BMSC-Derived Exosomal CircHIPK3 Promotes Osteogenic Differentiation of MC3T3-E1 Cells via Mitophagy
Abstract
1. Introduction
2. Results
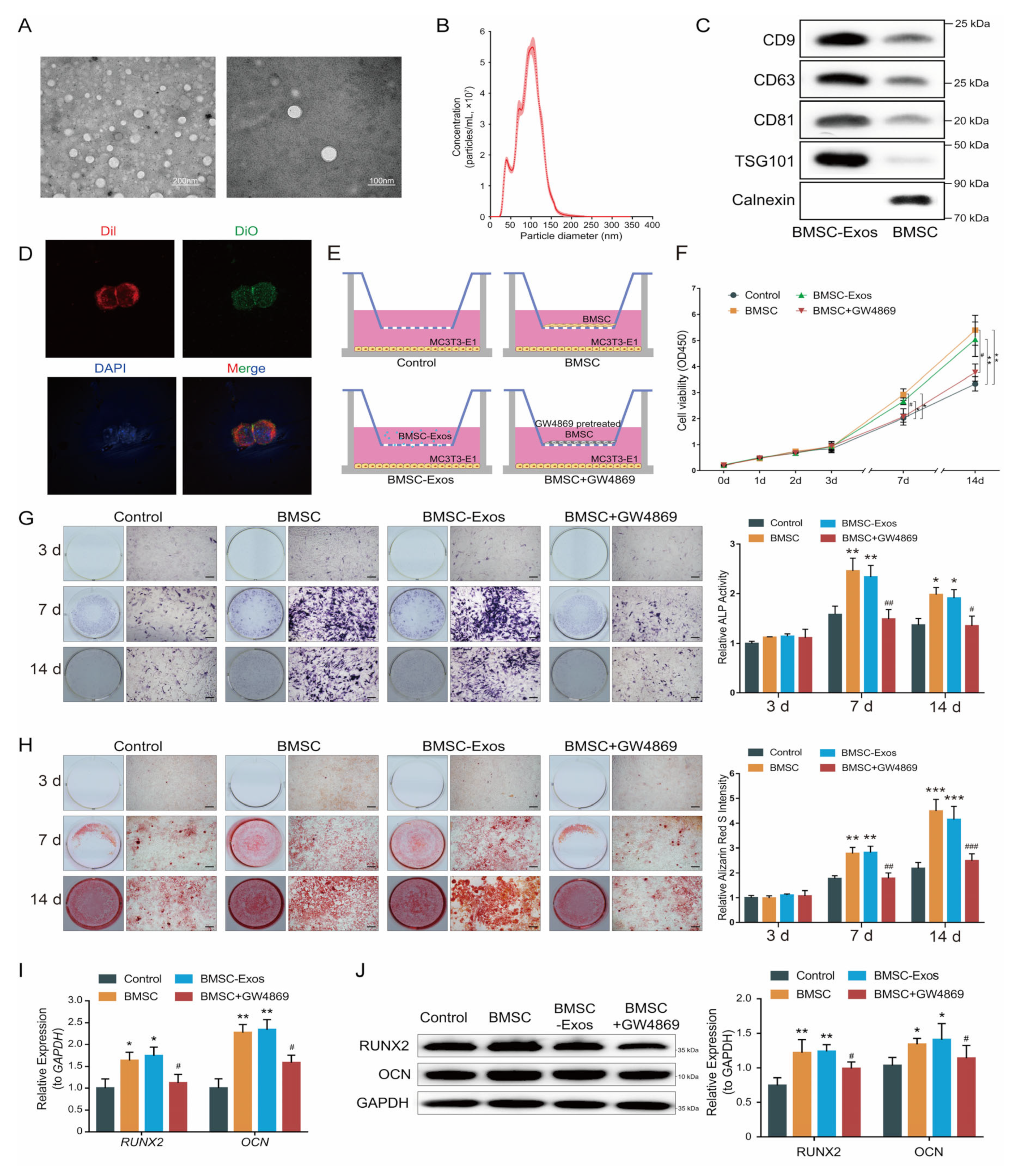
2.1. BMSC-Exos Enhances Osteogenic Differentiation of MC3T3-E1 Cells
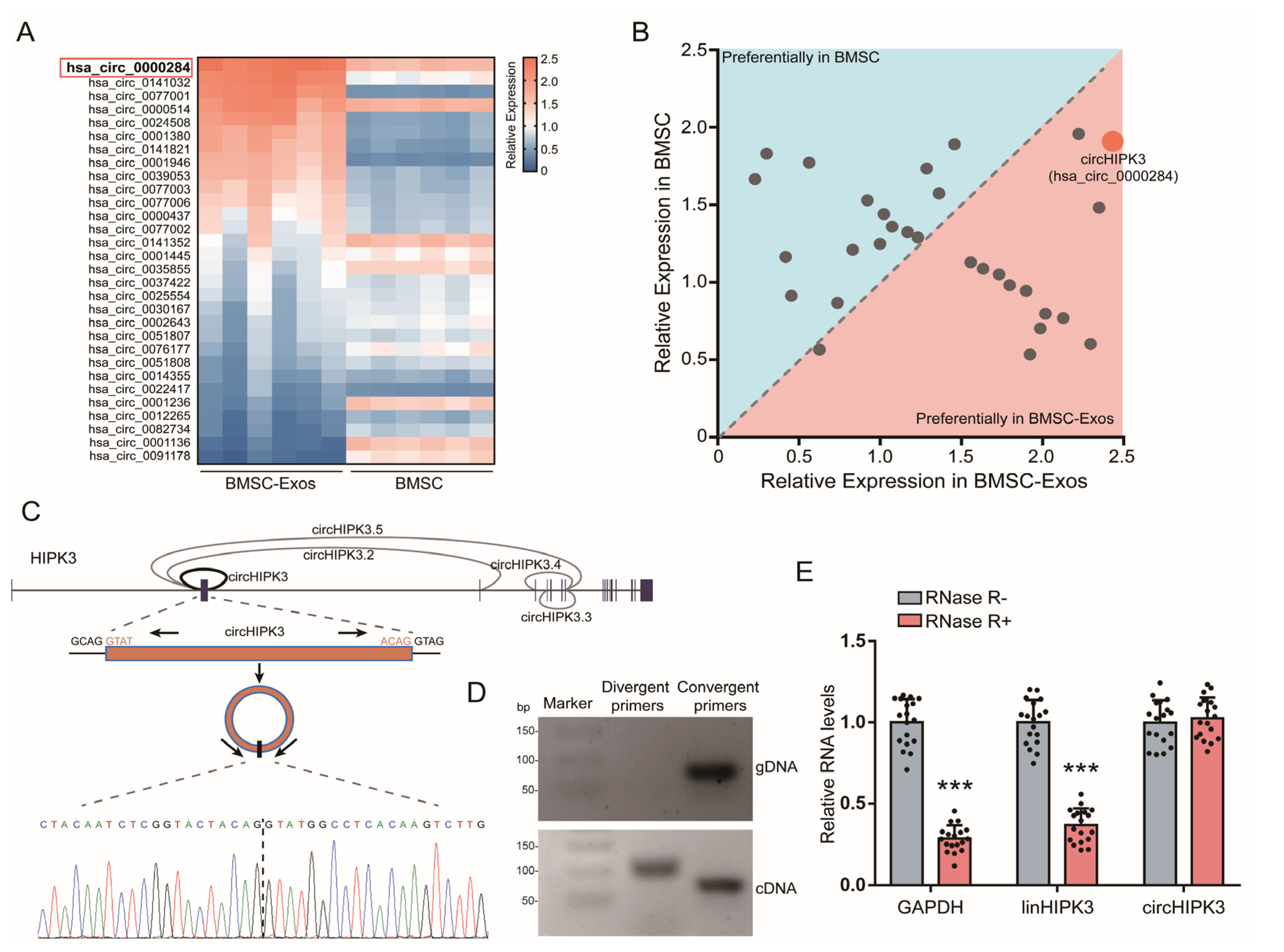
2.2. CircHIPK3 Is Enriched in BMSC-Exos
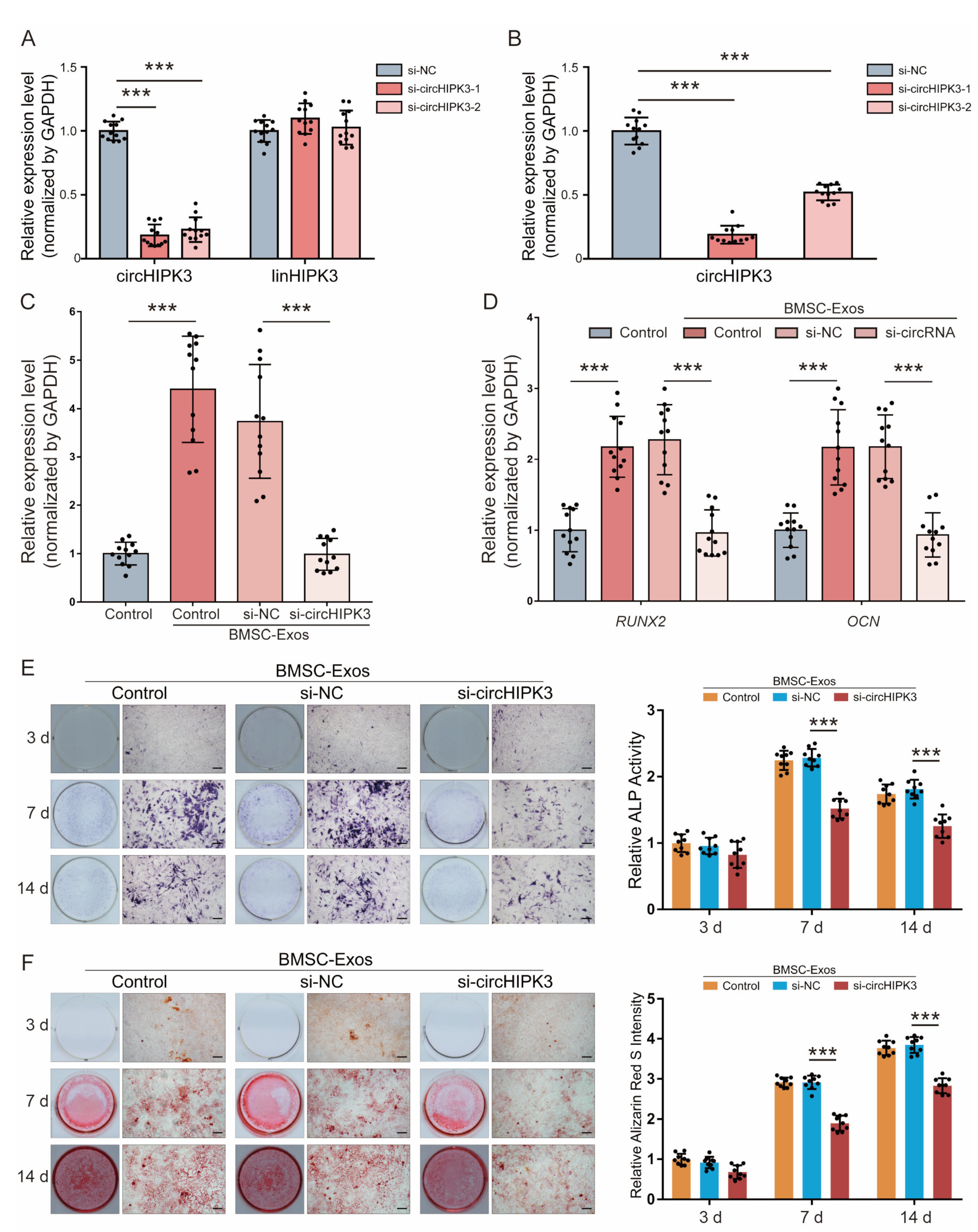
2.3. CircHIPK3 Is Critical for Pro-Osteogenic Effect of BMSC-Exos
2.4. CircHIPK3 Directly Binds to miR-29a-5p
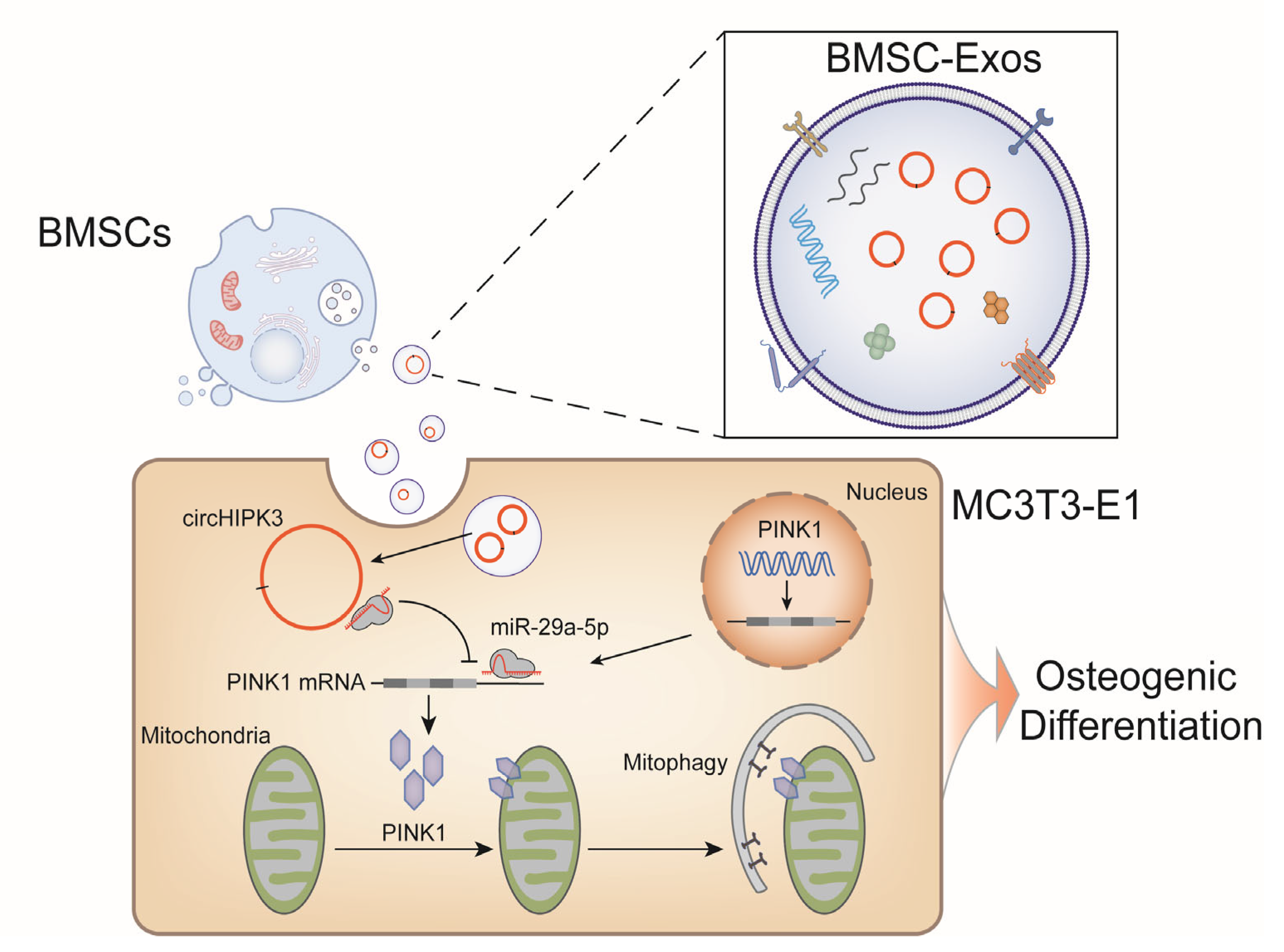
2.5. CircHIPK3 Promotes Mitophagy of MC3T3-E1 Cells through miR-29a-5p/PINK1 Axis
2.6. PINK1/Parkin-Mediated Mitophagy Contributed to the Pro-Osteogenic Effect of BMSC-Exos
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Flow Cytometry Analysis
4.3. Exosome Isolation
4.4. Exosome Characterization
4.5. Cellular Internalization of Exosomes
4.6. Transwell Coculture
4.7. Cell Proliferation
4.8. Quantitative Reverse Transcription-PCR (qRT-PCR)
4.9. Osteoblastic Differentiation Assays
4.10. PCR Amplification Using Divergent and Convergent Primers
4.11. RNase R Treatment
4.12. Small Interference (si) RNA Transfection
4.13. Dual-Luciferase Reporter Assay
4.14. Ago2-RIP Assay
4.15. CircRNA Pull-Down
4.16. LC3 Immunofluorescence Staining
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qin, Y.; Sun, R.; Wu, C.; Wang, L.; Zhang, C. Exosome: A novel approach to stimulate bone regeneration through regulation of osteogenesis and angiogenesis. Int. J. Mol. Sci. 2016, 17, 712. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.L.; Li, W.J. Identification of bone marrow-derived soluble factors regulating human mesenchymal stem cells for bone regeneration. Stem Cell Rep. 2017, 14, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Klimczak, A.; Kozlowska, U. Mesenchymal stromal cells and tissue-specific progenitor cells: Their role in tissue homeostasis. Stem Cells Int. 2016, 2016, 4285215. