Triple-Negative Breast Cancer and Predictive Markers of Response to Neoadjuvant Chemotherapy: A Systematic Review
Abstract
:1. Introduction
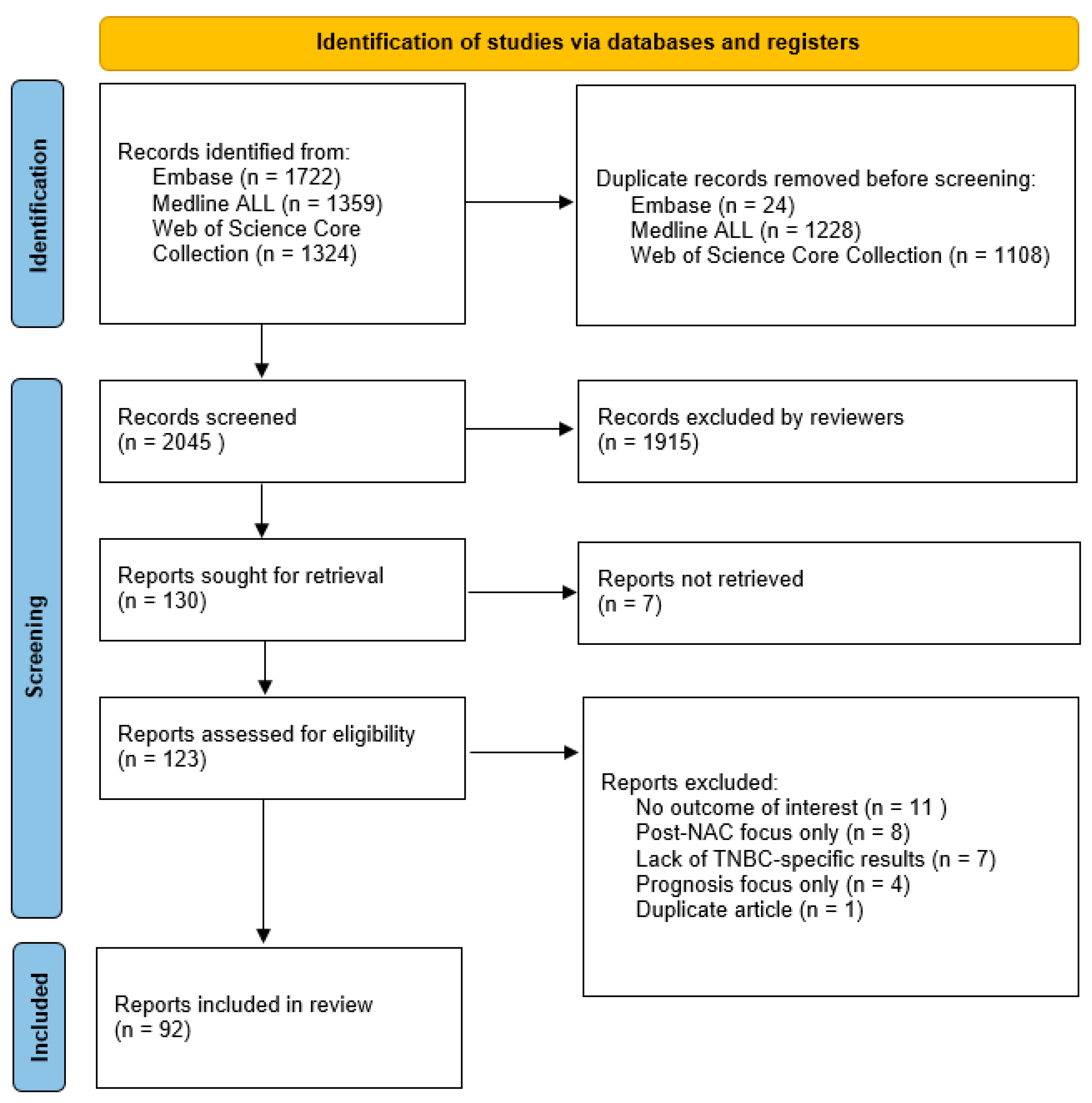
2. Methods
2.1. Literature Search
2.2. Inclusion and Exclusion
2.3. Synthesis Method
3. Results
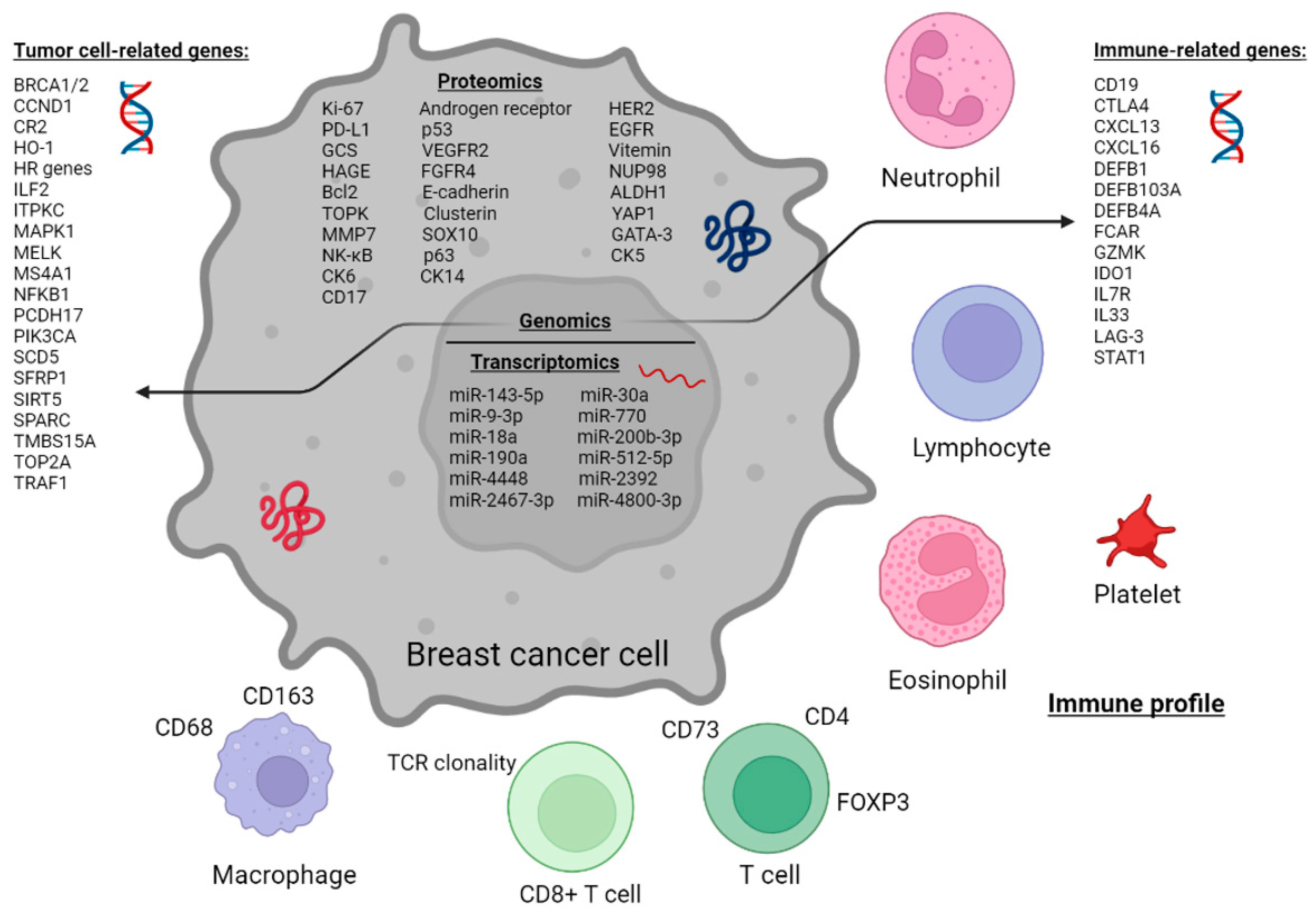
3.1. Proteomic Profile of Tumor Cells
3.2. Proteomic Profile of Tumor-Associated Immune Cells
3.3. Immune Cells in Peripheral Blood
3.4. Genomic Profile
3.5. Transcriptomic Profile
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALDH1 | Aldehyde dehydrogenase 1 family member A1 |
| AR | Androgen receptor |
| ATM | Ataxia telangiectasia-mutated |
| ATP | Adenosine 5′-triphosphate |
| Bcl2 | B-cell lymphoma 2 |
| BPESC1 | Blepharophimosis, epicanthus inversus and ptosis candidate 1 |
| BRCA | Breast cancer |
| CCND1 | Cyclin D1 |
| CK | Cytokeratine |
| CD19 | Cluster of differentiation 19 |
| CEP44 | Centrosomal protein 44 |
| CR2 | Complement component 3d receptor 2 |
| CREB1 | CAMP responsive element binding protein 1 |
| CTLA4 | Cluster of differentiation 152 |
| CXCL | Chemokine (C-X-C motif) ligand |
| DEFB | Defensin beta |
| DFS | Disease-free survival |
| EGFR | Epidermal growth factor receptor |
| ER | Estrogen receptor |
| FCAR | Fc fragment of IgA receptor |
| FOXP3 | Forkhead box P3 |
| FGFR4 | Fibroblast growth factor receptor 4 |
| GADD45A | Growth arrest and DNA damage inducible alpha |
| GATA3 | GATA binding protein 3 |
| GCS | Glucosylceramide synthase |
| GZMB | Granzyme B |
| GZMK | Granzyme K |
| HAGE | Helicase antigen |
| HER2 | Human epidermal growth factor 2 |
| HO-1 | Heme oxygenase 1 |
| IDO1 | Indoleamine 2,3-dioxygenase 1 |
| IL7R | Interleukin-7 receptor |
| IL33 | Interleukin 33 |
| ILF2 | Interleukin enhancer binding factor 2 |
| ITPKC | Inositol-trisphosphate 3-kinase C |
| LAG-3 | Lymphocyte activation gene-3 |
| LncRNA | Long non-coding RNA |
| MAPK1 | Mitogen-activated protein kinase 1 |
| MELK | Maternal embryonic leucine zipper kinase |
| MFS | Metastasis-free survival |
| miRNA | MicroRNA |
| MMP7 | Matrix metalloproteinase 7 |
| MS4A1 | Membrane spanning 4-domains A1 |
| NAC | Neoadjuvant chemotherapy |
| NFKB1 | Nuclear factor kappa B subunit 1 |
| NUP98 | Nucleoporin 98 and 96 precursor |
| OS | Overall survival |
| PCDH17 | Protocadherin 17 |
| pCR | Pathological complete response |
| PD-L1 | Programmed cell death protein ligand 1 |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| PR | Progesterone receptor |
| RCB | Residual cancer burden |
| RFS | Recurrence free survival |
| SCD5 | Stearoyl-CoA desaturase 5 |
| SFRP1 | Secreted frizzled related protein 1 |
| SIRT5 | Sirtuin 5 |
| SPARC | Secreted protein acidic and rich in cysteine |
| SOX10 | SRY-box transcription factor 10 |
| STAT1 | Signal transducer and activator of transcription 1 |
| (s)TIL | (Stromal) tumor-infiltrating lymphocyte |
| TCF3 | Transcription factor 3 |
| TCR | T cell receptor |
| TMBS15A | Thymosin beta-15A |
| TRAF1 | TNF receptor-associated factor 1 |
| TILV | Tumor-infiltrating lymphocyte volume |
| TP53 | Tumor protein 53 |
| TOP2A | Topoisomerase IIa |
| TOPK | T-LAK-cell originated protein kinase |
| Tregs | T regulator cells |
| VEGFR2 | Vascular endothelial growth factor receptor 2 |
| WDR72 | WD repeat domain 72 |
| YAP1 | Yes1-associated transcriptional regulator |
References
- International WCRF. Worldwide Cancer Data. 2020. Available online: https://www.wcrf.org/cancer-trends/worldwide-cancer-data/ (accessed on 3 November 2022).
- Rakha, E.A.; El-Sayed, M.E.; Green, A.R.; Lee, A.H.; Robertson, J.F.; Ellis, I.O. Prognostic markers in triple-negative breast cancer. Cancer 2007, 109, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef] [PubMed]
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J.; et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J. Clin. Oncol. 2020, 38, 1346–1366. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015, 24, S26–S35. [Google Scholar] [CrossRef] [PubMed]
- Bernemann, C.; Hülsewig, C.; Rückert, C.; Schäfer, S.; Blümel, L.; Hempel, G.; Götte, M.; Greve, B.; Barth, P.J.; Kiesel, L.; et al. Influence of secreted frizzled receptor protein 1 (SFRP1) on neoadjuvant chemotherapy in triple negative breast cancer does not rely on WNT signaling. Mol. Cancer 2014, 13, 174. [Google Scholar] [CrossRef]
- Zhu, M.; Liang, C.; Zhang, F.; Zhu, L.; Chen, D. A Nomogram to Predict Disease-Free Survival Following Neoadjuvant Chemotherapy for Triple Negative Breast Cancer. Front. Oncol. 2021, 11, 4399. [Google Scholar] [CrossRef]
- Abuhadra, N.; Stecklein, S.; Sharma, P.; Moulder, S. Early-stage Triple-negative Breast Cancer: Time to Optimize Personalized Strategies. Oncologist 2022, 27, 30–39. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef]
- Mittendorf, E.A.; Zhang, H.; Barrios, C.H.; Saji, S.; Jung, K.H.; Hegg, R.; Koehler, A.; Sohn, J.; Iwata, H.; Telli, M.L.; et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): A randomised, double-blind, phase 3 trial. Lancet 2020, 396, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Yost, S.E.; Yuan, Y. Neoadjuvant Treatment for Triple Negative Breast Cancer: Recent Progresses and Challenges. Cancers 2020, 12, 1404. [Google Scholar] [CrossRef] [PubMed]
- Holanek, M.; Selingerova, I.; Bilek, O.; Kazda, T.; Fabian, P.; Foretova, L.; Zvarikova, M.; Obermannova, R.; Kolouskova, I.; Coufal, O.; et al. Neoadjuvant Chemotherapy of Triple-Negative Breast Cancer: Evaluation of Early Clinical Response, Pathological Complete Response Rates, and Addition of Platinum Salts Benefit Based on Real-World Evidence. Cancers 2021, 13, 1586. [Google Scholar] [CrossRef]
- Paluch-Shimon, S.; Friedman, E.; Berger, R.; Papa, M.; Dadiani, M.; Friedman, N.; Shabtai, M.; Zippel, D.; Gutman, M.; Golan, T.; et al. Neo-adjuvant doxorubicin and cyclophosphamide followed by paclitaxel in triple-negative breast cancer among BRCA1 mutation carriers and non-carriers. Breast Cancer Res. Treat. 2016, 157, 157–165. [Google Scholar] [CrossRef]
- Bianchini, G.; De Angelis, C.; Licata, L.; Gianni, L. Treatment landscape of triple-negative breast cancer—Expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2022, 19, 91–113. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13 Pt 1, 4429–4434. [Google Scholar] [CrossRef]
