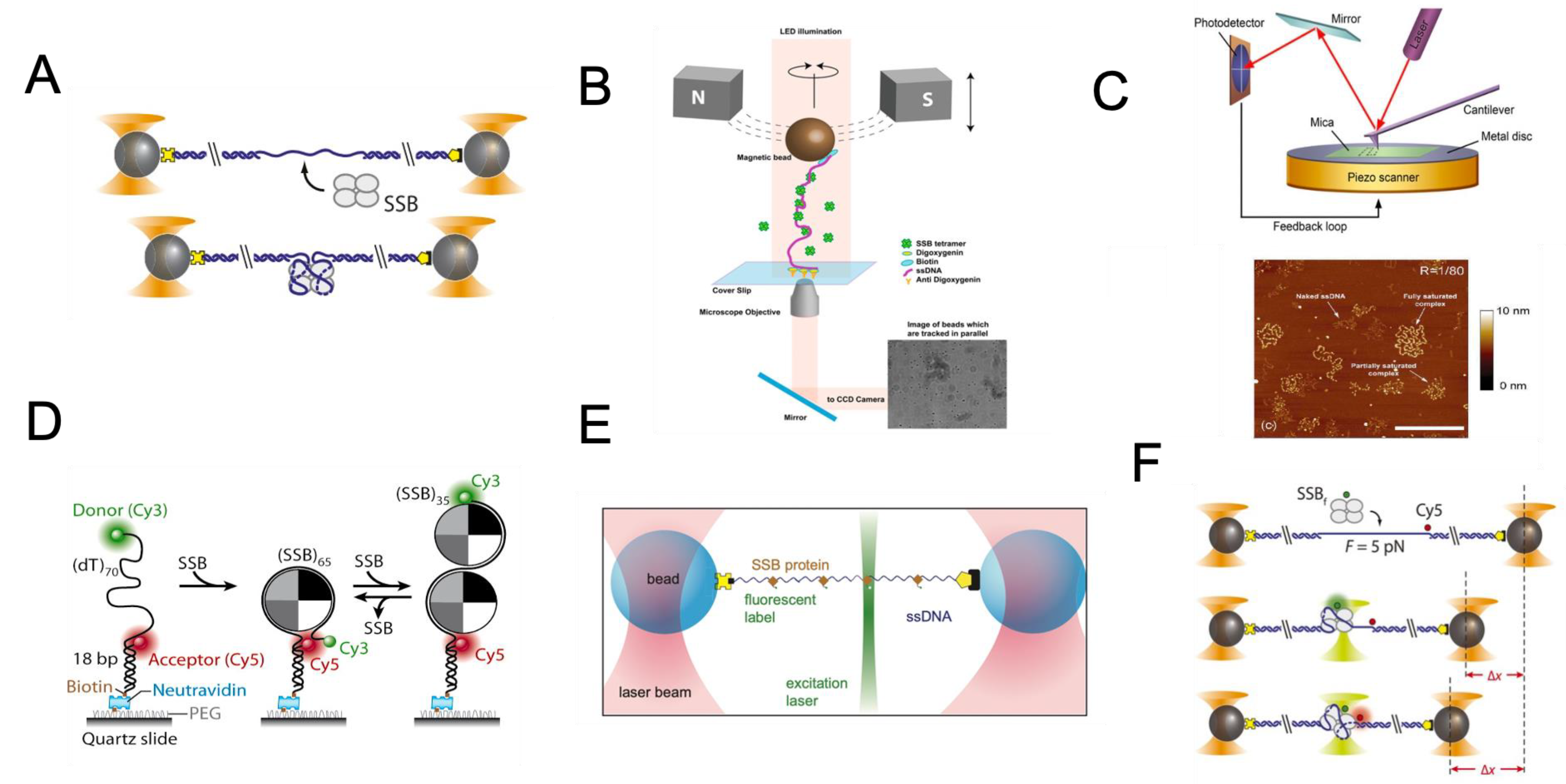
3.2. Image Measurement of SSB-ssDNA Complex
3.3. Hybrid Single-Molecule Tools
3.4. Example Output of Single-Molecule Studies
4. Examine the Interaction between ssDNA with SSB
4.1. General Binding Dynamics of SSB
4.2. Binding Dynamics of SSB to ssDNA under Tension
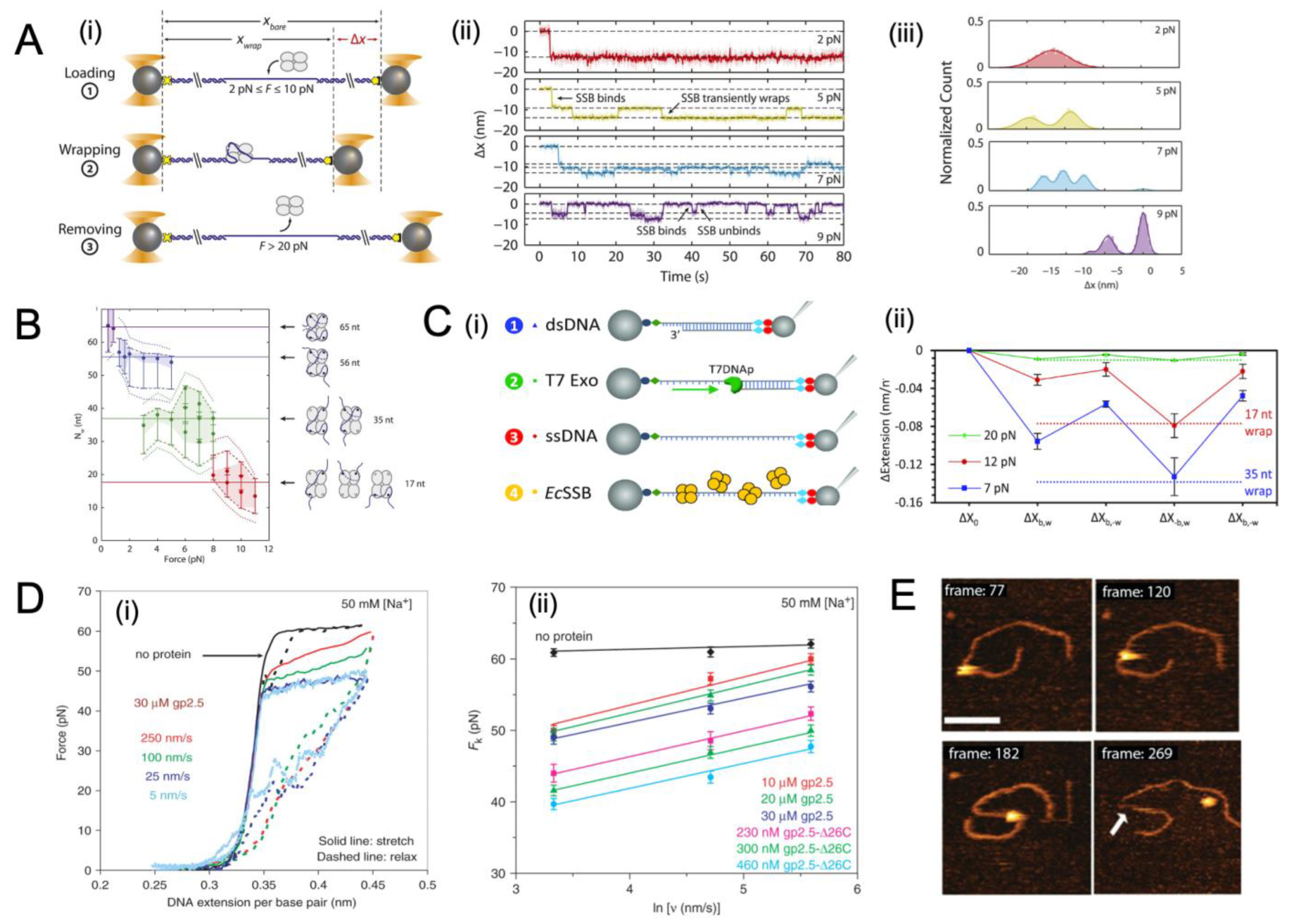
4.3. Movement of SSB on ssDNA Probed with Single-Molecule Approaches

4.4. Sequence-Dependent Properties of SSB
5. Coordination Role of SSB in DNA Metabolism
5.1. Overview of Single-Molecule Studies on SSB Interacting with Helicase
5.1.1. Interplay with Replicative Helicase CMG Complex

5.1.2. Interplay with Recombinational Repair Helicase XPD
5.1.3. Interplay with Recombinational Repair Helicase RecQ
5.1.4. Interplay with Replication Restart Helicase PriA
5.2. SSB Interacting with Replicative DNA Polymerase during Primer Extension

5.3. Single-Molecule Studies on SSB with DNA Polymerase during Strand Exchange
5.4. Single-Molecule Studies of SSB Interplay with Recombinase
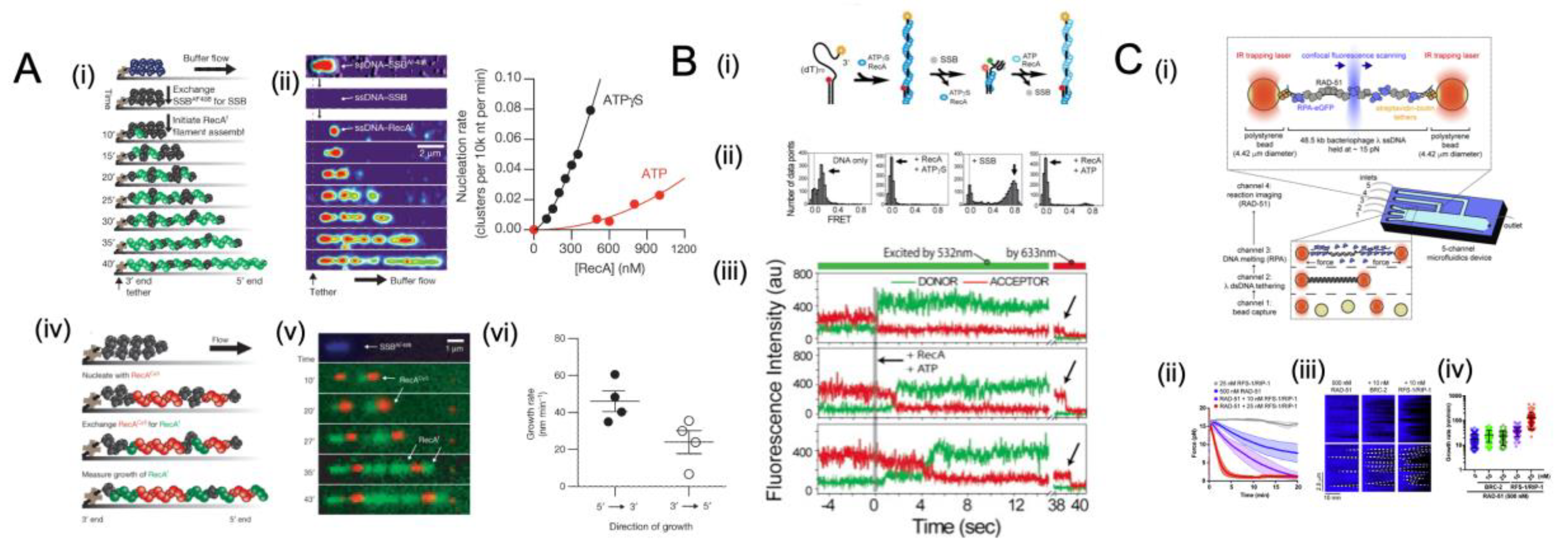
5.5. Chemo-Mechanical Pushing of E. coli SSB by a Translocating Protein Partner
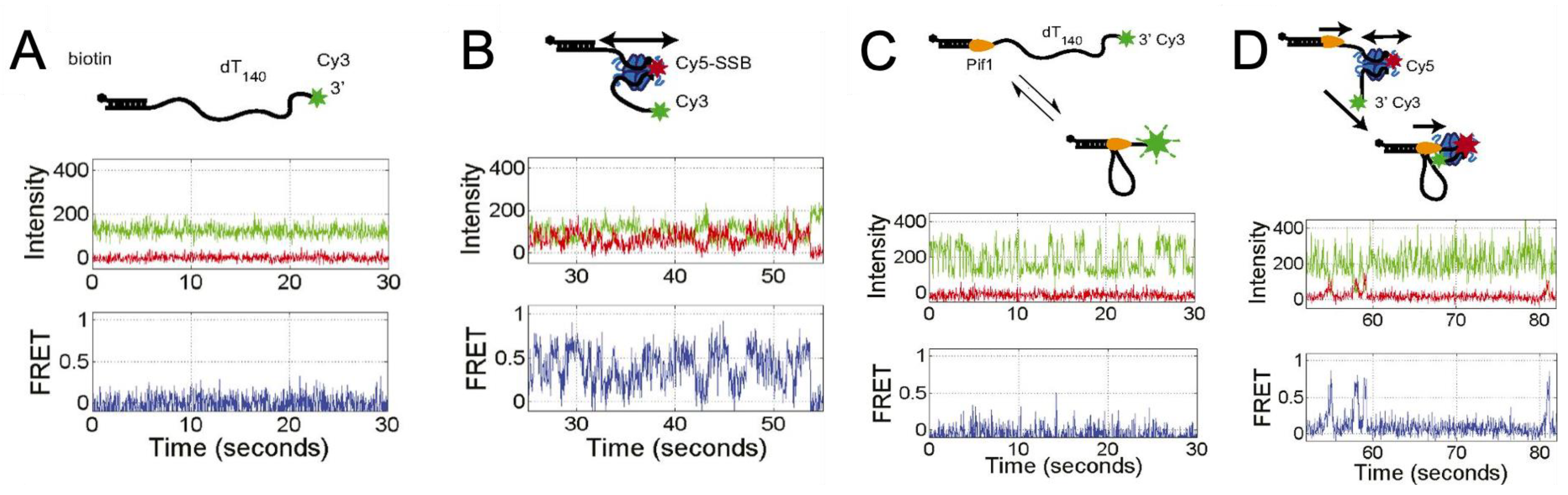
6. Conclusions
6.1. General Remarks on SSB
6.2. Potential Interesting Single-Molecule Experiments of SSBs
6.2.1. How the DNAp Displaces SSB from Different Organisms
6.2.2. Hybrid SSBs from Both Host and Viral Organisms Interacting with ssDNA
6.2.3. SSB Functions within a Complete Replisome
6.2.4. SSB as a Drug Target
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Vlaminck, I.; Vidic, I.; van Loenhout, M.T.J.; Kanaar, R.; Lebbink, J.H.G.; Dekker, C. Torsional regulation of hRPA-induced unwinding of double-stranded DNA. Nucleic Acids Res. 2010, 38, 4133–4142. [Google Scholar] [CrossRef] [PubMed]
- Hatch, K.; Danilowicz, C.; Coljee, V.; Prentiss, M. Direct measurements of the stabilization of single-stranded DNA under tension by single-stranded binding proteins. Phys. Rev. E 2007, 76, 021916. [Google Scholar] [CrossRef] [PubMed]
- Hatch, K.; Danilowicz, C.; Coljee, V.; Prentiss, M. Measurement of the salt-dependent stabilization of partially open DNA by Escherichia coli SSB protein. Nucleic Acids Res. 2007, 36, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Pant, K.; Karpel, R.L.; Rouzina, I.; Williams, M.C. Mechanical Measurement of Single-molecule Binding Rates: Kinetics of DNA Helix-destabilization by T4 Gene 32 Protein. J. Mol. Biol. 2004, 336, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Pant, K.; Karpel, R.L.; Rouzina, I.; Williams, M.C. Salt Dependent Binding of T4 Gene 32 Protein to Single and Double-stranded DNA: Single Molecule Force Spectroscopy Measurements. J. Mol. Biol. 2005, 349, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Shokri, L.; Marintcheva, B.; Eldib, M.; Hanke, A.; Rouzina, I.; Williams, M.C. Kinetics and thermodynamics of salt-dependent T7 gene 2.5 protein binding to single- and double-stranded DNA. Nucleic Acids Res. 2008, 36, 5668–5677. [Google Scholar] [CrossRef]
- Shokri, L.; Marintcheva, B.; Richardson, C.C.; Rouzina, I.; Williams, M.C. Single Molecule Force Spectroscopy of Salt-dependent Bacteriophage T7 Gene 2.5 Protein Binding to Single-stranded DNA. J. Biol. Chem. 2006, 281, 38689–38696. [Google Scholar] [CrossRef]
- Broderick, S.; Rehmet, K.; Concannon, C.; Nasheuer, H.-P. Eukaryotic Single-Stranded DNA Binding Proteins: Central Factors in Genome Stability. In Genome Stability and Human Diseases; Springer: Dordrecht, The Netherlands, 2009; Volume 50, pp. 143–163. [Google Scholar] [CrossRef]
- Szczepankowska, A.K.; Prestel, E.; Mariadassou, M.; Bardowski, J.K.; Bidnenko, E. Phylogenetic and Complementation Analysis of a Single-Stranded DNA Binding Protein Family from Lactococcal Phages Indicates a Non-Bacterial Origin. PLoS ONE 2011, 6, e26942. [Google Scholar] [CrossRef]
- Guo, J.-T.; Malik, F. Single-Stranded DNA Binding Proteins and Their Identification Using Machine Learning-Based Approaches. Biomolecules 2022, 12, 1187. [Google Scholar] [CrossRef]
- Cernooka, E.; Rumnieks, J.; Tars, K.; Kazaks, A. Structural Basis for DNA Recognition of a Single-stranded DNA-binding Protein from Enterobacter Phage Enc34. Sci. Rep. 2017, 7, 15529. [Google Scholar] [CrossRef]
- Ha, T.; Kozlov, A.G.; Lohman, T.M. Single-Molecule Views of Protein Movement on Single-Stranded DNA. Annu. Rev. Biophys. 2012, 41, 295–319. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.-W.; Lee, Y.-J.; Wang, C.-H.; Huang, H.; Sun, Y.-J. Single-Stranded DNA-Binding Protein Complex from Helicobacter pylori Suggests an ssDNA-Binding Surface. J. Mol. Biol. 2009, 388, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Saikrishnan, K.; Jeyakanthan, J.; Venkatesh, J.; Acharya, N.; Sekar, K.; Varshney, U.; Vijayan, M. Structure of Mycobacterium tuberculosis Single-stranded DNA-binding Protein. Variability in Quaternary Structure and Its Implications. J. Mol. Biol. 2003, 331, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Saikrishnan, K.; Manjunath, G.P.; Singh, P.; Jeyakanthan, J.; Dauter, Z.; Sekar, K.; Muniyappa, K.; Vijayan, M. Structure of Mycobacterium smegmatissingle-stranded DNA-binding protein and a comparative study involving homologus SSBs: Biological implications of structural plasticity and variability in quaternary association. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 1140–1148. [Google Scholar] [CrossRef]
- Suksombat, S.; Khafizov, R.; Kozlov, A.G.; Lohman, T.M.; Chemla, Y.R. Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways. eLife 2015, 4, e08193. [Google Scholar] [CrossRef]
- Bochkareva, E.; Korolev, S.; Lees-Miller, S.P.; Bochkarev, A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002, 21, 1855–1863. [Google Scholar] [CrossRef]
- Jose, D.; Weitzel, S.E.; Baase, W.A.; von Hippel, P.H. Mapping the interactions of the single-stranded DNA binding protein of bacteriophage T4 (gp32) with DNA lattices at single nucleotide resolution: gp32 monomer binding. Nucleic Acids Res. 2015, 43, 9276–9290. [Google Scholar] [CrossRef]
- Hernandez, A.J.; Richardson, C.C. Gp2.5, the multifunctional bacteriophage T7 single-stranded DNA binding protein. Semin. Cell Dev. Biol. 2018, 86, 92–101. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, Y.; Huang, D.; Liu, H.; Justin, N.; Zhao, W.; Liu, J.; Peng, Y. Structural features of the single-stranded DNA-binding protein MoSub1 from Magnaporthe oryzae. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1071–1076. [Google Scholar] [CrossRef]