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Ding, Y.; Zhang, Y.; Tse, H.F.; Lian, Q. Paracrine mechanisms of mesenchymal stem cell-based therapy: Current status and perspectives. Cell Transplant. 2014, 23, 1045–1059. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Li, H.; Chen, C.; Hu, B.; Niu, X.; Li, Q.; Zhao, B.; Xie, Z.; Wang, Y. Exosomes/tricalcium phosphate combination scaffolds can enhance bone regeneration by activating the PI3K/Akt signaling pathway. Stem Cell Res. Ther. 2016, 20, 136. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Liu, A.; Lin, D.; Zhao, H.; Chen, L.; Cai, B.; Lin, K.; Shen, S.G. Optimized BMSC-derived osteoinductive exosomes immobilized in hierarchical scaffold via lyophilization for bone repair through Bmpr2/Acvr2b competitive receptor-activated Smad pathway. Biomaterials 2021, 272, 120718. [Google Scholar] [CrossRef]
- Yang, X.; Yang, J.; Lei, P.; Wen, T. LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating microRNA-34c/SATB2 axis. Aging 2019, 11, 8777–8791. [Google Scholar] [CrossRef]
- Fang, S.; He, T.; Jiang, J.; Li, Y.; Chen, P. Osteogenic effect of tsRNA-10277-loaded exosome derived from bone mesenchymal stem cells on steroid-induced osteonecrosis of the femoral head. Drug Des. Dev. Ther. 2020, 14, 4579–4591. [Google Scholar] [CrossRef]
- Huang, X.; Cen, X.; Zhang, B.; Liao, Y.; Zhu, G.; Liu, J.; Zhao, Z. Prospect of circular RNA in osteogenesis: A novel orchestrator of signaling pathways. J. Cell. Physiol. 2019, 234, 21450–21459. [Google Scholar] [CrossRef]