- Oshi, M.; Newman, S.; Murthy, V.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. ITPKC as a Prognostic and Predictive Biomarker of Neoadjuvant Chemotherapy for Triple Negative Breast Cancer. Cancers 2020, 12, 2758. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Han, W.; Kim, M.K.; Lee, J.W.; Kim, J.; Ahn, S.K.; Lee, H.-B.; Moon, H.-G.; Lee, K.-H.; Kim, T.-Y.; et al. Predictive Significance of p53, Ki-67, and Bcl-2 Expression for Pathologic Complete Response after Neoadjuvant Chemotherapy for Triple-Negative Breast Cancer. J. Breast Cancer 2015, 18, 16–21. [Google Scholar] [CrossRef]
- Guestini, F.; Ono, K.; Miyashita, M.; Ishida, T.; Ohuchi, N.; Nakagawa, S.; Hirakawa, H.; Tamaki, K.; Ohi, Y.; Rai, Y.; et al. Impact of Topoisomerase IIα, PTEN, ABCC1/MRP1, and KI67 on triple-negative breast cancer patients treated with neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2019, 173, 275–288. [Google Scholar] [CrossRef]
- Mohammed, A.A.; Elsayed, F.M.; Algazar, M.; Rashed, H.E.; Anter, A.H. Neoadjuvant Chemotherapy in Triple Negative Breast Cancer: Correlation between Androgen Receptor Expression and Pathological Response. Asian Pac. J. Cancer Prev. 2020, 21, 563–568. [Google Scholar] [CrossRef]
- Zhu, M.; Yu, Y.; Shao, X.; Zhu, L.; Wang, L. Predictors of Response and Survival Outcomes of Triple Negative Breast Cancer Receiving Neoadjuvant Chemotherapy. Chemotherapy 2020, 65, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Kedzierawski, P.; Macek, P.; Ciepiela, I.; Kowalik, A.; Gozdz, S. Evaluation of Complete Pathological Regression after Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer Patients with BRCA1 Founder Mutation Aided Bayesian A/B Testing Approach. Diagnostics 2021, 11, 1144. [Google Scholar] [CrossRef] [PubMed]
- Bignon, L.; Fricker, J.P.; Nogues, C.; Mouret-Fourme, E.; Stoppa-Lyonnet, D.; Caron, O.; Lortholary, A.; Faivre, L.; Lasset, C.; Mari, V.; et al. Efficacy of anthracycline/taxane-based neo-adjuvant chemotherapy on triple-negative breast cancer in BRCA1/BRCA2 mutation carriers. Breast J. 2018, 24, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Masuda, N.; Kodama, Y.; Ogawa, M.; Karita, M.; Yamamura, J.; Tsukuda, K.; Doihara, H.; Miyoshi, S.; Mano, M.; et al. Predictive factors for the effectiveness of neoadjuvant chemotherapy and prognosis in triple-negative breast cancer patients. Cancer Chemother. Pharmacol. 2011, 67, 911–917. [Google Scholar] [CrossRef]
- Van Bockstal, M.R.; Noel, F.; Guiot, Y.; Duhoux, F.P.; Mazzeo, F.; Van Marcke, C.; Fellah, L.; Ledoux, B.; Berlière, M.; Galant, C. Predictive markers for pathological complete response after neo-adjuvant chemotherapy in triple-negative breast cancer. Ann. Diagn. Pathol. 2020, 49, 151634. [Google Scholar] [CrossRef]
- Tan, Q.; Qin, Q.; Huang, Z.; Lian, B.; Mo, Q.; Wei, C. Predictive and prognostic effect of HO-1 expression in breast cancer patients undergoing neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2022, 193, 393–403. [Google Scholar] [CrossRef]
- Guo, S.; Loibl, S.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Denkert, C. PIK3CA H1047R Mutation Associated with a Lower Pathological Complete Response Rate in Triple-Negative Breast Cancer Patients Treated with Anthracycline-Taxane-Based Neoadjuvant Chemotherapy. Cancer Res. Treat. 2020, 52, 689–696. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, J.J.; Song, I.H.; Park, I.A.; Kang, J.; Yu, J.H.; Ahn, J.H.; Gong, G. Prognostic and predictive value of NanoString-based immune-related gene signatures in a neoadjuvant setting of triple-negative breast cancer: Relationship to tumor-infiltrating lymphocytes. Breast Cancer Res. Treat. 2015, 151, 619–627. [Google Scholar] [CrossRef]
- Ye, J.H.; Wang, X.H.; Shi, J.J.; Yin, X.; Chen, C.; Chen, Y.; Wu, H.-Y.; Jiong, S.; Sun, Q.; Zhang, M.; et al. Tumor-associated macrophages are associated with response to neoadjuvant chemotherapy and poor outcomes in patients with triple-negative breast cancer. J. Cancer 2021, 12, 2886–2892. [Google Scholar] [CrossRef]
- Cerbelli, B.; Scagnoli, S.; Mezi, S.; De Luca, A.; Pisegna, S.; Amabile, M.I.; Roberto, M.; Fortunato, L.; Costarelli, L.; Pernazza, A.; et al. Tissue Immune Profile: A Tool to Predict Response to Neoadjuvant Therapy in Triple Negative Breast Cancer. Cancers 2020, 12, 2648. [Google Scholar] [CrossRef]
- Kong, D.D.; Fu, R.Z.; Li, L.; Wang, W.; Wang, S.B. Association between the methylation status of PCDH17 and the efficacy of neoadjuvant chemotherapy in triple-negative breast cancer. Oncol. Lett. 2020, 20, 1649–1656. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, S.; Wang, C.; Kong, X.; Sun, P. Prognostic value of using glucosylceramide synthase and cytochrome P450 family 1 subfamily A1 expression levels for patients with triple-negative breast cancer following neoadjuvant chemotherapy. Exp. Ther. Med. 2021, 21, 247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X.I.; Zhang, S. Tumor-infiltrating lymphocyte volume is a better predictor of neoadjuvant therapy response and overall survival in triple-negative invasive breast cancer. Hum. Pathol. 2018, 80, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- García-Vazquez, R.; Ruiz-García, E.; Meneses Garcia, A.; Astudillo-De La Vega, H.; Lara-Medina, F.; Alvarado-Miranda, A.; Maldonado-Martínez, H.; González-Barrios, J.A.; Campos-Parra, A.D.; Rodriguez Cuevas, S.; et al. A microRNA signature associated with pathological complete response to novel neoadjuvant therapy regimen in triple-negative breast cancer. Tumor Biol. 2017, 39, 1010428317702899. [Google Scholar] [CrossRef]
- Miyashita, M.; Sasano, H.; Tamaki, K.; Chan, M.; Hirakawa, H.; Suzuki, A.; Tada, H.; Watanabe, G.; Nemoto, N.; Nakagawa, S.; et al. Tumor-infiltrating CD8+ and FOXP3+ lymphocytes in triple-negative breast cancer: Its correlation with pathological complete response to neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2014, 148, 525–534. [Google Scholar] [CrossRef]
- Gluz, O.; Kolberg-Liedtke, C.; Prat, A.; Christgen, M.; Gebauer, D.; Kates, R.; Paré, L.; Grischke, E.M.; Forstbauer, H.; Braun, M.; et al. Efficacy of deescalated chemotherapy according to PAM50 subtypes, immune and proliferation genes in triple-negative early breast cancer: Primary translational analysis of the WSG-ADAPT-TN trial. Int J Cancer. 2020, 146, 262–271. [Google Scholar] [CrossRef]
- Zuo, K.; Yuan, X.; Liang, X.; Sun, X.; Liu, S.; Connell, P.P.; Li, X.; Yang, W. qRT-PCR-based DNA homologous recombination-associated 4-gene score predicts pathologic complete response to platinum-based neoadjuvant chemotherapy in triple-negative breast cancer. Breast Cancer Res. Treat. 2022, 191, 335–344. [Google Scholar] [CrossRef]