- Qian, Y.; Johnson, K.A. The human mitochondrial single-stranded DNA-binding protein displays distinct kinetics and thermodynamics of DNA binding and exchange. J. Biol. Chem. 2017, 292, 13068–13084. [Google Scholar] [CrossRef]
- Kozlov, A.G.; Weiland, E.; Mittal, A.; Waldman, V.; Antony, E.; Fazio, N.; Pappu, R.V.; Lohman, T.M. Intrinsically Disordered C-Terminal Tails of E. coli Single-Stranded DNA Binding Protein Regulate Cooperative Binding to Single-Stranded DNA. J. Mol. Biol. 2015, 427, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Marintcheva, B.; Takahashi, M.; Richardson, C.C. C-terminal Phenylalanine of Bacteriophage T7 Single-stranded DNA-binding Protein Is Essential for Strand Displacement Synthesis by T7 DNA Polymerase at a Nick in DNA. J. Biol. Chem. 2009, 284, 30339–30349. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.G.; Cox, M.M.; Lohman, T.M. Regulation of Single-stranded DNA Binding by the C Termini of Escherichia coli Single-stranded DNA-binding (SSB) Protein. J. Biol. Chem. 2010, 285, 17246–17252. [Google Scholar] [CrossRef] [PubMed]
- Perumal, S.K.; Nelson, S.W.; Benkovic, S.J. Interaction of T4 UvsW Helicase and Single-Stranded DNA Binding Protein gp32 through Its Carboxy-Terminal Acidic Tail. J. Mol. Biol. 2013, 425, 2823–2839. [Google Scholar] [CrossRef]
- Mer, G.; Bochkarev, A.; Gupta, R.; Bochkareva, E.; Frappier, L.; Ingles, C.; Edwards, A.M.; Chazin, W.J. Structural Basis for the Recognition of DNA Repair Proteins UNG2, XPA, and RAD52 by Replication Factor RPA. Cell 2000, 103, 449–456. [Google Scholar] [CrossRef]
- Sharma, N.; Chakravarthy, S.; Longley, M.J.; Copeland, W.; Prakash, A. The C-terminal tail of the NEIL1 DNA glycosylase interacts with the human mitochondrial single-stranded DNA binding protein. DNA Repair 2018, 65, 11–19. [Google Scholar] [CrossRef]
- Lindner, C.; Nijland, R.; van Hartskamp, M.; Bron, S.; Hamoen, L.W.; Kuipers, O.P. Differential Expression of Two Paralogous Genes of Bacillus subtilis Encoding Single-Stranded DNA Binding Protein. J. Bacteriol. 2004, 186, 1097–1105. [Google Scholar] [CrossRef]
- Frickey, T.; Lupas, A. CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics 2004, 20, 3702–3704. [Google Scholar] [CrossRef]
- Antony, E.; Lohman, T.M. Dynamics of E. coli single stranded DNA binding (SSB) protein-DNA complexes. Semin. Cell Dev. Biol. 2019, 86, 102–111. [Google Scholar] [CrossRef]
- Haseltine, C. Single-Stranded DNA-Binding Proteins. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 450–452. ISBN 978-0-08-096156-9. [Google Scholar]
- Zhou, R.; Kozlov, A.G.; Roy, R.; Zhang, J.; Korolev, S.; Lohman, T.M.; Ha, T. SSB Functions as a Sliding Platform that Migrates on DNA via Reptation. Cell 2011, 146, 222–232. [Google Scholar] [CrossRef]
- Honda, M.; Park, J.; Pugh, R.A.; Ha, T.; Spies, M. Single-Molecule Analysis Reveals Differential Effect of ssDNA-Binding Proteins on DNA Translocation by XPD Helicase. Mol. Cell 2009, 35, 694–703. [Google Scholar] [CrossRef]
- Naufer, M.N.; Morse, M.; Möller, G.B.; McIsaac, J.; Rouzina, I.; Beuning, P.J.; Williams, M.C. Multiprotein E. coli SSB–ssDNA complex shows both stable binding and rapid dissociation due to interprotein interactions. Nucleic Acids Res. 2021, 49, 1532–1549. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, D.; Zhang, W.; Hodeib, S.; Ducos, B.; Croquette, V.; Manosas, M. Magnetic Tweezers-Based Single-Molecule Assays to Study Interaction of E. coli SSB with DNA and RecQ Helicase. Methods Mol. Biol. 2021, 2281, 93–115. [Google Scholar] [CrossRef] [PubMed]
- Hamon, L.; Pastré, D.; Dupaigne, P.; Le Breton, C.; Le Cam, E.; Piétrement, O. High-resolution AFM imaging of single-stranded DNA-binding (SSB) protein--DNA complexes. Nucleic Acids Res. 2007, 35, e58. [Google Scholar] [CrossRef] [PubMed]
- Shlyakhtenko, L.S.; Lushnikov, A.Y.; Miyagi, A.; Lyubchenko, Y.L. Specificity of Binding of Single-Stranded DNA-Binding Protein to Its Target. Biochemistry 2012, 51, 1500–1509. [Google Scholar] [CrossRef]
- Kose, H.B.; Xie, S.; Cameron, G.; Strycharska, M.S.; Yardimci, H. Duplex DNA engagement and RPA oppositely regulate the DNA-unwinding rate of CMG helicase. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Bell, J.C.; Plank, J.L.; Dombrowski, C.C.; Kowalczykowski, S.C. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature 2012, 491, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Marshall, M.M.; Ruzicka, J.; Zahid, O.K.; Henrich, V.C.; Taylor, E.W.; Hall, A.R. Nanopore Analysis of Single-Stranded Binding Protein Interactions with DNA. Langmuir 2015, 31, 4582–4588. [Google Scholar] [CrossRef] [PubMed]
- Bryant, Z.; Stone, M.D.; Gore, J.; Smith, S.B.; Cozzarelli, N.R.; Bustamante, C. Structural transitions and elasticity from torque measurements on DNA. Nature 2003, 424, 338–341. [Google Scholar] [CrossRef]
- La Porta, A.; Wang, M.D. Optical Torque Wrench: Angular Trapping, Rotation, and Torque Detection of Quartz Microparticles. Phys. Rev. Lett. 2004, 92, 190801. [Google Scholar] [CrossRef]
- Lang, M.J.; Asbury, C.L.; Shaevitz, J.W.; Block, S.M. An Automated Two-Dimensional Optical Force Clamp for Single Molecule Studies. Biophys. J. 2002, 83, 491–501. [Google Scholar] [CrossRef]
- Hohng, S.; Zhou, R.; Nahas, M.K.; Yu, J.; Schulten, K.; Lilley, D.M.J.; Ha, T. Fluorescence-Force Spectroscopy Maps Two-Dimensional Reaction Landscape of the Holliday Junction. Science 2007, 318, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Gross, P.; Farge, G.; Peterman, E.J.; Wuite, G.J. Combining Optical Tweezers, Single-Molecule Fluorescence Microscopy, and Microfluidics for Studies of DNA–Protein Interactions. Methods Enzymol. 2010, 475, 427–453. [Google Scholar] [CrossRef]
- Schakenraad, K.; Biebricher, A.S.; Sebregts, M.; ten Bensel, B.; Peterman, E.J.G.; Wuite, G.J.L.; Heller, I.; Storm, C.; van der Schoot, P. Hyperstretching DNA. Nat. Commun. 2017, 8, 2197. [Google Scholar] [CrossRef] [PubMed]
- van Mameren, J.; Modesti, M.; Kanaar, R.; Wyman, C.; Wuite, G.J.; Peterman, E.J. Dissecting Elastic Heterogeneity along DNA Molecules Coated Partly with Rad51 Using Concurrent Fluorescence Microscopy and Optical Tweezers. Biophys. J. 2006, 91, L78–L80. [Google Scholar] [CrossRef] [PubMed]
- Greenleaf, W.J.; Woodside, M.T.; Abbondanzieri, E.A.; Block, S.M. Passive All-Optical Force Clamp for High-Resolution Laser Trapping. Phys. Rev. Lett. 2005, 95, 208102. [Google Scholar] [CrossRef] [PubMed]