- Liu, C.X.; Chen, L.L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.; Zhang, J.; Yang, L.; Chen, L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016, 15, 611–624. [Google Scholar] [CrossRef]
- Qian, D.Y.; Yan, G.B.; Bai, B.; Chen, Y.; Zhang, S.J.; Yao, Y.C.; Xia, H. Differential circRNA expression profiles during the BMP2-induced osteogenic differentiation of MC3T3-E1 cells. Biomed. Pharmacother. 2017, 90, 492–499. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, X.; Huang, Y.; Jia, L.; Li, W. The circular RNA landscape of periodontal ligament stem cells during osteogenesis. J. Periodontol. 2017, 88, 906–914. [Google Scholar] [CrossRef]
- Zhang, D.; Ni, N.; Wang, Y.; Tang, Z.; Gao, H.; Ju, Y.; Sun, N.; He, X.; Gu, P.; Fan, X. CircRNA-vgll3 promotes osteogenic differentiation of adipose-derived mesenchymal stem cells via modulating miRNA-dependent integrin α5 expression. Cell Death Differ. 2021, 28, 283–302. [Google Scholar] [CrossRef]
- Li, X.; Zheng, Y.; Zheng, Y.; Huang, Y.; Zhang, Y.; Jia, L.; Li, W. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res. Ther. 2018, 9, 232. [Google Scholar] [CrossRef]
- Ouyang, Z.; Tan, T.; Zhang, X.; Wan, J.; Zhou, Y.; Jiang, G.; Yang, D.; Guo, X.; Liu, T. CircRNA hsa_circ_0074834 promotes the osteogenesis-angiogenesis coupling process in bone mesenchymal stem cells (BMSCs) by acting as a ceRNA for miR-942-5p. Cell Death Dis. 2019, 10, 932. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Ma, J.; Sun, T.; Zhou, Q.; Wang, W.; Wang, G.; Wu, P.; Wang, H.; Jiang, L.; et al. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer 2019, 18, 116. [Google Scholar] [CrossRef]
- Onishi, M.; Yamano, K.; Sato, M.; Matsuda, N.; Okamoto, K. Molecular mechanisms and physiological functions of mitophagy. Embo J. 2021, 40, e104705. [Google Scholar] [CrossRef]