- Baba, M.; Takahashi, M.; Yamashiro, K.; Yokoo, H.; Fukai, M.; Sato, M.; Hosoda, M.; Kamiyama, T.; Taketomi, A.; Yamashita, H. Strong cytoplasmic expression of NF-κB/p65 correlates with a good prognosis in patients with triple-negative breast cancer. Surg. Today 2016, 46, 843–851. [Google Scholar] [CrossRef]
- Kraus, J.A.; Beriwal, S.; Dabbs, D.J.; Ahrendt, G.M.; McGuire, K.P.; Johnson, R.R.; Badve, P.; Puhalla, S.L.; Bhargava, R. Predictors of pathologic complete response after standard neoadjuvant chemotherapy in triple-negative breast carcinoma. Appl. Immunohistochem. Mol. Morphol. 2012, 20, 334–339. [Google Scholar] [CrossRef]
- Kawate, T.; Iwaya, K.; Kikuchi, R.; Kaise, H.; Oda, M.; Sato, E.; Hiroi, S.; Matsubara, O.; Kohno, N. DJ-1 protein expression as a predictor of pathological complete remission after neoadjuvant chemotherapy in breast cancer patients. Breast Cancer Res. Treat. 2013, 139, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, K.; Kurosumi, M.; Oba, H.; Kobayashi, Y.; Takei, H.; Inoue, K.; Tabei, T.; Oyama, T. Pathological tumor response to neoadjuvant chemotherapy using anthracycline and taxanes in patients with triple-negative breast cancer. Exp. Ther. Med. 2011, 2, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Di Leone, A.; Fragomeni, S.M.; Scardina, L.; Ionta, L.; Mulè, A.; Magno, S.; Terribile, D.; Masetti, R.; Franceschini, G. Androgen receptor expression and outcome of neoadjuvant chemotherapy in triple-negative breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1910–1915. [Google Scholar]
- Sridhar, N.; Glisch, C.; Jawa, Z.; Chaudhary, L.N.; Kamaraju, S.; Burfeind, J.; Charlson, J.; Chitambar, C.R.; Jorns, J.M.; Cheng, Y.C. Androgen receptor expression in patients with triple negative breast cancer treated with neoadjuvant chemotherapy: A single institution study. J. Cancer 2022, 13, 2472–2476. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Li, J.; Li, P.; Wang, L.; Di, L. An Enhancer-Based Analysis Revealed a New Function of Androgen Receptor in Tumor Cell Immune Evasion. Front. Genet. 2020, 11, 595550. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, C.; Schiff, R. HER 2, biology, detection, and clinical implications. Arch. Pathol. Lab. Med. 2011, 135, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Seither, F.; Schneeweiss, A.; Link, T.; Blohmer, J.-U.; Just, M.; Wimberger, P.; Forberger, A.; Tesch, H.; Jackisch, C.; et al. Clinical and molecular characteristics of HER2-low-positive breast cancer: Pooled analysis of individual patient data from four prospective, neoadjuvant clinical trials. Lancet Oncol. 2021, 22, 1151–1161. [Google Scholar] [CrossRef]
- Ende, N.S.v.D.; Smid, M.; Timmermans, A.; van Brakel, J.B.; Hansum, T.; Foekens, R.; Trapman, A.M.A.C.; Heemskerk-Gerritsen, B.A.M.; Jager, A.; Martens, J.W.M.; et al. HER2-low breast cancer shows a lower immune response compared to HER2-negative cases. Sci. Rep. 2022, 12, 12974. [Google Scholar] [CrossRef]
- Baez-Navarro, X.; Salgado, R.; Denkert, C.; Lennerz, J.K.; Penault-Llorca, F.; Viale, G.; Bartlett, J.M.; van Deurzen, C.H. Selecting patients with HER2-low breast cancer: Getting out of the tangle. Eur. J. Cancer 2022, 175, 187–192. [Google Scholar] [CrossRef]
- Duffy, M.J.; Synnott, N.C.; Crown, J. Mutant p53 in breast cancer: Potential as a therapeutic target and biomarker. Breast Cancer Res. Treat. 2018, 170, 213–219. [Google Scholar] [CrossRef]
- Liao, G.; Jiang, Z.; Yang, Y.; Zhang, C.; Jiang, M.; Zhu, J.; Xu, L.; Xie, A.; Yan, M.; Zhang, Y.; et al. Combined homologous recombination repair deficiency and immune activation analysis for predicting intensified responses of anthracycline, cyclophosphamide and taxane chemotherapy in triple-negative breast cancer. BMC Med. 2021, 19, 190. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.Y.; Lee, J.H.; Bae, J.W.; Jung, S.P. Differences in prognosis by p53 expression after neoadjuvant chemotherapy in triple-negative breast cancer. Ann. Surg. Treat. Res. 2020, 98, 291–298. [Google Scholar] [CrossRef]
- Ademuyiwa, F.O.; Chen, I.; Luo, J.; Rimawi, M.F.; Hagemann, I.S.; Fisk, B.; Jeffers, G.; Skidmore, Z.L.; Basu, A.; Richters, M.; et al. Immunogenomic profiling and pathological response results from a clinical trial of docetaxel and carboplatin in triple-negative breast cancer. Breast Cancer Res. Treat. 2021, 189, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhu, L.; Li, Y.; Ji, J.; Li, J.; Yuan, F.; Wang, D.; Chen, W.; Huang, O.; Chen, X.; et al. Overexpression of epithelial growth factor receptor (EGFR) predicts better response to neo-adjuvant chemotherapy in patients with triple-negative breast cancer. J. Transl. Med. 2012, 10, S4. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, A.E.; Rashed, H.E.; Abdelgawad, M.; Abdelhamid, M.I. Prognostic impact of EGFR and cytokeratin 5/6 immunohistochemical expression in triple-negative breast cancer. Ann. Diagn. Pathol. 2017, 28, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Babyshkina, N.; Zavyalova, M.; Tarabanovskaya, N.; Dronova, T.; Krakhmal, N.; Slonimskaya, E.; Kzhyshkowska, J.; Choynzonov, E.; Cherdyntseva, N. Predictive value of vascular endothelial growth factor receptor type 2 in triple-negative breast cancer patients treated with neoadjuvant chemotherapy. Mol. Cell. Biochem. 2018, 444, 197–206. [Google Scholar] [CrossRef]
- Abdel-Fatah, T.M.; McArdle, S.E.; Agarwal, D.; Moseley, P.M.; Green, A.R.; Ball, G.R.; Pockley, A.G.; Ellis, I.O.; Rees, R.C.; Chan, S.Y. HAGE in Triple-Negative Breast Cancer Is a Novel Prognostic, Predictive, and Actionable Biomarker: A Transcriptomic and Protein Expression Analysis. Clin. Cancer Res. 2016, 22, 905–914. [Google Scholar] [CrossRef]
- Mullan, P.B.; Bingham, V.; Haddock, P.; Irwin, G.W.; Kay, E.; McQuaid, S.; Buckley, N.E. NUP98—A novel predictor of response to anthracycline-based chemotherapy in triple negative breast cancer. BMC Cancer. 2019, 19, 236. [Google Scholar] [CrossRef]
- Abdel-Fatah, T.; Perry, C.; Dickinson, P.; Ball, G.; Moseley, P.; Madhusudan, S.; Ellis, I.; Chan, S. Bcl2 is an independent prognostic marker of triple negative breast cancer (TNBC) and predicts response to anthracycline combination (ATC) chemotherapy (CT) in adjuvant and neoadjuvant settings. Ann. Oncol. 2013, 24, 2801–2807. [Google Scholar] [CrossRef]
- Kida, K.; Ishikawa, T.; Yamada, A.; Shimada, K.; Narui, K.; Sugae, S.; Shimizu, D.; Tanabe, M.; Sasaki, T.; Ichikawa, Y.; et al. Effect of ALDH1 on prognosis and chemoresistance by breast cancer subtype. Breast Cancer Res. Treat. 2016, 156, 261–269. [Google Scholar] [CrossRef]
- Wang, Y.; Brodsky, A.S.; Xiong, J.; Lopresti, M.L.; Yang, D.; Resnick, M.B. Stromal Clusterin Expression Predicts Therapeutic Response to Neoadjuvant Chemotherapy in Triple Negative Breast Cancer. Clin. Breast Cancer 2018, 18, e373–e379. [Google Scholar] [CrossRef]