- Candelli, A.; Hoekstra, T.P.; Farge, G.; Gross, P.; Peterman, E.J.G.; Wuite, G.J.L. A toolbox for generating single-stranded DNA in optical tweezers experiments. Biopolymers 2013, 99, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Sarlós, K.; Biebricher, A.S.; Bizard, A.H.; Bakx, J.A.M.; Ferreté-Bonastre, A.G.; Modesti, M.; Paramasivam, M.; Yao, Q.; Peterman, E.J.G.; Wuite, G.J.L.; et al. Reconstitution of anaphase DNA bridge recognition and disjunction. Nat. Struct. Mol. Biol. 2018, 25, 868–876. [Google Scholar] [CrossRef]
- Candelli, A.; Wuite, G.J.L.; Peterman, E.J.G. Combining optical trapping, fluorescence microscopy and micro-fluidics for single molecule studies of DNA–protein interactions. Phys. Chem. Chem. Phys. 2011, 13, 7263–7272. [Google Scholar] [CrossRef]
- King, G.A.; Burla, F.; Peterman, E.J.G.; Wuite, G.J.L. Supercoiling DNA optically. Proc. Natl. Acad. Sci. USA 2019, 116, 26534–26539. [Google Scholar] [CrossRef]
- Strick, T.R.; Allemand, J.-F.; Bensimon, D.; Croquette, V. The Elasticity of a Single Supercoiled DNA Molecule. Science 1996, 271, 1835–1837. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Lipfert, J.; Hao, X.; Dekker, N.H. Quantitative Modeling and Optimization of Magnetic Tweezers. Biophys. J. 2009, 96, 5040–5049. [Google Scholar] [CrossRef] [PubMed]
- Gosse, C.; Croquette, V. Magnetic Tweezers: Micromanipulation and Force Measurement at the Molecular Level. Biophys. J. 2002, 82, 3314–3329. [Google Scholar] [CrossRef]
- Sarkar, R.; Rybenkov, V.V. A Guide to Magnetic Tweezers and Their Applications. Front. Phys. 2016, 4, 48. [Google Scholar] [CrossRef]
- Ristic, D.; Modesti, M.; Van Der Heijden, T.; Van Noort, J.; Dekker, C.; Kanaar, R.; Wyman, C. Human Rad51 filaments on double- and single-stranded DNA: Correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005, 33, 3292–3302. [Google Scholar] [CrossRef]
- Hansma, H.G.; Sinsheimer, R.L.; Groppe, J.; Bruice, T.C.; Elings, V.; Gurley, G.; Bezanilla, M.; Mastrangelo, I.A.; Hough, P.V.C.; Hansma, P.K. Recent advances in atomic force microscopy of DNA. Scanning 1993, 15, 296–299. [Google Scholar] [CrossRef]
- Lyubchenko, Y.L.; Jacobs, B.L.; Lindsay, S.M.; Stasiak, A. Atomic force microscopy of nucleoprotein complexes. Scanning Microsc. 1995, 9, 705–724; discussion 724–727. [Google Scholar]
- Uchihashi, T.; Ando, T. High-Speed Atomic Force Microscopy and Biomolecular Processes. In Atomic Force Microscopy in Biomedical Research: Methods and Protocols; Braga, P.C., Ricci, D., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; pp. 285–300. ISBN 978-1-61779-105-5. [Google Scholar]
- Lyubchenko, Y.L.; Shlyakhtenko, L.S.; Ando, T. Imaging of nucleic acids with atomic force microscopy. Methods 2011, 54, 274–283. [Google Scholar] [CrossRef]
- Roy, R.; Kozlov, A.G.; Lohman, T.M.; Ha, T. Dynamic Structural Rearrangements between DNA Binding Modes of E. coli SSB Protein. J. Mol. Biol. 2007, 369, 1244–1257. [Google Scholar] [CrossRef] [PubMed]
- Hellenkamp, B.; Schmid, S.; Doroshenko, O.; Opanasyuk, O.; Kühnemuth, R.; Adariani, S.R.; Ambrose, B.; Aznauryan, M.; Barth, A.; Birkedal, V.; et al. Precision and accuracy of single-molecule FRET measurements—A multi-laboratory benchmark study. Nat. Methods 2018, 15, 669–676. [Google Scholar] [CrossRef]
- Maleki, P.; Budhathoki, J.B.; Roy, W.A.; Balci, H. A practical guide to studying G-quadruplex structures using single-molecule FRET. Mol. Genet. Genom. 2017, 292, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.; Enderle, T.; Ogletree, D.F.; Chemla, D.S.; Selvin, P.R.; Weiss, S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA 1996, 93, 6264–6268. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Kozlov, A.G.; Lohman, T.M.; Ha, T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 2009, 461, 1092–1097. [Google Scholar] [CrossRef]
- Kozlov, A.G.; Lohman, T.M. Stopped-Flow Studies of the Kinetics of Single-Stranded DNA Binding and Wrapping around the Escherichia coli SSB Tetramer. Biochemistry 2002, 41, 6032–6044. [Google Scholar] [CrossRef]
- Kuznetsov, S.V.; Kozlov, A.G.; Lohman, T.M.; Ansari, A. Microsecond Dynamics of Protein–DNA Interactions: Direct Observation of the Wrapping/Unwrapping Kinetics of Single-stranded DNA around the E. coli SSB Tetramer. J. Mol. Biol. 2006, 359, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Roemer, R.; Schomburg, U.; Krauss, G.; Maass, G. Escherichia coli single-stranded DNA binding protein is mobile on DNA: Proton NMR study of its interaction with oligo- and polynucleotides. Biochemistry 1984, 23, 6132–6137. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.G.; Lohman, T.M. Kinetic Mechanism of Direct Transfer of Escherichia coli SSB Tetramers between Single-Stranded DNA Molecules. Biochemistry 2002, 41, 11611–11627. [Google Scholar] [CrossRef]
- Grieb, M.S.; Nivina, A.; Cheeseman, B.L.; Hartmann, A.; Mazel, D.; Schlierf, M. Dynamic stepwise opening of integron attC DNA hairpins by SSB prevents toxicity and ensures functionality. Nucleic Acids Res. 2017, 45, 10555–10563. [Google Scholar] [CrossRef]
- Heller, I.; Sitters, G.; Broekmans, O.D.; Farge, G.; Menges, C.; Wende, W.; Hell, S.W.; Peterman, E.J.G.; Wuite, G.J.L. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nat. Methods 2013, 10, 910–916. [Google Scholar] [CrossRef]
- Brouwer, I.; Sitters, G.; Candelli, A.; Heerema, S.J.; Heller, I.; De, A.J.M.; Zhang, H.; Normanno, D.; Modesti, M.; Peterman, E.J.G.; et al. Sliding sleeves of XRCC4–XLF bridge DNA and connect fragments of broken DNA. Nature 2016, 535, 566–569. [Google Scholar] [CrossRef]
- Pant, K.; Karpel, R.L.; Williams, M.C. Kinetic Regulation of Single DNA Molecule Denaturation by T4 Gene 32 Protein Structural Domains. J. Mol. Biol. 2003, 327, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Morten, M.J.; Peregrina, J.R.; Figueira-Gonzalez, M.; Ackermann, K.; Bode, B.E.; White, M.F.; Penedo, J.C. Binding dynamics of a monomeric SSB protein to DNA: A single-molecule multi-process approach. Nucleic Acids Res. 2015, 43, 10907–10924. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.C.; Liu, B.; Kowalczykowski, S.C. Imaging and energetics of single SSB-ssDNA molecules reveal intramolecular condensation and insight into RecOR function. eLife 2015, 4, e08646. [Google Scholar] [CrossRef]
- Kozlov, A.G.; Shinn, M.K.; Weiland, E.A.; Lohman, T.M. Glutamate promotes SSB protein–protein Interactions via intrinsically disordered regions. J. Mol. Biol. 2017, 429, 2790–2801. [Google Scholar] [CrossRef] [PubMed]
- Salerno, D.; Beretta, G.L.; Zanchetta, G.; Brioschi, S.; Cristofalo, M.; Missana, N.; Nardo, L.; Cassina, V.; Tempestini, A.; Giovannoni, R.; et al. Platinum-Based Drugs and DNA Interactions Studied by Single-Molecule and Bulk Measurements. Biophys. J. 2016, 110, 2151–2161. [Google Scholar] [CrossRef]