- Pei, D.D.; Sun, J.L.; Zhu, C.H.; Tian, F.C.; Jiao, K.; Anderson, M.R.; Yiu, C.; Huang, C.; Jin, C.X.; Bergeron, B.E.; et al. Contribution of mitophagy to cell-mediated mineralization: Revisiting a 50-year-old conundrum. Adv. Sci. 2018, 5, 1800873. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.C.; Tang, X.Y.; Li, J.; Gao, P.; Wang, F.; Shen, M.J.; Gu, J.; Tay, F.; Chen, J.H.; Niu, L.N.; et al. Upregulation of mitochondrial dynamics is responsible for osteogenic differentiation of mesenchymal stem cells cultured on self-mineralized collagen membranes. Acta Biomater. 2021, 136, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, X.; Xie, J.; Weng, S.J.; Xie, Z.J.; Tang, J.H.; Yan, D.Y.; Wang, B.Z.; Fang, K.H.; Hong, C.X.; et al. Apelin-13 induces mitophagy in bone marrow mesenchymal stem cells to suppress intracellular oxidative stress and ameliorate osteoporosis by activation of AMPK signaling pathway. Free Radic. Biol. Med. 2021, 163, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; An, H.J.; Kim, J.M.; Sung, M.J.; Kim, D.K.; Kim, H.K.; Oh, J.; Jeong, H.Y.; Lee, Y.H.; Yang, T.; et al. PINK1 deficiency impairs osteoblast differentiation through aberrant mitochondrial homeostasis. Stem Cell Res. Ther. 2021, 12, 589. [Google Scholar] [CrossRef]
- Li, G.; Jian, Z.; Wang, H.; Xu, L.; Zhang, T.; Song, J. Irisin promotes osteogenesis by modulating oxidative stress and mitophagy through SIRT3 signaling under diabetic conditions. Oxidative Med. Cell. Longev. 2022, 2022, 3319056. [Google Scholar] [CrossRef]