- Wang, K.; Chai, J.; Xu, J.; Wei, J.; Li, P.; Liu, Y.; Ma, J.; Xu, T.; Zhao, D.; Yu, K.; et al. TOPK: A new predictor of the therapeutic response to neoadjuvant chemotherapy and prognosis in triple-negative breast cancer. Pathol.-Res. Pract. 2021, 226, 153603. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.-Q.; Zhang, K.-J.; Wang, S.-M.; Guo, L. YAP1/MMP7/CXCL16 axis affects efficacy of neoadjuvant chemotherapy via tumor environment immunosuppression in triple-negative breast cancer. Gland Surg. 2021, 10, 2799–2814. [Google Scholar] [CrossRef] [PubMed]
- Na, T.-Y.; Schecterson, L.; Mendonsa, A.M.; Gumbiner, B.M. The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. Proc. Natl. Acad. Sci. USA 2020, 117, 5931–5937. [Google Scholar] [CrossRef]
- Guo, L.; Chen, Y.; Luo, J.; Zheng, J.; Shao, G. YAP1 overexpression is associated with poor prognosis of breast cancer patients and induces breast cancer cell growth by inhibiting PTEN. FEBS Open Bio 2019, 9, 437–445. [Google Scholar] [CrossRef]
- Soleyman-Jahi, S.; Nedjat, S.; Abdirad, A.; Hoorshad, N.; Heidari, R.; Zendehdel, K. Prognostic Significance of Matrix Metalloproteinase-7 in Gastric Cancer Survival: A Meta-Analysis. PLoS ONE 2015, 10, e0122316. [Google Scholar] [CrossRef]
- Hammerl, D.; Massink, M.P.; Smid, M.; van Deurzen, C.H.; Meijers-Heijboer, H.E.; Waisfisz, Q.; Debets, R.; Martens, J.W. Clonality, Antigen Recognition, and Suppression of CD8(+) T Cells Differentially Affect Prognosis of Breast Cancer Subtypes. Clin. Cancer Res. 2020, 26, 505–517. [Google Scholar] [CrossRef]
- Wang, Y.; Zong, B.; Yu, Y.; Wang, Y.; Tang, Z.; Chen, R.; Huang, M.; Liu, S. Ki67 Index Changes and Tumor-Infiltrating Lymphocyte Levels Impact the Prognosis of Triple-Negative Breast Cancer Patients with Residual Disease after Neoadjuvant Chemotherapy. Front. Oncol. 2021, 11, 668610. [Google Scholar] [CrossRef]
- Savas, P.; Salgado, R.; Loi, S. Seeing the forest and the tree: TILs and PD-L1 as immune biomarkers. Breast Cancer Res. Treat. 2021, 189, 599–606. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Pénault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef]
- Loi, S.; Drubay, D.; Adams, S.; Pruneri, G.; Francis, P.A.; Lacroix-Triki, M.; Joensuu, H.; Dieci, M.V.; Badve, S.; Demaria, S.; et al. Tumor-Infiltrating Lymphocytes and Prognosis: A Pooled Individual Patient Analysis of Early-Stage Triple-Negative Breast Cancers. J. Clin. Oncol. 2019, 37, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jonas, S.F.; Bataillon, G.; Criscitiello, C.; Salgado, R.; Loi, S.; Viale, G.; Lee, H.J.; Dieci, M.V.; Kim, S.B.; et al. Prognostic value of tumor-infiltrating lymphocytes in patients with early-stage triple-negative breast cancers (TNBC) who did not receive adjuvant chemotherapy. Ann. Oncol. 2019, 30, 1941–1949. [Google Scholar] [CrossRef] [PubMed]
- de Jong, V.M.; Wang, Y.; ter Hoeve, N.D.; Opdam, M.; Stathonikos, N.; Jóźwiak, K.; Hauptmann, M.; Cornelissen, S.; Vreuls, W.; Rosenberg, E.H.; et al. Prognostic Value of Stromal Tumor-Infiltrating Lymphocytes in Young, Node-Negative, Triple-Negative Breast Cancer Patients Who Did Not Receive (neo)Adjuvant Systemic Therapy. J. Clin. Oncol. 2022, 40, 2361–2374. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- Foldi, J.; Silber, A.; Reisenbichler, E.; Singh, K.; Fischbach, N.; Persico, J.; Adelson, K.; Katoch, A.; Horowitz, N.; Lannin, D.; et al. Neoadjuvant durvalumab plus weekly nab-paclitaxel and dose-dense doxorubicin/cyclophosphamide in triple-negative breast cancer. NPJ Breast Cancer 2021, 7, 9. [Google Scholar] [CrossRef]
- Lusho, S.; Durando, X.; Mouret-Reynier, M.A.; Kossai, M.; Lacrampe, N.; Molnar, I.; Penault-Llorca, F.; Radosevic-Robin, N.; Abrial, C. Platelet-to-Lymphocyte Ratio Is Associated with Favorable Response to Neoadjuvant Chemotherapy in Triple Negative Breast Cancer: A Study on 120 Patients. Front. Oncol. 2021, 11, 678315. [Google Scholar] [CrossRef]
- Ochi, T.; Bianchini, G.; Ando, M.; Nozaki, F.; Kobayashi, D.; Criscitiello, C.; Curigliano, G.; Iwamoto, T.; Niikura, N.; Takei, H.; et al. Predictive and prognostic value of stromal tumour-infiltrating lymphocytes before and after neoadjuvant therapy in triple negative and HER2-positive breast cancer. Eur. J. Cancer. 2019, 118, 41–48. [Google Scholar] [CrossRef]
- Rao, N.; Qiu, J.; Wu, J.; Zeng, H.; Su, F.; Qiu, K.; Wu, J.; Yao, H. Significance of Tumor-Infiltrating Lymphocytes and the Expression of Topoisomerase IIα in the Prediction of the Clinical Outcome of Patients with Triple-Negative Breast Cancer after Taxane-Anthracycline-Based Neoadjuvant Chemotherapy. Chemotherapy 2017, 62, 246–255. [Google Scholar] [CrossRef]
- Herrero-Vicent, C.; Guerrero, A.; Gavilá, J.; Gozalbo, F.; Hernandez, A.; Sandiego, S.; Algarra, M.A.; Calatrava, A.; Guillem-Porta, V.; Ruiz-Simón, A. Predictive and prognostic impact of tumour-infiltrating lymphocytes in triple-negative breast cancer treated with neoadjuvant chemotherapy. Ecancermedicalscience 2017, 11, 759. [Google Scholar] [CrossRef]
- Ono, M.; Tsuda, H.; Shimizu, C.; Yamamoto, S.; Shibata, T.; Yamamoto, H.; Hirata, T.; Yonemori, K.; Ando, M.; Tamura, K.; et al. Tumor-infiltrating lymphocytes are correlated with response to neoadjuvant chemotherapy in triple-negative breast cancer. Breast Cancer Res. Treat. 2012, 132, 793–805. [Google Scholar] [CrossRef]
- Castaneda, C.A.; Mittendorf, E.; Casavilca, S.; Wu, Y.; Castillo, M.; Arboleda, P.; Nunez, T.; Guerra, H.; Barrionuevo, C.; Dolores-Cerna, K.; et al. Tumor infiltrating lymphocytes in triple negative breast cancer receiving neoadjuvant chemotherapy. World J. Clin. Oncol. 2016, 7, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Cerbelli, B.; Botticelli, A.; Pisano, A.; Pernazza, A.; Campagna, D.; De Luca, A.; Ascierto, P.A.; Pignataro, M.G.; Pelullo, M.; Della Rocca, C.; et al. CD73 expression and pathologic response to neoadjuvant chemotherapy in triple negative breast cancer. Virchows Arch. 2020, 476, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, A.E.; Rashed, H.E.; Toam, M.; Omar, A.; Abdelhamid, M.I.; Matar, I. Clinicopathological significance of the immunologic signature (PDL1, FOXP3+ Tregs, TILs) in early stage triple-negative breast cancer treated with neoadjuvant chemotherapy. Ann. Diagn. Pathol. 2021, 51, 151676. [Google Scholar] [CrossRef] [PubMed]