- Soengas, M.; Esteban, J.A.; Salas, M.; Gutierrez, C. Complex Formation between Phage Phi φ29 Single-stranded DNA Binding Protein and DNA. J. Mol. Biol. 1994, 239, 213–226. [Google Scholar] [CrossRef]
- Rényi, A. On a one-dimensional problem concerning random space filling. Publ. Math. Inst. Hung. Acad. Sci. 1958, 3, 109–127. [Google Scholar]
- Clementi, E.; Raimondi, D.L.; Reinhardt, W.P. Atomic Screening Constants from SCF Functions. II. Atoms with 37 to 86 Electrons. J. Chem. Phys. 1967, 47, 1300–1307. [Google Scholar] [CrossRef]
- Kriegel, F.; Ermann, N.; Forbes, R.; Dulin, D.; Dekker, N.; Lipfert, J. Probing the salt dependence of the torsional stiffness of DNA by multiplexed magnetic torque tweezers. Nucleic Acids Res. 2017, 45, 5920–5929. [Google Scholar] [CrossRef]
- Anthony, P.C.; Sim, A.Y.L.; Chu, V.B.; Doniach, S.; Block, S.M.; Herschlag, D. Electrostatics of Nucleic Acid Folding under Conformational Constraint. J. Am. Chem. Soc. 2012, 134, 4607–4614. [Google Scholar] [CrossRef]
- Bizarro, C.V.; Alemany, A.; Ritort, F. Non-specific binding of Na + and Mg 2+ to RNA determined by force spectroscopy methods. Nucleic Acids Res. 2012, 40, 6922–6935. [Google Scholar] [CrossRef] [PubMed]
- Baumann, C.G.; Smith, S.B.; Bloomfield, V.A.; Bustamante, C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA 1997, 94, 6185–6190. [Google Scholar] [CrossRef] [PubMed]
- Todd, B.A.; Rau, D.C. Interplay of ion binding and attraction in DNA condensed by multivalent cations. Nucleic Acids Res. 2007, 36, 501–510. [Google Scholar] [CrossRef]
- Meijer, W.J.J.; Horcajadas, J.A.; Salas, M. φ29 Family of Phages. Microbiol. Mol. Biol. Rev. 2001, 65, 261–287. [Google Scholar] [CrossRef]
- McGhee, J.D.; von Hippel, P.H. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
- Lechuga, A.; Kazlauskas, D.; Salas, M.; Redrejo-Rodríguez, M. Unlimited Cooperativity of Betatectivirus SSB, a Novel DNA Binding Protein Related to an Atypical Group of SSBs From Protein-Primed Replicating Bacterial Viruses. Front. Microbiol. 2021, 12, 699140. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.G.; Galletto, R.; Lohman, T.M. SSB–DNA Binding Monitored by Fluorescence Intensity and Anisotropy. Methods Mol. Biol. 2012, 922, 55–83. [Google Scholar] [CrossRef] [PubMed]
- von Hippel, P.H.; Marcus, A.H. The Many Roles of Binding Cooperativity in the Control of DNA Replication. Biophys. J. 2019, 117, 2043–2046. [Google Scholar] [CrossRef]
- Kozlov, A.G.; Shinn, M.K.; Lohman, T.M. Regulation of Nearest-Neighbor Cooperative Binding of E. coli SSB Protein to DNA. Biophys. J. 2019, 117, 2120–2140. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, R.; Inoue, J.; Mikawa, T.; Ha, T. Single molecule analysis of Thermus thermophilus SSB protein dynamics on single-stranded DNA. Nucleic Acids Res. 2013, 42, 3821–3832. [Google Scholar] [CrossRef]
- Hollis, T.; Stattel, J.M.; Walther, D.S.; Richardson, C.C.; Ellenberger, T. Structure of the Gene 2.5 Protein, a Single-Stranded DNA Binding Protein Encoded by Bacteriophage T7. Proc. Natl. Acad. Sci. USA 2001, 98, 9557–9562. [Google Scholar] [CrossRef]
- Bujalowski, W.; Lohman, T.M. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry 1986, 25, 7799–7802. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, S.; Kozlov, A.G.; Lohman, T.M.; Waksman, G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Mol. Biol. 2000, 7, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Initiation of translation in prokaryotes and eukaryotes. Gene 1999, 234, 187–208. [Google Scholar] [CrossRef] [PubMed]
- Woodside, M.T.; Behnke-Parks, W.M.; Larizadeh, K.; Travers, K.; Herschlag, D.; Block, S.M. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. USA 2006, 103, 6190–6195. [Google Scholar] [CrossRef]
- Doma, M.K.; Parker, R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 2006, 440, 561–564. [Google Scholar] [CrossRef]
- Kurkcuoglu, O.; Doruker, P.; Sen, T.; Kloczkowski, A.; Jernigan, R.L. The ribosome structure controls and directs mRNA entry, translocation and exit dynamics. Phys. Biol. 2008, 5, 046005. [Google Scholar] [CrossRef]
- McGlynn, P. Helicases at the Replication Fork. Adv. Exp. Med. Biol. 2012, 767, 97–121. [Google Scholar] [CrossRef]
- Daley, J.M.; Niu, H.; Sung, P. Roles of DNA Helicases in the Mediation and Regulation of Homologous Recombination. Adv. Exp. Med. Biol. 2012, 767, 185–202. [Google Scholar] [CrossRef]
- Kuper, J.; Kisker, C. DNA Helicases in NER, BER, and MMR. Adv. Exp. Med. Biol. 2012, 767, 203–224. [Google Scholar] [CrossRef]
- Stekas, B.; Yeo, S.; Troitskaia, A.; Honda, M.; Sho, S.; Spies, M.; Chemla, Y.R. Switch-like control of helicase processivity by single-stranded DNA binding protein. eLife 2021, 10, e60515. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Bueno, C.; Lu, W.; Wang, Q.; Chen, M.; Chen, X.; Wolynes, P.G.; Gao, Y. Computationally exploring the mechanism of bacteriophage T7 gp4 helicase translocating along ssDNA. Proc. Natl. Acad. Sci. USA 2022, 119, e2202239119. [Google Scholar] [CrossRef] [PubMed]
- Lohman, T.M.; Tomko, E.J.; Wu, C.G. Non-hexameric DNA helicases and translocases: Mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008, 9, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.G.; Spies, M. Overview: What Are Helicases? Adv. Exp. Med. Biol. 2012, 767, 1–16. [Google Scholar] [CrossRef]
- Beyer, D.C.; Ghoneim, M.K.; Spies, M. Structure and Mechanisms of SF2 DNA Helicases. Adv. Exp. Med. Biol. 2012, 767, 47–73. [Google Scholar] [CrossRef]
- Umezu, K.; Nakayama, H. RecQ DNA Helicase of Escherichia coli: Characterization of the Helix-unwinding Activity with Emphasis on the Effect of Single-stranded DNA-binding Protein. J. Mol. Biol. 1993, 230, 1145–1150. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Chastain, P.; Griffith, J.D.; Richardson, C.C. Lagging strand synthesis in coordinated DNA synthesis by bacteriophage T7 replication proteins. J. Mol. Biol. 2002, 316, 19–34. [Google Scholar] [CrossRef]
- Harmon, F.G.; Kowalczykowski, S.C. RecQ Helicase, in Concert with RecA and SSB Proteins, Initiates and Disrupts DNA Recombination. Genes Dev. 1998, 12, 1134–1144. [Google Scholar] [CrossRef]
- Rajagopal, V.; Patel, S.S. Single Strand Binding Proteins Increase the Processivity of DNA Unwinding by the Hepatitis C Virus Helicase. J. Mol. Biol. 2008, 376, 69–79. [Google Scholar] [CrossRef]
- Fu, Y.V.; Yardimci, H.; Long, D.T.; Ho, T.V.; Guainazzi, A.; Bermudez, V.P.; Hurwitz, J.; van Oijen, A.; Schärer, O.D.; Walter, J.C. Selective Bypass of a Lagging Strand Roadblock by the Eukaryotic Replicative DNA Helicase. Cell 2011, 146, 931–941. [Google Scholar] [CrossRef]