- Tang, S.; Tang, T.; Gao, G.; Wei, Q.; Sun, K.; Huang, W. Bone marrow mesenchymal stem cell-derived exosomes inhibit chondrocyte apoptosis and the expression of MMPs by regulating Drp1-mediated mitophagy. Acta Histochem. 2021, 123, 151796. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Gao, Z.; Wu, J.; Zheng, X.; Jing, S.; Wang, W. MSC-derived extracellular vesicles activate mitophagy to alleviate renal ischemia/reperfusion injury via the miR-223-3p/NLRP3 axis. Stem Cells Int. 2022, 2022, 6852661. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, B.; Wang, Z.; Wang, D.; Ni, H.; Zhang, L.; Wang, Y. Exosomes from nicotine-stimulated macrophages accelerate atherosclerosis through miR-21-3p/PTEN-mediated VSMC migration and proliferation. Theranostics 2019, 9, 6901–6919. [Google Scholar] [CrossRef]
- Guo, B.B.; Bellingham, S.A.; Hill, A.F. The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. J. Biol. Chem. 2021, 290, 3455–3467. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, Y.; Wang, Z.; Xu, C.; Qiao, S.; Liu, T.; Qi, K.; Tong, D.; Li, C. Bone marrow mesenchymal stem cell exosome attenuates inflammasome-related pyroptosis via delivering circ_003564 to improve the recovery of spinal cord injury. Mol. Neurobiol. 2022, 59, 6771–6789. [Google Scholar] [CrossRef]
- Mao, G.; Xu, Y.; Long, D.; Sun, H.; Li, H.; Xin, R.; Zhang, Z.; Li, Z.; Yang, Z.; Kang, Y. Exosome-transported circRNA_0001236 enhances chondrogenesis and suppress cartilage degradation via the miR-3677-3p/Sox9 axis. Stem Cell Res. Ther. 2021, 12, 389. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Mao, Y.; Wu, D.; Zhu, Y.; Lu, J.; Huang, Y.; Guo, Y.; Wang, Z.; Zhu, S.; Li, X.; et al. Exosomal circ_0030167 derived from BM-MSCs inhibits the invasion, migration, proliferation and stemness of pancreatic cancer cells by sponging miR-338-5p and targeting the Wif1/Wnt8/beta-catenin axis. Cancer Lett. 2021, 512, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef] [PubMed]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef]