- Würfel, F.; Erber, R.; Huebner, H.; Hein, A.; Lux, M.P.; Jud, S.; Kremer, A.; Kranich, H.; Mackensen, A.; Häberle, L.; et al. TILGen: A Program to Investigate Immune Targets in Breast Cancer Patients—First Results on the Influence of Tumor-Infiltrating Lymphocytes. Breast Care 2018, 13, 8–15. [Google Scholar] [CrossRef]
- Ruan, M.; Tian, T.; Rao, J.; Xu, X.; Yu, B.; Yang, W.; Shui, R. Predictive value of tumor-infiltrating lymphocytes to pathological complete response in neoadjuvant treated triple-negative breast cancers. Diagn. Pathol. 2018, 13, 66. [Google Scholar] [CrossRef]
- Khoury, T.; Aljabab, S.; Yao, S.; Ambrosone, C.; Omilian, A.; Attwood, K.; Ji, W.; Gandhi, S. Tumor-associated mononuclear cells in the tumor bed of triple-negative breast cancer associate with clinical outcomes in the post-neoadjuvant chemotherapy setting. Breast Cancer Res. Treat. 2022, 194, 531–540. [Google Scholar] [CrossRef]
- Goda, N.; Nakashima, C.; Nagamine, I.; Otagaki, S. The Effect of Intratumoral Interrelation among FOXP3+ Regulatory T Cells on Treatment Response and Survival in Triple-Negative Breast Cancer. Cancers 2022, 14, 2138. [Google Scholar] [CrossRef]
- Symmans, W.F.; Yau, C.; Chen, Y.Y.; Balassanian, R.; Klein, M.E.; Pusztai, L.; Nanda, R.; Parker, B.A.; Datnow, B.; Krings, G.; et al. Assessment of Residual Cancer Burden and Event-Free Survival in Neoadjuvant Treatment for High-risk Breast Cancer: An Analysis of Data From the I-SPY2 Randomized Clinical Trial. JAMA Oncol. 2021, 7, 1654–1663. [Google Scholar] [CrossRef]
- Yau, C.; Osdoit, M.; van der Noordaa, M.; Shad, S.; Wei, J.; de Croze, D.; Hamy, A.-S.; Laé, M.; Reyal, F.; Sonke, G.S.; et al. Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: A multicentre pooled analysis of 5161 patients. Lancet Oncol. 2022, 23, 149–160. [Google Scholar] [CrossRef]
- Hammerl, D.; Martens, J.W.; Timmermans, M.; Smid, M.; Trapman-Jansen, A.M.; Foekens, R.; Isaeva, O.I.; Voorwerk, L.; Balcioglu, H.E.; Wijers, R.; et al. Spatial immunophenotypes predict response to anti-PD1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat. Commun. 2021, 12, 5668. [Google Scholar] [CrossRef]
- El Bairi, K.; Haynes, H.R.; Blackley, E.; Fineberg, S.; Shear, J.; Turner, S.; de Freitas, J.R.; Sur, D.; Amendola, L.C.; Gharib, M.; et al. The tale of TILs in breast cancer: A report from The International Immuno-Oncology Biomarker Working Group. NPJ Breast Cancer 2021, 7, 150. [Google Scholar] [CrossRef]
- Arole, V.; Nitta, H.; Wei, L.; Shen, T.; Parwani, A.V.; Li, Z. M2 tumor-associated macrophages play important role in predicting response to neoadjuvant chemotherapy in triple-negative breast carcinoma. Breast Cancer Res. Treat. 2021, 188, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, T.; Xuan, Q.; Zhao, H.; Qin, L.; Zhang, Q. Lymphocyte-Activation Gene-3 Expression and Prognostic Value in Neoadjuvant-Treated Triple-Negative Breast Cancer. J. Breast Cancer 2018, 21, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Iwase, T.; Blenman, K.R.M.; Li, X.; Reisenbichler, E.; Seitz, R.; Hout, D.; Nielsen, T.J.; Schweitzer, B.L.; Bailey, D.B.; Shen, Y.; et al. A Novel Immunomodulatory 27-Gene Signature to Predict Response to Neoadjuvant Immunochemotherapy for Primary Triple-Negative Breast Cancer. Cancers 2021, 13, 4839. [Google Scholar] [CrossRef]
- Ueno, T.; Kitano, S.; Masuda, N.; Ikarashi, D.; Yamashita, M.; Chiba, T.; Kadoya, T.; Bando, H.; Yamanaka, T.; Ohtani, S.; et al. Immune microenvironment, homologous recombination deficiency, and therapeutic response to neoadjuvant chemotherapy in triple-negative breast cancer: Japan Breast Cancer Research Group (JBCRG)22 TR. BMC Med. 2022, 20, 136. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Spaapen, R.M.; Zha, Y.; Williams, J.; Meng, Y.; Ha, T.T.; Gajewski, T.F. Up-regulation of PD-L1, IDO and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci. Transl. Med. 2013, 5, 200ra116. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, J.; Chatterjee, M.; Ganguly, S.; Datta, A.; Biswas, B.; Mukherjee, G.; Agarwal, S.; Ahmed, R.; Chatterjee, S.; Dabkara, D. PDL1 expression and its correlation with outcomes in non-metastatic triple-negative breast cancer (TNBC). Ecancermedicalscience 2021, 15, 1217. [Google Scholar] [CrossRef]
- Yam, C.; Yen, E.-Y.; Chang, J.T.; Bassett, J.R.L.; Alatrash, G.; Garber, H.; Huo, L.; Yang, F.; Philips, A.V.; Ding, Q.-Q.; et al. Immune Phenotype and Response to Neoadjuvant Therapy in Triple-Negative Breast Cancer. Clin. Cancer Res. 2021, 27, 5365–5375. [Google Scholar] [CrossRef]
- Cerbelli, B.; Pernazza, A.; Botticelli, A.; Fortunato, L.; Monti, M.; Sciattella, P.; Campagna, D.; Mazzuca, F.; Mauri, M.; Naso, G.; et al. PD-L1 Expression in TNBC: A Predictive Biomarker of Response to Neoadjuvant Chemotherapy? BioMed Res. Int. 2017, 2017, 1750925. [Google Scholar] [CrossRef]
- Sabatier, R.; Finetti, P.; Mamessier, E.; Adelaide, J.; Chaffanet, M.; Ali, H.R.; Viens, P.; Caldas, C.; Birnbaum, D.; Bertucci, F. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget 2015, 6, 5449–5464. [Google Scholar] [CrossRef]
- Bianchini, G. Single-cell spatial analysis by imaging mass cytometry and immunotherapy response in triple-negative breast cancer (TNBC) in the NeoTRIPaPDL1 trial. In Proceedings of the SABCS 2021, San Antonio, TX, USA, 8–11 December 2021. [Google Scholar]
- Long, W.; Chen, J.; Gao, C.; Lin, Z.; Xie, X.; Dai, H. Brief review on the roles of neutrophils in cancer development. J. Leukoc. Biol. 2021, 109, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Tokumaru, Y.; Oshi, M.; Murthy, V.; Tian, W.; Yan, L.; Angarita, F.A.; Nagahashi, M.; Matsuhashi, N.; Futamura, M.; Yoshida, K.; et al. Low intratumoral genetic neutrophil-to-lymphocyte ratio (NLR) is associated with favorable tumor immune microenvironment and with survival in triple negative breast cancer (TNBC). Am. J. Cancer Res. 2021, 11, 5743–5755. [Google Scholar] [PubMed]
- Asano, Y.; Kashiwagi, S.; Onoda, N.; Noda, S.; Kawajiri, H.; Takashima, T.; Ohsawa, M.; Kitagawa, S.; Hirakawa, K. Predictive Value of Neutrophil/Lymphocyte Ratio for Efficacy of Preoperative Chemotherapy in Triple-Negative Breast Cancer. Ann. Surg. Oncol. 2016, 23, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Chae, S.; Kang, K.M.; Kim, H.J.; Kang, E.; Park, S.Y.; Kim, J.H.; Kim, S.H.; Kim, E.K. Neutrophil-lymphocyte ratio predicts response to chemotherapy in triple-negative breast cancer. Curr. Oncol. 2018, 25, e113–e119. [Google Scholar] [CrossRef]
- Ocana, A.; Chacon, J.I.; Calvo, L.; Anton, A.; Mansutti, M.; Albanell, J.; Martinez, M.T.; Lahuerta, A.; Bisagni, G.; Bermejo, B.; et al. Derived Neutrophil-to-Lymphocyte Ratio Predicts Pathological Complete Response to Neoadjuvant Chemotherapy in Breast Cancer. Front. Oncol. 2022, 11, 5834. [Google Scholar] [CrossRef]