- Wasserman, M.R.; Schauer, G.D.; O’Donnell, M.E.; Liu, S. Replication Fork Activation Is Enabled by a Single-Stranded DNA Gate in CMG Helicase. Cell 2019, 178, 600–611.e16. [Google Scholar] [CrossRef] [PubMed]
- Raghuraman, M.K.; Winzeler, E.A.; Collingwood, D.; Hunt, S.; Wodicka, L.; Conway, A.; Lockhart, D.J.; Davis, R.W.; Brewer, B.J.; Fangman, W.L. Replication Dynamics of the Yeast Genome. Science 2001, 294, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Anglana, M.; Apiou, F.; Bensimon, A.; Debatisse, M. Dynamics of DNA Replication in Mammalian Somatic Cells: Nucleotide Pool Modulates Origin Choice and Interorigin Spacing. Cell 2003, 114, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Kose, H.B.; Larsen, N.B.; Duxin, J.P.; Yardimci, H. Dynamics of the Eukaryotic Replicative Helicase at Lagging-Strand Protein Barriers Support the Steric Exclusion Model. Cell Rep. 2019, 26, 2113–2125.e6. [Google Scholar] [CrossRef]
- Manosas, M.; Spiering, M.M.; Ding, F.; Croquette, V.; Benkovic, S.J. Collaborative coupling between polymerase and helicase for leading-strand synthesis. Nucleic Acids Res. 2012, 40, 6187–6198. [Google Scholar] [CrossRef]
- Spinks, R.R.; Spenkelink, L.M.; Dixon, N.E.; van Oijen, A.M. Single-Molecule Insights Into the Dynamics of Replicative Helicases. Front. Mol. Biosci. 2021, 8, 741718. [Google Scholar] [CrossRef]
- Fan, L.; Fuss, J.O.; Cheng, Q.; Arvai, A.S.; Hammel, M.; Roberts, V.A.; Cooper, P.K.; Tainer, J.A. XPD Helicase Structures and Activities: Insights into the Cancer and Aging Phenotypes from XPD Mutations. Cell 2008, 133, 789–800. [Google Scholar] [CrossRef]
- Bhattacharyya, B.; George, N.P.; Thurmes, T.M.; Zhou, R.; Jani, N.; Wessel, S.R.; Sandler, S.J.; Ha, T.; Keck, J.L. Structural mechanisms of PriA-mediated DNA replication restart. Proc. Natl. Acad. Sci. USA 2013, 111, 1373–1378. [Google Scholar] [CrossRef]
- E Fairman-Williams, M.; Guenther, U.-P.; Jankowsky, E. SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef]
- Byrd, A.K.; Raney, K.D. Superfamily 2 Helicases. Front. Biosci. (Landmark Ed.) 2012, 17, 2070–2088. [Google Scholar] [CrossRef]
- White, M.F.; Dillingham, M.S. Iron–sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 2012, 22, 94–100. [Google Scholar] [CrossRef]
- van Brabant, A.J.; Stan, R.; Ellis, N.A. DNA Helicases, Genomic Instability, and Human Genetic Disease. Annu. Rev. Genom. Hum. Genet. 2000, 1, 409–459. [Google Scholar] [CrossRef]
- Egly, J.-M.; Coin, F. A history of TFIIH: Two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair 2011, 10, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Fuss, J.O.; Tainer, J.A. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair 2011, 10, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Kuper, J.; Braun, C.; Elias, A.; Michels, G.; Sauer, F.; Schmitt, D.R.; Poterszman, A.; Egly, J.-M.; Kisker, C. In TFIIH, XPD Helicase Is Exclusively Devoted to DNA Repair. PLoS Biol. 2014, 12, e1001954. [Google Scholar] [CrossRef] [PubMed]
- Van Houten, B.; Kuper, J.; Kisker, C. Role of XPD in cellular functions: To TFIIH and beyond. DNA Repair 2016, 44, 136–142. [Google Scholar] [CrossRef]
- Ito, S.; Tan, L.J.; Andoh, D.; Narita, T.; Seki, M.; Hirano, Y.; Narita, K.; Kuraoka, I.; Hiraoka, Y.; Tanaka, K. MMXD, a TFIIH-Independent XPD-MMS19 Protein Complex Involved in Chromosome Segregation. Mol. Cell 2010, 39, 632–640. [Google Scholar] [CrossRef]
- Yoder, K.; Sarasin, A.; Kraemer, K.; McIlhatton, M.; Bushman, F.; Fishel, R. The DNA repair genes XPB and XPD defend cells from retroviral infection. Proc. Natl. Acad. Sci. USA 2006, 103, 4622–4627. [Google Scholar] [CrossRef]
- Harmon, F.G.; Kowalczykowski, S.C. Biochemical Characterization of the DNA Helicase Activity of the Escherichia coli RecQ Helicase. J. Biol. Chem. 2001, 276, 232–243. [Google Scholar] [CrossRef]
- Cadman, C.J.; McGlynn, P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004, 32, 6378–6387. [Google Scholar] [CrossRef]
- Cui, S.; Arosio, D.; Doherty, K.M.; Brosh, R.M.; Falaschi, A.; Vindigni, A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004, 32, 2158–2170. [Google Scholar] [CrossRef]
- Pugh, R.A.; Lin, Y.; Eller, C.; Leesley, H.; Cann, I.K.; Spies, M. J. Mol. Biol. 2008, 383, 982–998. [CrossRef] [PubMed]
- Handa, N.; Morimatsu, K.; Lovett, S.T.; Kowalczykowski, S.C. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev. 2009, 23, 1234–1245. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Ukita, T.; Kohno, Y.; Saito, K.; Kato, J.-I.; Ikeda, H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl. Acad. Sci. USA 1997, 94, 3860–3865. [Google Scholar] [CrossRef] [PubMed]
- Harami, G.M.; Seol, Y.; In, J.; Ferencziová, V.; Martina, M.; Gyimesi, M.; Sarlós, K.; Kovács, Z.J.; Nagy, N.T.; Sun, Y.; et al. Shuttling along DNA and directed processing of D-loops by RecQ helicase support quality control of homologous recombination. Proc. Natl. Acad. Sci. USA 2017, 114, E466–E475. [Google Scholar] [CrossRef] [PubMed]
- Mohaghegh, P.; Karow, J.K.; Brosh, R.M.; Bohr, V.A.; Hickson, I.D. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001, 29, 2843–2849. [Google Scholar] [CrossRef]
- Hishida, T.; Han, Y.-W.; Shibata, T.; Kubota, Y.; Ishino, Y.; Iwasaki, H.; Shinagawa, H. Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev. 2004, 18, 1886–1897. [Google Scholar] [CrossRef]
- Courcelle, J.; Hanawalt, P.C. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Genet. Genom. 1999, 262, 543–551. [Google Scholar] [CrossRef]
- Bachrati, C.Z.; Hickson, I.D. RecQ helicases: Guardian angels of the DNA replication fork. Chromosoma 2008, 117, 219–233. [Google Scholar] [CrossRef]
- Bagchi, D.; Manosas, M.; Zhang, W.; Manthei, K.A.; Hodeib, S.; Ducos, B.; Keck, J.L.; Croquette, V. Single molecule kinetics uncover roles for E. coli RecQ DNA helicase domains and interaction with SSB. Nucleic Acids Res. 2018, 46, 8500–8515. [Google Scholar] [CrossRef]
- Nakayama, H.; Nakayama, K.; Nakayama, R.; Irino, N.; Nakayama, Y.; Hanawalt, P.C. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: Identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol. Genet. Genom. 1984, 195, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Harami, G.M.; Nagy, N.T.; Martina, M.; Neuman, K.; Kovács, M. The HRDC domain of E. coli RecQ helicase controls single-stranded DNA translocation and double-stranded DNA unwinding rates without affecting mechanoenzymatic coupling. Sci. Rep. 2015, 5, 11091. [Google Scholar] [CrossRef] [PubMed]
- Greenleaf, W.J.; Woodside, M.T.; Block, S.M. High-Resolution, Single-Molecule Measurements of Biomolecular Motion. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 171–190. [Google Scholar] [CrossRef] [PubMed]