- Zhou, X.; Cao, H.; Guo, J.; Yuan, Y.; Ni, G. Effects of BMSC-derived EVs on bone metabolism. Pharmaceutics 2022, 14, 1012. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Z.; Gao, B.; Zhang, L. Exosome mediated biological functions within skeletal microenvironment. Front. Bioeng. Biotechnol. 2022, 10, 953916. [Google Scholar] [CrossRef]
- Shao, Q.; Huang, Y.; Zhang, C.; Gao, X.; Gao, S. Emerging landscape of circHIPK3 and its role in cancer and other diseases (Review). Mol. Med. Rep. 2021, 23, 409. [Google Scholar] [CrossRef]
- Zhuang, Z.; Jin, C.; Li, X.; Han, Y.; Yang, Q.; Huang, Y.; Zheng, Y.; Li, W. Knockdown of circHIPK3 promotes the osteogenic differentiation of human bone marrow mesenchymal stem cells through activating the autophagy flux. FASEB J. 2022, 36, e22590. [Google Scholar] [CrossRef]
- Sun, X.; Li, J.; Sun, Y.; Zhang, Y.; Dong, L.; Shen, C.; Yang, L.; Yang, M.; Li, Y.; Shen, G.; et al. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget 2016, 7, 53558–53570. [Google Scholar] [CrossRef]
- Ding, K.; Liao, Y.; Chen, Y.; Gong, D.; Zhao, X.; Ji, W. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem. Biophys. Res. Commun. 2018, 503, 863–869. [Google Scholar]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, S.; Liao, X.; Li, M.; Chen, S.; Li, X.; Wu, X.; Yang, M.; Tang, M.; Hu, Y.; et al. Circular RNA circIKBKB promotes breast cancer bone metastasis through sustaining NF-κB/bone remodeling factors signaling. Mol. Cancer 2021, 20, 98. [Google Scholar] [CrossRef]