- Lou, C.; Jin, F.; Zhao, Q.; Qi, H. Correlation of serum, N.L.R.; PLR and HALP with efficacy of neoadjuvant chemotherapy and prognosis of triple-negative breast cancer. Am. J. Transl. Res. 2022, 14, 3240–3246. [Google Scholar]
- Pang, J.; Zhou, H.; Dong, X.; Wang, S.; Xiao, Z. Relationship Between the Neutrophil to Lymphocyte Ratio, Stromal Tumor-infiltrating Lymphocytes, and the Prognosis and Response to Neoadjuvant Chemotherapy in Triple-negative Breast Cancer. Clin. Breast Cancer 2021, 21, e681–e687. [Google Scholar] [CrossRef]
- Onesti, C.E.; Josse, C.; Poncin, A.; Frères, P.; Poulet, C.; Bours, V.; Jerusalem, G. Predictive and prognostic role of peripheral blood eosinophil count in triple-negative and hormone receptor-negative/HER2-positive breast cancer patients undergoing neoadjuvant treatment. Oncotarget 2018, 9, 33719–33733. [Google Scholar] [CrossRef]
- Hu, H.; Zhu, J.; Zhong, Y.; Geng, R.; Ji, Y.; Guan, Q.; Hong, C.; Wei, Y.; Min, N.; Qi, A.; et al. PIK3CA mutation confers resistance to chemotherapy in triple-negative breast cancer by inhibiting apoptosis and activating the PI3K/AKT/mTOR signaling pathway. Ann. Transl. Med. 2021, 9, 410. [Google Scholar] [CrossRef]
- Loibl, S.; Treue, D.; Budczies, J.; Weber, K.; Stenzinger, A.; Schmitt, W.D.; Weichert, W.; Jank, P.; Furlanetto, J.; Klauschen, F.; et al. Mutational Diversity and Therapy Response in Breast Cancer: A Sequencing Analysis in the Neoadjuvant GeparSepto Trial. Clin. Cancer Res. 2019, 25, 3986–3995. [Google Scholar] [CrossRef]
- Atchley, D.P.; Albarracin, C.T.; Lopez, A.; Valero, V.; Amos, C.I.; Gonzalez-Angulo, A.M.; Hortobagyi, G.N.; Arun, B.K. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J. Clin. Oncol. 2008, 26, 4282–4288. [Google Scholar] [CrossRef]
- Huang, L.; Lang, G.T.; Liu, Q.; Shi, J.X.; Shao, Z.M.; Cao, A.Y. A predictor of pathological complete response to neoadjuvant chemotherapy in triple-negative breast cancer patients with the DNA repair genes. Ann. Transl. Med. 2021, 9, 301. [Google Scholar] [CrossRef]
- Telli, M.L.; Hellyer, J.; Audeh, W.; Jensen, K.C.; Bose, S.; Timms, K.M.; Gutin, A.; Abkevich, V.; Peterson, R.N.; Neff, C.; et al. Pathological complete response rate and survival in patients with BRCA-associated triple-negative breast cancer after 12 weeks of de-escalated neoadjuvant chemotherapy: Translational results of the WSG-ADAPT TN randomized phase II trial (NCT01815242). J. Clin. Oncol. 2021, 39 (Suppl. 15), 579. [Google Scholar]
- Richters, L.K.K.; Gluz, O.; Weber-Lassalle, N.; Christgen, M.; Haverkamp, H.; Kuemmel, S.; Kayali, M.; Kates, R.E.; Grischke, E.M.; Braun, M.; et al. Prevalence of BRCA1 mutations and responses to neoadjuvant chemotherapy among BRCA1 carriers and non-carriers with triple-negative breast cancer. Ann. Oncol. 2015, 26, 523–528. [Google Scholar]
- Telli, M.L.; Hellyer, J.; Audeh, W.; Jensen, K.C.; Bose, S.; Timms, K.M.; Gutin, A.; Abkevich, V.; Peterson, R.N.; Neff, C.; et al. Homologous recombination deficiency (HRD) status predicts response to standard neoadjuvant chemotherapy in patients with triple-negative or BRCA1/2 mutation-associated breast cancer. Breast Cancer Res. Treat. 2018, 168, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, J.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Fan, T.; Lin, B.; Xie, Y. Incidence of BRCA1 somatic mutations and response to neoadjuvant chemotherapy in Chinese women with triple-negative breast cancer. Gene 2016, 584, 26–30. [Google Scholar] [CrossRef]
- Watanabe, Y.; Maeda, I.; Oikawa, R.; Wu, W.; Tsuchiya, K.; Miyoshi, Y.; Itoh, F.; Tsugawa, K.-I.; Ohta, T. Aberrant DNA methylation status of DNA repair genes in breast cancer treated with neoadjuvant chemotherapy. Genes Cells 2013, 18, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Clifton, S.; Locke, W.; Luu, P.-L.; Du, Q.; Lam, D.; Armstrong, N.J.; Kumar, B.; Deng, N.; Harvey, K.; et al. Identification of DNA methylation biomarkers with potential to predict response to neoadjuvant chemotherapy in triple-negative breast cancer. Clin. Epigenet. 2021, 13, 226. [Google Scholar] [CrossRef]
- Akashi-Tanaka, S.; Watanabe, C.; Takamaru, T.; Kuwayama, T.; Ikeda, M.; Ohyama, H.; Mori, M.; Yoshida, R.; Hashimoto, R.; Terumasa, S.; et al. BRCAness predicts resistance to taxane-containing regimens in triple negative breast cancer during neoadjuvant chemotherapy. Clin. Breast Cancer 2015, 15, 80–85. [Google Scholar] [CrossRef]
- Nogi, H.; Uchida, K.; Kamio, M.; Kato, K.; Toriumi, Y.; Akiba, T.; Morikawa, T.; Suzuki, M.; Kobayashi, T.; Takeyama, H. Triple-negative breast cancer exhibits a favorable response to neoadjuvant chemotherapy independent of the expression of topoisomerase IIα. Mol. Clin. Oncol. 2016, 4, 383–389. [Google Scholar] [CrossRef]
- Wang, Q.; Li, C.; Tang, P.; Ji, R.; Chen, S.; Wen, J. A Minimal lncRNA-mRNA Signature Predicts Sensitivity to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Cell. Physiol. Biochem. 2018, 48, 2539–2548. [Google Scholar] [CrossRef]
- Liebermann, D.A. Gadd45 in Normal Hematopoiesis and Leukemia. Adv. Exp. Med. Biol. 2022, 1360, 41–54. [Google Scholar]
- Zheng, T.; Pang, Z.; Zhao, Z. A gene signature predicts response to neoadjuvant chemotherapy in triple-negative breast cancer patients. Biosci. Rep. 2019, 39, BSR20190414. [Google Scholar] [CrossRef]
- Hoeffler, J.P.; Meyer, T.E.; Yun, Y.; Jameson, J.L.; Habener, J.F. Cyclic AMP-Responsive DNA-Binding Protein: Structure Based on a Cloned Placental cDNA. Science 1988, 242, 1430–1433. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Yi, G. Development of Gene Expression-Based Random Forest Model for Predicting Neoadjuvant Chemotherapy Response in Triple-Negative Breast Cancer. Cancers 2022, 14, 881. [Google Scholar] [CrossRef]
- Darb-Esfahani, S.; Kronenwett, R.; von Minckwitz, G.; Denkert, C.; Gehrmann, M.; Rody, A.; Budczies, J.; Brase, J.C.; Mehta, M.K.; Bojar, H.; et al. Thymosin beta 15A (TMSB15A) is a predictor of chemotherapy response in triple-negative breast cancer. Br. J. Cancer 2012, 107, 1892–1900. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Che, X.; Wu, Y.; Song, N.; Shi, S.; Wang, S.; Li, C.; Zhang, L.; Zhang, X.; Qu, X.; et al. SIRT5 as a biomarker for response to anthracycline-taxane-based neoadjuvant chemotherapy in triple-negative breast cancer. Oncol. Rep. 2018, 39, 2315–2323. [Google Scholar] [CrossRef]