- Tanner, N.A.; van Oijen, A.M. Visualizing DNA Replication at the Single-Molecule Level. Methods Enzymol. 2010, 475, 259–278. [Google Scholar] [CrossRef] [PubMed]
- Dessinges, M.-N.; Lionnet, T.; Xi, X.G.; Bensimon, D.; Croquette, V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Natl. Acad. Sci. USA 2004, 101, 6439–6444. [Google Scholar] [CrossRef]
- Lionnet, T.; Spiering, M.M.; Benkovic, S.J.; Bensimon, D.; Croquette, V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. USA 2007, 104, 19790–19795. [Google Scholar] [CrossRef]
- Manosas, M.; Xi, X.G.; Bensimon, D.; Croquette, V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010, 38, 5518–5526. [Google Scholar] [CrossRef]
- Manosas, M.; Spiering, M.M.; Zhuang, Z.; Benkovic, S.J.; Croquette, V. Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome. Nat. Chem. Biol. 2009, 5, 904–912. [Google Scholar] [CrossRef]
- Manosas, M.; Perumal, S.K.; Croquette, V.; Benkovic, S.J. Direct Observation of Stalled Fork Restart via Fork Regression in the T4 Replication System. Science 2012, 338, 1217–1220. [Google Scholar] [CrossRef]
- Cox, M.M.; Goodman, M.F.; Kreuzer, K.N.; Sherratt, D.J.; Sandler, S.J.; Marians, K.J. The importance of repairing stalled replication forks. Nature 2000, 404, 37–41. [Google Scholar] [CrossRef]
- Heller, R.C.; Marians, K.J. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 2006, 7, 932–943. [Google Scholar] [CrossRef]
- Merrikh, H.; Zhang, Y.; Grossman, A.D.; Wang, J.D. Replication–transcription conflicts in bacteria. Nat. Rev. Microbiol. 2012, 10, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.; Poli, J.; Marians, K.J.; Pasero, P. Rescuing Stalled or Damaged Replication Forks. Cold Spring Harb. Perspect. Biol. 2013, 5, a012815. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; A Al-Deib, A.; Liu, J.; Marians, K.J.; Lloyd, R.G. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J. Mol. Biol. 1997, 270, 212–221. [Google Scholar] [CrossRef]
- Nurse, P.; Liu, J.; Marians, K.J. Two Modes of PriA Binding to DNA. J. Biol. Chem. 1999, 274, 25026–25032. [Google Scholar] [CrossRef] [PubMed]
- Manhart, C.M.; McHenry, C.S. The PriA Replication Restart Protein Blocks Replicase Access Prior to Helicase Assembly and Directs Template Specificity through Its ATPase Activity. J. Biol. Chem. 2013, 288, 3989–3999. [Google Scholar] [CrossRef]
- Lee, M.S.; Marians, K.J. Escherichia coli replication factor Y, a component of the primosome, can act as a DNA helicase. Proc. Natl. Acad. Sci. USA 1987, 84, 8345–8349. [Google Scholar] [CrossRef]
- Lasken, R.S.; Kornberg, A. The primosomal protein n′ of Escherichia coli is a DNA helicase. J. Biol. Chem. 1988, 263, 5512–5518. [Google Scholar] [CrossRef]
- Lecointe, F.; Sérèna, C.; Velten, M.; Costes, A.; McGovern, S.; Meile, J.-C.; Errington, J.; Ehrlich, S.D.; Noirot, P.; Polard, P. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J. 2007, 26, 4239–4251. [Google Scholar] [CrossRef]
- Kozlov, A.G.; Jezewska, M.J.; Bujalowski, W.; Lohman, T.M. Binding Specificity of Escherichia coli Single-Stranded DNA Binding Protein for the χ Subunit of DNA pol III Holoenzyme and PriA Helicase. Biochemistry 2010, 49, 3555–3566. [Google Scholar] [CrossRef]
- Wessel, S.R.; Marceau, A.H.; Massoni, S.C.; Zhou, R.; Ha, T.; Sandler, S.J.; Keck, J.L. PriC-mediated DNA Replication Restart Requires PriC Complex Formation with the Single-stranded DNA-binding Protein. J. Biol. Chem. 2013, 288, 17569–17578. [Google Scholar] [CrossRef] [PubMed]
- Ciesielski, G.L.; Bermek, O.; Rosado-Ruiz, F.A.; Hovde, S.L.; Neitzke, O.J.; Griffith, J.D.; Kaguni, L.S. Mitochondrial Single-stranded DNA-binding Proteins Stimulate the Activity of DNA Polymerase γ by Organization of the Template DNA. J. Biol. Chem. 2015, 290, 28697–28707. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.J.; Kaguni, L.S. Stimulation of Drosophila Mitochondrial DNA Polymerase by Single-stranded DNA-binding Protein. J. Biol. Chem. 1995, 270, 860–865. [Google Scholar] [CrossRef] [PubMed]
- A Korhonen, J.; Pham, X.H.; Pellegrini, M.; Falkenberg, M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004, 23, 2423–2429. [Google Scholar] [CrossRef]
- Ghosh, S.; Hamdan, S.; Richardson, C.C. Two Modes of Interaction of the Single-stranded DNA-binding Protein of Bacteriophage T7 with the DNA Polymerase-Thioredoxin Complex. J. Biol. Chem. 2010, 285, 18103–18112. [Google Scholar] [CrossRef]
- Kim, Y.; Tabor, S.; Churchich, J.; Richardson, C. Interactions of gene 2.5 protein and DNA polymerase of bacteriophage T7. J. Biol. Chem. 1992, 267, 15032–15040. [Google Scholar] [CrossRef]
- Shereda, R.D.; Kozlov, A.G.; Lohman, T.M.; Cox, M.M.; Keck, J.L. SSB as an Organizer/Mobilizer of Genome Maintenance Complexes. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 289–318. [Google Scholar] [CrossRef]
- Cerrón, F.; de Lorenzo, S.; Lemishko, K.M.; Ciesielski, G.L.; Kaguni, L.S.; Cao, F.J.; Ibarra, B. Replicative DNA polymerases promote active displacement of SSB proteins during lagging strand synthesis. Nucleic Acids Res. 2019, 47, 5723–5734. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J. The Effect of Single-Strand DNA-Binding Proteins (SSB Proteins) on the Structure of Single-Stranded DNA. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002; Figure 5–17. [Google Scholar]
- Lee, Y.-S.; Kennedy, W.D.; Yin, Y.W. Structural Insight into Processive Human Mitochondrial DNA Synthesis and Disease-Related Polymerase Mutations. Cell 2009, 139, 312–324. [Google Scholar] [CrossRef]
- Yang, C.; Curth, U.; Urbanke, C.; Kang, C. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 Å resolution. Nat. Struct. Biol. 1997, 4, 153–157. [Google Scholar] [CrossRef]
- Manosas, M.; Spiering, M.M.; Ding, F.; Bensimon, D.; Allemand, J.-F.; Benkovic, S.J.; Croquette, V. Mechanism of strand displacement synthesis by DNA replicative polymerases. Nucleic Acids Res. 2012, 40, 6174–6186. [Google Scholar] [CrossRef]
- Ismael, P.-G.A.; Lemishko, K.M.; Crespo, R.; Truong, T.Q.; Kaguni, L.S.; Cao-García, F.J.; Ciesielski, G.L.; Ibarra, B. Mechanism of Strand Displacement DNA Synthesis by the Coordinated Activities of Human Mitochondrial DNA Polymerase and SSB. bioRxiv 2022. [Google Scholar] [CrossRef]
- Sullivan, E.D.; Longley, M.J.; Copeland, W.C. Polymerase γ efficiently replicates through many natural template barriers but stalls at the HSP1 quadruplex. J. Biol. Chem. 2020, 295, 17802–17815. [Google Scholar] [CrossRef]