- He, M.; Lei, H.; He, X.; Liu, Y.; Wang, A.; Ren, Z.; Liu, X.; Yan, G.; Wang, W.; Wang, Y.; et al. METTL14 regulates osteogenesis of bone marrow mesenchymal stem cells via inducing autophagy through m6A/IGF2BPs/Beclin-1 signal axis. Stem Cells Transl. Med. 2022, 11, 987–1001. [Google Scholar] [CrossRef]
- Fei, D.; Xia, Y.; Zhai, Q.; Wang, Y.; Zhou, F.; Zhao, W.; He, X.; Wang, Q.; Jin, Y.; Li, B. Exosomes regulate interclonal communication on osteogenic differentiation among heterogeneous osteogenic single-cell clones through PINK1/Parkin-mediated mitophagy. Front. Cell Dev. Biol. 2021, 9, 687258. [Google Scholar] [CrossRef]
- Xia, W.; Xie, J.; Cai, Z.; Liu, X.; Wen, J.; Cui, Z.K.; Zhao, R.; Zhou, X.; Chen, J.; Mao, X.; et al. Damaged brain accelerates bone healing by releasing small extracellular vesicles that target osteoprogenitors. Nat. Commun. 2021, 12, 6043. [Google Scholar] [CrossRef]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Wu, H.; Zhou, J.; Mei, S.; Wu, D.; Mu, Z.; Chen, B.; Xie, Y.; Ye, Y.; Liu, J. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J. Cell. Mol. Med. 2017, 21, 1228–1236. [Google Scholar] [CrossRef]
- Ma, S.; Tong, C.; Ibeagha-Awemu, E.M.; Zhao, X. Identification and characterization of differentially expressed exosomal microRNAs in bovine milk infected with Staphylococcus aureus. BMC Genomics 2019, 20, 934. [Google Scholar] [CrossRef]
- Fang, J.H.; Zhang, Z.J.; Shang, L.R.; Luo, Y.W.; Lin, Y.F.; Yuan, Y.; Zhaung, S.M. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology 2018, 68, 1459–1475. [Google Scholar] [CrossRef]
- Yan, Y.; Jiang, W.; Tan, Y.; Zuo, S.; Zhang, H.; Mao, F.; Gong, A.; Qian, H.; Xu, W. hucMSC exosome-derived GPX1 is required for the recovery of hepatic oxidant injury. Mol. Ther. 2017, 25, 465–479. [Google Scholar] [CrossRef]