- Lindner, J.; Loibl, S.; Denkert, C.; Ataseven, B.; Fasching, P.; Pfitzner, B.; Gerber, B.; Gade, S.; Darb-Esfahani, S.; Sinn, B.; et al. Expression of secreted protein acidic and rich in cysteine (SPARC) in breast cancer and response to neoadjuvant chemotherapy. Ann. Oncol. 2015, 26, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Orozco, J.I.J.; Grumley, J.G.; Matsuba, C.; Manughian-Peter, A.O.; Chang, S.-C.; Chang, G.; Gago, F.E.; Salomon, M.P.; Marzese, D.M. Clinical Implications of Transcriptomic Changes after Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Ann. Surg. Oncol. 2019, 26, 3185–3193. [Google Scholar] [CrossRef]
- Oshi, M.; Gandhi, S.; Huyser, M.R.; Tokumaru, Y.; Yan, L.; Yamada, A.; Matsuyama, R.; Endo, I.; Takabe, K. MELK expression in breast cancer is associated with infiltration of immune cell and pathological compete response (pCR) after neoadjuvant chemotherapy. Am. J. Cancer Res. 2021, 11, 4421–4437. [Google Scholar] [CrossRef]
- Echavarria Diaz-Guardamino, I.; Lopez-Tarruella Cobo, S.; Del Monte-Millan, M.; Alvarez, E.; Jerez, Y.; Moreno Anton, F.; Lopez-Tarruella Cobo, J.Á.; Massarrah, T.; Ocaña, I.; Cebollero, M.; et al. 141MO—Pathological response and early survival data according to TNBCtype4 classifier in operable triple-negative breast cancer (TNBC) treated with neoadjuvant carboplatin and docetaxel. In Proceedings of the ESMO Congress, Paris, France, 9–13 September 2022. [Google Scholar]
- Pérez-Pena, J.; Tibor Fekete, J.; Páez, R.; Baliu-Piqué, M.; García-Saenz, J.Á.; García-Barberán, V.; Manzano, A.; Pérez-Segura, P.; Esparis-Ogando, A.; Pandiella, A.; et al. A Transcriptomic Immunologic Signature Predicts Favorable Outcome in Neoadjuvant Chemotherapy Treated Triple Negative Breast Tumors. Front. Immunol. 2019, 10, 2802. [Google Scholar] [CrossRef]
- Sha, L.-Y.; Zhang, Y.; Wang, W.; Sui, X.; Liu, S.-K.; Wang, T.; Zhang, H. MiR-18a upregulation decreases Dicer expression and confers paclitaxel resistance in triple negative breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2201–2208. [Google Scholar] [PubMed]
- García-García, F.; Salinas-Vera, Y.M.; García-Vázquez, R.; Marchat, L.A.; Rodríguez-Cuevas, S.; López-González, J.S.; Carlos-Reyes, Á.; Ramos-Payán, R.; Aguilar-Medina, M.; Pérez-Plasencia, C.; et al. miR-145–5p is associated with pathological complete response to neoadjuvant chemotherapy and impairs cell proliferation by targeting TGFβR2 in breast cancer. Oncol. Rep. 2019, 41, 3527–3534. [Google Scholar] [PubMed]
- Kolacinska, A.; Morawiec, J.; Fendler, W.; Malachowska, B.; Morawiec, Z.; Szemraj, J.; Pawlowska, Z.; Chowdhury, D.; Choi, Y.E.; Kubiak, R.; et al. Association of microRNAs and pathologic response to preoperative chemotherapy in triple negative breast cancer: Preliminary report. Mol. Biol. Rep. 2014, 41, 2851–2857. [Google Scholar] [CrossRef]
- Sueta, A.; Fujiki, Y.; Goto-Yamaguchi, L.; Tomiguchi, M.; Yamamoto-Ibusuki, M.; Iwase, H.; Yamamoto, Y. Exosomal miRNA profiles of triple-negative breast cancer in neoadjuvant treatment. Oncol. Lett. 2021, 22, 819. [Google Scholar] [CrossRef] [PubMed]
- Graeser, M.; Feuerhake, F.; Gluz, O.; Volk, V.; Hauptmann, M.; Jozwiak, K.; Christgen, M.; Kuemmel, S.; Grischke, E.M.; Forstbauer, H.; et al. Immune cell composition and functional marker dynamics from multiplexed immunohistochemistry to predict response to neoadjuvant chemotherapy in the WSG-ADAPT-TN trial. J. Immunother. Cancer. 2021, 9, e002198. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A. Chemotherapy and tumor immunity: An unexpected collaboration. Front. Biosci. 2008, 13, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Voorwerk, L.; Slagter, M.; Horlings, H.M.; Sikorska, K.; van de Vijver, K.K.; de Maaker, M.; Nederlof, I.; Kluin, R.J.; Warren, S.; Ong, S.; et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: The TONIC trial. Nat. Med. 2019, 25, 920–928. [Google Scholar] [CrossRef]
- Buisseret, L.; Pommey, S.; Allard, B.; Garaud, S.; Bergeron, M.; Cousineau, I.; Ameye, L.; Bareche, Y.; Paesmans, M.; Crown, J.P.A.; et al. Clinical significance of CD73 in triple-negative breast cancer: Multiplex analysis of a phase III clinical trial. Ann. Oncol. 2018, 29, 1056–1062. [Google Scholar] [CrossRef]
- Loi, S.; Pommey, S.; Haibe-Kains, B.; Beavis, P.A.; Darcy, P.K.; Smyth, M.J.; Stagg, J. CD73 promotes anthracycline resistance and poor prognosis in triple negative breast cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 11091–11096. [Google Scholar] [CrossRef]
- Jinushi, M.; Chiba, S.; Yoshiyama, H.; Masutomi, K.; Kinoshita, I.; Dosaka-Akita, H.; Yagita, H.; Takaoka, A.; Tahara, H. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc. Natl. Acad. Sci. USA 2011, 108, 12425–12430. [Google Scholar] [CrossRef]
- Von Minckwitz, G.; Untch, M.; Blohmer, J.U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and Impact of Pathologic Complete Response on Prognosis After Neoadjuvant Chemotherapy in Various Intrinsic Breast Cancer Subtypes. J. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine Committee on Qualification of Biomarkers and Surrogate Endpoints in Chronic Disease. References. In Evaluation of Biomarkers and Surrogate Endpoints in Chronic Disease; Micheel, C.M., Ball, J.R., Eds.; National Academies Press: Washington, DC, USA, 2010; ISBN 978-0-309-15130-6. [Google Scholar]
- Simon, R. Sensitivity, Specificity, PPV and NPV for Predictive Biomarkers. J. Natl. Cancer Inst. 2015, 107, djv153. [Google Scholar] [CrossRef]
- Brower, V. Biomarkers: Portents of malignancy. Nature 2011, 471, S19–S20. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yang, Y.; Wei, Y.; He, P.; Chen, J.; Zheng, Z.; Bu, H. Deep learning-based predictive biomarker of pathological complete response to neoadjuvant chemotherapy from histological images in breast cancer. J. Transl. Med. 2021, 19, 348. [Google Scholar] [CrossRef] [PubMed]
| Author | Year of Publication | Study Size | Cut-Off Value High vs. Low | p-Value of the Association between Ki-67 and pCR | General Result |
|---|---|---|---|---|---|
| Masuda et al. [25] | 2011 | N = 33 | 50% | p = 0.03 | The percentage of patients with a pCR who had high Ki-67 expression was 50%, whereas 15% had low Ki-67 expression |
| Sakuma et al. [43] | 2011 | N = 44 | 50% | p > 0.9999 | The percentage of patients with a high Ki-67 expression who had a pCR was 39%, compared to 36% of patients with a low Ki-67 expression |
| Kraus et al. [41] | 2012 | N = 56 | 25% | p = 0.542 | Patients with a pCR had a mean Ki-67 expression of 61% (range 35–90%), compared to 58% (range 5–95%) in patients without a pCR |