- Fusté, J.M.; Shi, Y.; Wanrooij, S.; Zhu, X.; Jemt, E.; Persson, Ö.; Sabouri, N.; Gustafsson, C.M.; Falkenberg, M. In Vivo Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication. PLoS Genet. 2014, 10, e1004832. [Google Scholar] [CrossRef]
- Takamatsu, C.; Umeda, S.; Ohsato, T.; Ohno, T.; Abe, Y.; Fukuoh, A.; Shinagawa, H.; Hamasaki, N.; Kang, D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002, 3, 451–456. [Google Scholar] [CrossRef]
- Nicholls, T.J.; Zsurka, G.; Peeva, V.; Schöler, S.; Szczesny, R.J.; Cysewski, D.; Reyes, A.; Kornblum, C.; Sciacco, M.; Moggio, M.; et al. Linear mtDNA fragments and unusual mtDNA rearrangements associated with pathological deficiency of MGME1 exonuclease. Hum. Mol. Genet. 2014, 23, 6147–6162. [Google Scholar] [CrossRef] [PubMed]
- Uhler, J.P.; Thörn, C.; Nicholls, T.J.; Matic, S.; Milenkovic, D.; Gustafsson, C.M.; Falkenberg, M. MGME1 processes flaps into ligatable nicks in concert with DNA polymerase γ during mtDNA replication. Nucleic Acids Res. 2016, 44, 5861–5871. [Google Scholar] [CrossRef]
- Zheng, L.; Zhou, M.; Guo, Z.; Lu, H.; Qian, L.; Dai, H.; Qiu, J.; Yakubovskaya, E.; Bogenhagen, D.F.; Demple, B.; et al. Human DNA2 Is a Mitochondrial Nuclease/Helicase for Efficient Processing of DNA Replication and Repair Intermediates. Mol. Cell 2008, 32, 325–336. [Google Scholar] [CrossRef]
- He, Q.; Shumate, C.K.; A White, M.; Molineux, I.J.; Yin, Y.W. Exonuclease of human DNA polymerase gamma disengages its strand displacement function. Mitochondrion 2013, 13, 592–601. [Google Scholar] [CrossRef]
- Farge, G.; Pham, X.H.; Holmlund, T.; Khorostov, I.; Falkenberg, M. The accessory subunit B of DNA polymerase is required for mitochondrial replisome function. Nucleic Acids Res. 2007, 35, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Farr, C.L.; Wang, Y.; Kaguni, L.S. Functional Interactions of Mitochondrial DNA Polymerase and Single-stranded DNA-binding Protein: Template-primer DNA binding and initiation and elongation of DNA strand synthesis. J. Biol. Chem. 1999, 274, 14779–14785. [Google Scholar] [CrossRef] [PubMed]
- Macao, B.; Uhler, J.P.; Siibak, T.; Zhu, X.; Shi, Y.; Sheng, W.; Olsson, M.; Stewart, J.B.; Gustafsson, C.M.; Falkenberg, M. The exonuclease activity of DNA polymerase γ is required for ligation during mitochondrial DNA replication. Nat. Commun. 2015, 6, 7303. [Google Scholar] [CrossRef] [PubMed]
- Canceill, D.; Viguera, E.; Ehrlich, S.D. Replication Slippage of Different DNA Polymerases Is Inversely Related to Their Strand Displacement Efficiency. J. Biol. Chem. 1999, 274, 27481–27490. [Google Scholar] [CrossRef]
- Stano, N.M.; Jeong, Y.-J.; Donmez, I.; Tummalapalli, P.; Levin, M.K.; Patel, S.S. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature 2005, 435, 370–373. [Google Scholar] [CrossRef]
- Yuan, Q.; McHenry, C.S. Strand Displacement by DNA Polymerase III Occurs through a τ-ψ-χ Link to Single-stranded DNA-binding Protein Coating the Lagging Strand Template. J. Biol. Chem. 2009, 284, 31672–31679. [Google Scholar] [CrossRef]
- Stephens, K.M.; McMacken, R. Functional Properties of Replication Fork Assemblies Established by the Bacteriophage λ O and P Replication Proteins. J. Biol. Chem. 1997, 272, 28800–28813. [Google Scholar] [CrossRef]
- Nandakumar, D.; Pandey, M.; Patel, S.S. Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other. eLife 2015, 4, e06562. [Google Scholar] [CrossRef]
- Koc, K.N.; Stodola, J.L.; Burgers, P.M.; Galletto, R. Regulation of yeast DNA polymerase δ-mediated strand displacement synthesis by 5′-flaps. Nucleic Acids Res. 2015, 43, 4179–4190. [Google Scholar] [CrossRef]
- McInerney, P.; O’Donnell, M. Replisome Fate upon Encountering a Leading Strand Block and Clearance from DNA by Recombination Proteins. J. Biol. Chem. 2007, 282, 25903–25916. [Google Scholar] [CrossRef]
- Heller, R.C.; Marians, K.J. The Disposition of Nascent Strands at Stalled Replication Forks Dictates the Pathway of Replisome Loading during Restart. Mol. Cell 2005, 17, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro-Stone, M.; Makhov, A.M.; Zaritskaya, L.S.; Griffith, J.D. Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol. 1999, 289, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, K.; Katayama, T.; Iwai, S.; Hidaka, M.; Horiuchi, T.; Maki, H. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells 2003, 8, 437–449. [Google Scholar] [CrossRef]
- Kowalezykowski, S.C. Biochemistry of Genetic Recombination: Energetics and Mechanism of DNA Strand Exchange. Annu. Rev. Biophys. Biophys. Chem. 1991, 20, 539–575. [Google Scholar] [CrossRef] [PubMed]
- Kowalczykowski, S.C.; Clow, J.; Somani, R.; Varghese, A. Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA: Demonstration of competitive binding and the lack of a specific protein-protein interaction. J. Mol. Biol. 1987, 193, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Thresher, R.J.; Christiansen, G.; Griffith, J.D. Assembly of presynaptic filaments: Factors affecting the assembly of RecA protein onto single-stranded DNA. J. Mol. Biol. 1988, 201, 101–113. [Google Scholar] [CrossRef]
- Cazenave, C.; Toulmé, J.; Hélène, C. Binding of RecA protein to single-stranded nucleic acids: Spectroscopic studies using fluorescent polynucleotides. EMBO J. 1983, 2, 2247–2251. [Google Scholar] [CrossRef]
- Shivashankar, G.V.; Feingold, M.; Krichevsky, O.; Libchaber, A. RecA polymerization on double-stranded DNA by using single-molecule manipulation: The role of ATP hydrolysis. Proc. Natl. Acad. Sci. USA 1999, 96, 7916–7921. [Google Scholar] [CrossRef]
- Galletto, R.; Amitani, I.; Baskin, R.J.; Kowalczykowski, S.C. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 2006, 443, 875–878. [Google Scholar] [CrossRef]
- Joo, C.; McKinney, S.A.; Nakamura, M.; Rasnik, I.; Myong, S.; Ha, T. Real-Time Observation of RecA Filament Dynamics with Single Monomer Resolution. Cell 2006, 126, 515–527. [Google Scholar] [CrossRef]
- Handa, N.; Amitani, I.; Gumlaw, N.; Sandler, S.J.; Kowalczykowski, S.C. Single Molecule Analysis of a Red Fluorescent RecA Protein Reveals a Defect in Nucleoprotein Filament Nucleation That Relates to Its Reduced Biological Functions. J. Biol. Chem. 2009, 284, 18664–18673. [Google Scholar] [CrossRef] [PubMed]
- Modesti, M.; Ristic, D.; van der Heijden, T.; Dekker, C.; van Mameren, J.; Peterman, E.J.; Wuite, G.J.; Kanaar, R.; Wyman, C. Fluorescent Human RAD51 Reveals Multiple Nucleation Sites and Filament Segments Tightly Associated along a Single DNA Molecule. Structure 2007, 15, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heijden, T.; Seidel, R.; Modesti, M.; Kanaar, R.; Wyman, C.; Dekker, C. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007, 35, 5646–5657. [Google Scholar] [CrossRef] [PubMed]