- Krell, J.; Stebbing, J.; Carissimi, C.; Dabrowska, A.F.; de Giorgio, A.; Frampton, A.E.; Harding, V.; Fulci, V.; Macino, G.; Colombo, T.; et al. TP53 regulates miRNA association with AGO2 to remodel the miRNA-mRNA interaction network. Genome Res. 2016, 26, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Hasanain, M.; Bhattacharjee, A.; Pandey, P.; Ashraf, R.; Singh, N.; Sharma, S.; Vishwakarma, A.L.; Datta, D.; Mitra, K.; Sarkar, J. α-Solanine induces ROS-mediated autophagy through activation of endoplasmic reticulum stress and inhibition of Akt/mTOR pathway. Cell Death Dis. 2015, 6, e1860. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, S.; Li, S.; Zhang, Y.; Nie, J.; Cao, J.; Li, A.; Li, Y.; Pei, D. BMSC-Derived Exosomal CircHIPK3 Promotes Osteogenic Differentiation of MC3T3-E1 Cells via Mitophagy. Int. J. Mol. Sci. 2023, 24, 2785. https://doi.org/10.3390/ijms24032785
Ma S, Li S, Zhang Y, Nie J, Cao J, Li A, Li Y, Pei D. BMSC-Derived Exosomal CircHIPK3 Promotes Osteogenic Differentiation of MC3T3-E1 Cells via Mitophagy. International Journal of Molecular Sciences. 2023; 24(3):2785. https://doi.org/10.3390/ijms24032785
Chicago/Turabian StyleMa, Shaoyang, Sijia Li, Yuchen Zhang, Jiaming Nie, Jiao Cao, Ang Li, Ye Li, and Dandan Pei. 2023. "BMSC-Derived Exosomal CircHIPK3 Promotes Osteogenic Differentiation of MC3T3-E1 Cells via Mitophagy" International Journal of Molecular Sciences 24, no. 3: 2785. https://doi.org/10.3390/ijms24032785
APA StyleMa, S., Li, S., Zhang, Y., Nie, J., Cao, J., Li, A., Li, Y., & Pei, D. (2023). BMSC-Derived Exosomal CircHIPK3 Promotes Osteogenic Differentiation of MC3T3-E1 Cells via Mitophagy. International Journal of Molecular Sciences, 24(3), 2785. https://doi.org/10.3390/ijms24032785