| Kawate et al. [42] | 2013 | N = 205 | 14% | p = 0.1011 | The percentage of patients with a high Ki-67 expression who had a pCR was 28%, compared to 16% of the patients with a low Ki-67 expression |
| Miyashita et al. [37] | 2014 | N = 110 | 57.5% | p = 0.002 | High Ki-67 expression was associated with a pCR rate of 70%, while low Ki-67 expression was associated with a pCR rate of 5% |
| Kim et al. [19] | 2015 | N = 198 | 10% | p = 0.056 | High Ki-67 expression was associated with a pCR rate of 19.5%, while low Ki-67 expression was associated with a pCR rate of 6% |
| Baba et al. [40] | 2016 | N = 34 | 35% | p = 0.66 | In patients with a pCR, the mean Ki-67 expression was 55% ± 30 standard deviation. In patients without a pCR, the mean Ki-67 expression was 80% ± 12 standard deviation for partial response, 73% ± 8 standard deviation for stable disease and 55% ± 41 standard deviation for progressive disease |
| Garcia-Vazquez et al. [36] | 2017 | N = 18 | Mean expression of Ki-67 | 64% expression of Ki-67 in pCR vs. 51% expression of Ki-67 in non-pCR | In patients with a pCR, the median Ki-67 expression was 71% (range 30–90), compared to 62% in the non-pCR group (range 15–90) |
| Bignon et al. [24] | 2018 | N = 53 | Mean pCR vs. non-pCR | p = 0.48 | The mean Ki-67 expression in the pCR group was 68%, compared to 64% in the non-pCR group |
| Guestini et al. [20] | 2019 | N = 148 | 53% | p = 0.023 | Patients with a pCR had higher expression levels of Ki-67 |
| Gluz et al. [38] | 2020 | N = 336 | Not mentioned | p < 0.001 | n/a |
| Kong et al. [32] | 2020 | N = 280 | 20% | p = 0.451 | n/a |
| Van Bockstal et al. [26] | 2020 | N = 35 | 20% | p = 1.000 | In the pCR group, no patients had a low Ki-67 expression. In the non-pCR group, 4% had low Ki-67 expression, while 96% had high Ki-67 expression |
| Zuo et al. [39] | 2022 | N = 127 | 40% | p = 0.028 | n/a |
| Author | Year of Publication | Study Size | Cut-Off Value High vs. Low | Location of TILs: Stromal (s) or Tumoral | p-Value of the Association between pCR and Density of TILs | General Result |
|---|---|---|---|---|---|---|
| Ono et al. [81] | 2012 | N = 102 | 50% | sTILs | p = 0.05 | TIL-high patients had a pCR rate of 37%, compared to 16% in TIL-low patients |
| Abdel-Fatah et al. [58] | 2016 | N = 110 | 60% | sTILs and intratumoral TILs | p = 0.005 | TIL-high patients had a pCR rate of 53%, compared to 15% in TIL-low patients |
| Castaneda et al. [82] | 2016 | N = 98 | Continuous scale (per 10% increment) | sTILs | p = 0.0251 | A higher median TIL percentage of 40% ± 17.5 interquartile deviation was associated with a pCR, compared to a median of 30% ± 20 interquartile deviation TIL percentage in the non-pCR patients. When a cut-off of 50% was used for TIL-high vs. TIL-low, no significant association was found for pCR rate (p = 0.16) |
| Rao et al. [79] | 2017 | N = 52 | 35% (CD4) and 15% (CD8) | sTILs | p = 0.004 (CD4+ TILs) and p = 0.006 (CD8+ TILs) | When CD4+ TILs were high, 41.9% achieved a pCR, compared to 4.8% in cases with low CD4+ TILs. When CD8+ TIL levels were high, 47.6% achieved a pCR, compared to 12.9% of cases with low CD8+ TIL levels. Patients with both high CD4+ and CD8+ TIL levels had a pCR rate of 71.4% |
| Herrero-Vicent et al. [80] | 2017 | N = 164 | 40% | sTILs | p = 0.001 | TIL-high cases had a pCR rate of 88%, compared to 9% of the TIL-low cases |
| Würfel et al. [85] | 2018 | N = 146 | 50% | sTILs | p < 0.01 | TIL-high cases had a pCR rate of 67%, compared to 33% of the TIL-low cases |
| Ruan et al. [86] | 2018 | N = 166 | 20% for sTIL and 10% for intratumoral TILs | sTILs and intratumoral TILs | p = 0.006 (stromal) and p = 0.04 (intratumoral) | n/a |
| Zhang et al. [34] | 2018 | N = 58 | 60% | sTILs | p = 0.01 | The percentage of the pCR cases that were TIL-high was 46%, compared to 16% in the non-pCR group |
| Ochi et al. [78] | 2019 | N = 80 | 9% | sTILs | p < 0.001 | TIL-high cases had a pCR rate of 44%, compared to 4% of TIL-low cases |
| Van Bockstal et al. [26] | 2020 | N = 35 | 40% | sTILs | p = 0.002 (2-tier) p = 0.013 (continuous percentage) | The percentage of the pCR cases that were TIL-high was 62%, compared to 9% in the non-pCR group |
| Cerbelli et al. [83] | 2020 | N = 75 | Low: ≤9% Intermediate: ≥10–49% High: ≥50% | sTILs | p = 0.037 | TIL-high cases had a pCR rate of 76.5%, compared to 16% in the TIL-intermediate cases and 42% in the TIL-low cases |
| Foldi et al. [76] | 2021 | N = 69 | Low: ≤9% Intermediate: ≥10–29% High: ≥30% | sTILs | p = 0.0167 | TIL-high cases has a pCR rate of 57%, compared to 60% in the TIL-intermediate cases and 29% in the TIL-low cases |
| Abdelrahman et al. [84] | 2021 | N = 50 | 50% | sTILs | p < 0.02 | TIL-high cases had a pCR rate of 71%, compared to 28% of TIL-low cases |
| Ademuyiwa et al. [54] | 2021 | N = 127 | Continuous scale (per 10% increment) | sTILs | p = 0.05 | n/a |
| Lusho et al. [77] | 2021 | N = 120 | 30% | Not mentioned | p = 0.007 | TIL-high cases had a pCR rate of 54%, compared to 24% in TIL-low cases |
| Yuan et al. [64] | 2021 | N = 433 | 20% | sTILs | p = 0.014 | n/a |
| Goda et al. [88] | 2022 | N = 20 | 50% | sTILs | p = 0.002 | The percentage of the pCR cases that were TIL-high was 67%, and 18% of the non-pCR cases were TIL-high |
| Khoury et al. [87] | 2022 | N = 129 | Mean pCR vs. non-pCR | sTILs | p = 0.0003 | n/a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van den Ende, N.S.; Nguyen, A.H.; Jager, A.; Kok, M.; Debets, R.; van Deurzen, C.H.M. Triple-Negative Breast Cancer and Predictive Markers of Response to Neoadjuvant Chemotherapy: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 2969. https://doi.org/10.3390/ijms24032969
van den Ende NS, Nguyen AH, Jager A, Kok M, Debets R, van Deurzen CHM. Triple-Negative Breast Cancer and Predictive Markers of Response to Neoadjuvant Chemotherapy: A Systematic Review. International Journal of Molecular Sciences. 2023; 24(3):2969. https://doi.org/10.3390/ijms24032969
Chicago/Turabian Stylevan den Ende, Nadine S., Anh H. Nguyen, Agnes Jager, Marleen Kok, Reno Debets, and Carolien H. M. van Deurzen. 2023. "Triple-Negative Breast Cancer and Predictive Markers of Response to Neoadjuvant Chemotherapy: A Systematic Review" International Journal of Molecular Sciences 24, no. 3: 2969. https://doi.org/10.3390/ijms24032969
APA Stylevan den Ende, N. S., Nguyen, A. H., Jager, A., Kok, M., Debets, R., & van Deurzen, C. H. M. (2023). Triple-Negative Breast Cancer and Predictive Markers of Response to Neoadjuvant Chemotherapy: A Systematic Review. International Journal of Molecular Sciences, 24(3), 2969. https://doi.org/10.3390/ijms24032969