- Hilario, J.; Amitani, I.; Baskin, R.J.; Kowalczykowski, S.C. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl. Acad. Sci. USA 2009, 106, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.D.; Harris, L.D.; Register, J. Visualization of SSB-ssDNA Complexes Active in the Assembly of Stable RecA-DNA Filaments. Cold Spring Harb. Symp. Quant. Biol. 1984, 49, 553–559. [Google Scholar] [CrossRef]
- Kowalczykowski, S.; Steinhardt, J. Kinetics of hemoglobin S gelation followed by continuously sensitive low-shear viscosity: Changes in viscosity and volume on aggregation. J. Mol. Biol. 1977, 115, 201–213. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, H.; Pavletich, N.P. Mechanism of homologous recombination from the RecA–ssDNA/dsDNA structures. Nature 2008, 453, 489–494. [Google Scholar] [CrossRef]
- Amundsen, S.K.; Smith, G.R. Interchangeable Parts of the Escherichia coli Recombination Machinery. Cell 2003, 112, 741–744. [Google Scholar] [CrossRef]
- Kowalczykowski, S.C.; Krupp, R.A. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein: Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J. Mol. Biol. 1987, 193, 97–113. [Google Scholar] [CrossRef]
- Candelli, A.; Holthausen, J.T.; Depken, M.; Brouwer, I.; Franker, M.A.M.; Marchetti, M.; Heller, I.; Bernard, S.; Garcin, E.B.; Modesti, M.; et al. Visualization and quantification of nascent RAD51 filament formation at single-monomer resolution. Proc. Natl. Acad. Sci. USA 2014, 111, 15090–15095. [Google Scholar] [CrossRef]
- Belan, O.; Barroso, C.; Kaczmarczyk, A.; Anand, R.; Federico, S.; O’Reilly, N.; Newton, M.D.; Maeots, E.; Enchev, R.I.; Martinez-Perez, E.; et al. Single-molecule analysis reveals cooperative stimulation of Rad51 filament nucleation and growth by mediator proteins. Mol. Cell 2021, 81, 1058–1073.e7. [Google Scholar] [CrossRef]
- Hegner, M.; Smith, S.B.; Bustamante, C. Polymerization and mechanical properties of single RecA–DNA filaments. Proc. Natl. Acad. Sci. USA 1999, 96, 10109–10114. [Google Scholar] [CrossRef]
- Lohman, T.M.; Ferrari, M.E. Escherichia coli single-stranded DNA-binding protein: Multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994, 63, 527–570. [Google Scholar] [CrossRef]
- Crozat, E.; Grainge, I. FtsK DNA Translocase: The Fast Motor That Knows Where It’s Going. Chembiochem 2010, 11, 2232–2243. [Google Scholar] [CrossRef]
- Fishburn, J.; Tomko, E.; Galburt, E.; Hahn, S. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc. Natl. Acad. Sci. USA 2015, 112, 3961–3966. [Google Scholar] [CrossRef]
- Sokoloski, J.E.; Kozlov, A.G.; Galletto, R.; Lohman, T.M. Chemo-mechanical pushing of proteins along single-stranded DNA. Proc. Natl. Acad. Sci. USA 2016, 113, 6194–6199. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.J.; Wetmur, J.G. Kinetics of transfer of Escherichia coli single strand DNA binding protein between single-stranded DNA molecules. Biochemistry 1982, 21, 608–615. [Google Scholar] [CrossRef]
- Lee, K.S.; Marciel, A.B.; Kozlov, A.G.; Schroeder, C.M.; Lohman, T.M.; Ha, T. Ultrafast Redistribution of E. coli SSB along Long Single-Stranded DNA via Intersegment Transfer. J. Mol. Biol. 2014, 426, 2413–2421. [Google Scholar] [CrossRef] [PubMed]
- Takayama, Y.; Clore, G.M. Intra- and intermolecular translocation of the bi-domain transcription factor Oct1 characterized by liquid crystal and paramagnetic NMR. Proc. Natl. Acad. Sci. USA 2011, 108, E169–E176. [Google Scholar] [CrossRef]
- van Oijen, A.M.; Loparo, J.J. Single-Molecule Studies of the Replisome. Annu. Rev. Biophys. 2010, 39, 429–448. [Google Scholar] [CrossRef] [PubMed]
- Geertsema, H.J.; Kulczyk, A.W.; Richardson, C.C.; van Oijen, A.M. Single-molecule studies of polymerase dynamics and stoichiometry at the bacteriophage T7 replication machinery. Proc. Natl. Acad. Sci. USA 2014, 111, 4073–4078. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, S.; van Oijen, A.M. Timing, Coordination, and Rhythm: Acrobatics at the DNA Replication Fork. J. Biol. Chem. 2010, 285, 18979–18983. [Google Scholar] [CrossRef] [PubMed]
- Tanner, N.A.; Loparo, J.J.; Van Oijen, A.M. Visualizing Single-molecule DNA Replication with Fluorescence Microscopy. J. Vis. Exp. 2009, 44, 1529. [Google Scholar] [CrossRef]
- A Tanner, N.; Hamdan, S.M.; Jergic, S.; Loscha, K.V.; Schaeffer, P.M.; E Dixon, N.; van Oijen, A.M. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat. Struct. Mol. Biol. 2008, 15, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Tanner, N.A.; Loparo, J.J.; Hamdan, S.M.; Jergic, S.; Dixon, N.E.; van Oijen, A.M. Real-time single-molecule observation of rolling-circle DNA replication. Nucleic Acids Res. 2009, 37, e27. [Google Scholar] [CrossRef]
| Phenomenon | Quantities | Structural Insights | Example Study |
|---|---|---|---|
| Binding kinetics | Time constants of binding (kon and koff) | Binding steps, timescales of binding processes | [76] |
| Binding footprint | Binding footprint from density | DNA binding pocket | [77] |
| Binding thermodynamics | Differential stability is based on temperature or applied force. Possible to calculate by FEC hysteresis | Binding stability and reaction energetics | [2,77] |
| Diffusion | Diffusion constant, velocity, direction | Directionality of movement, interaction with DNA (wrapping, base interference, etc.) | [33] |
| Cooperativity | Cooperativity score, based on concentration-dependent binding affinities | Interactions between SSB units | [22,78] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Halma, M.T.J.; Wuite, G.J.L. Unravelling How Single-Stranded DNA Binding Protein Coordinates DNA Metabolism Using Single-Molecule Approaches. Int. J. Mol. Sci. 2023, 24, 2806. https://doi.org/10.3390/ijms24032806
Xu L, Halma MTJ, Wuite GJL. Unravelling How Single-Stranded DNA Binding Protein Coordinates DNA Metabolism Using Single-Molecule Approaches. International Journal of Molecular Sciences. 2023; 24(3):2806. https://doi.org/10.3390/ijms24032806
Chicago/Turabian StyleXu, Longfu, Matthew T. J. Halma, and Gijs J. L. Wuite. 2023. "Unravelling How Single-Stranded DNA Binding Protein Coordinates DNA Metabolism Using Single-Molecule Approaches" International Journal of Molecular Sciences 24, no. 3: 2806. https://doi.org/10.3390/ijms24032806
APA StyleXu, L., Halma, M. T. J., & Wuite, G. J. L. (2023). Unravelling How Single-Stranded DNA Binding Protein Coordinates DNA Metabolism Using Single-Molecule Approaches. International Journal of Molecular Sciences, 24(3), 2806. https://doi.org/10.3390/ijms24032806








