Unveiling New Druggable Pockets in Influenza Non-Structural Protein 1: NS1–Host Interactions as Antiviral Targets for Flu
Abstract
1. Introduction
2. Results and Discussion
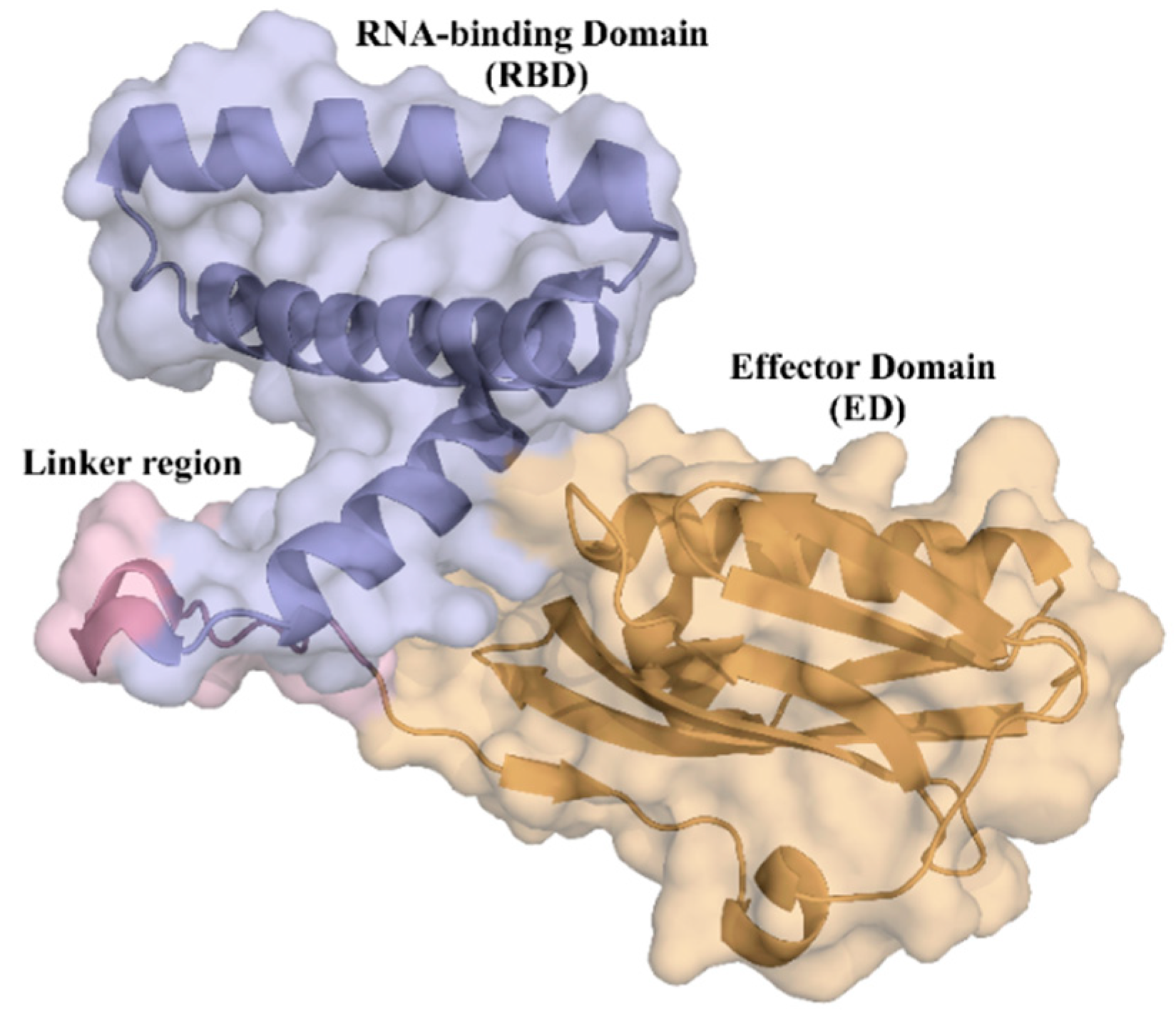
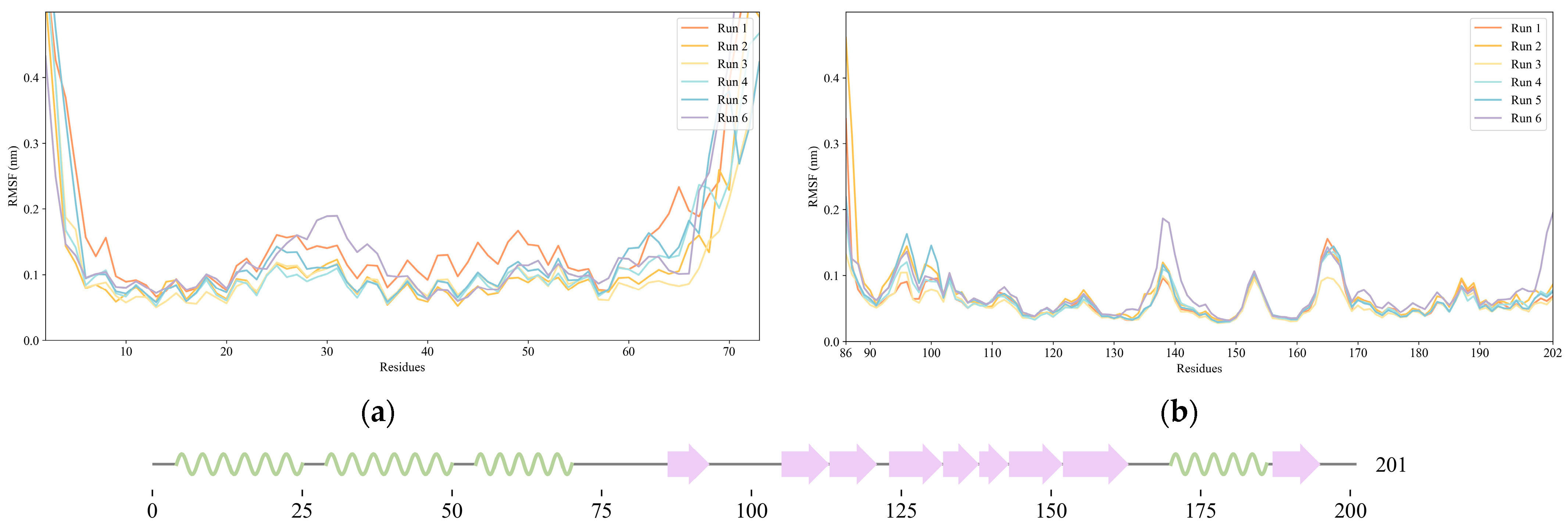
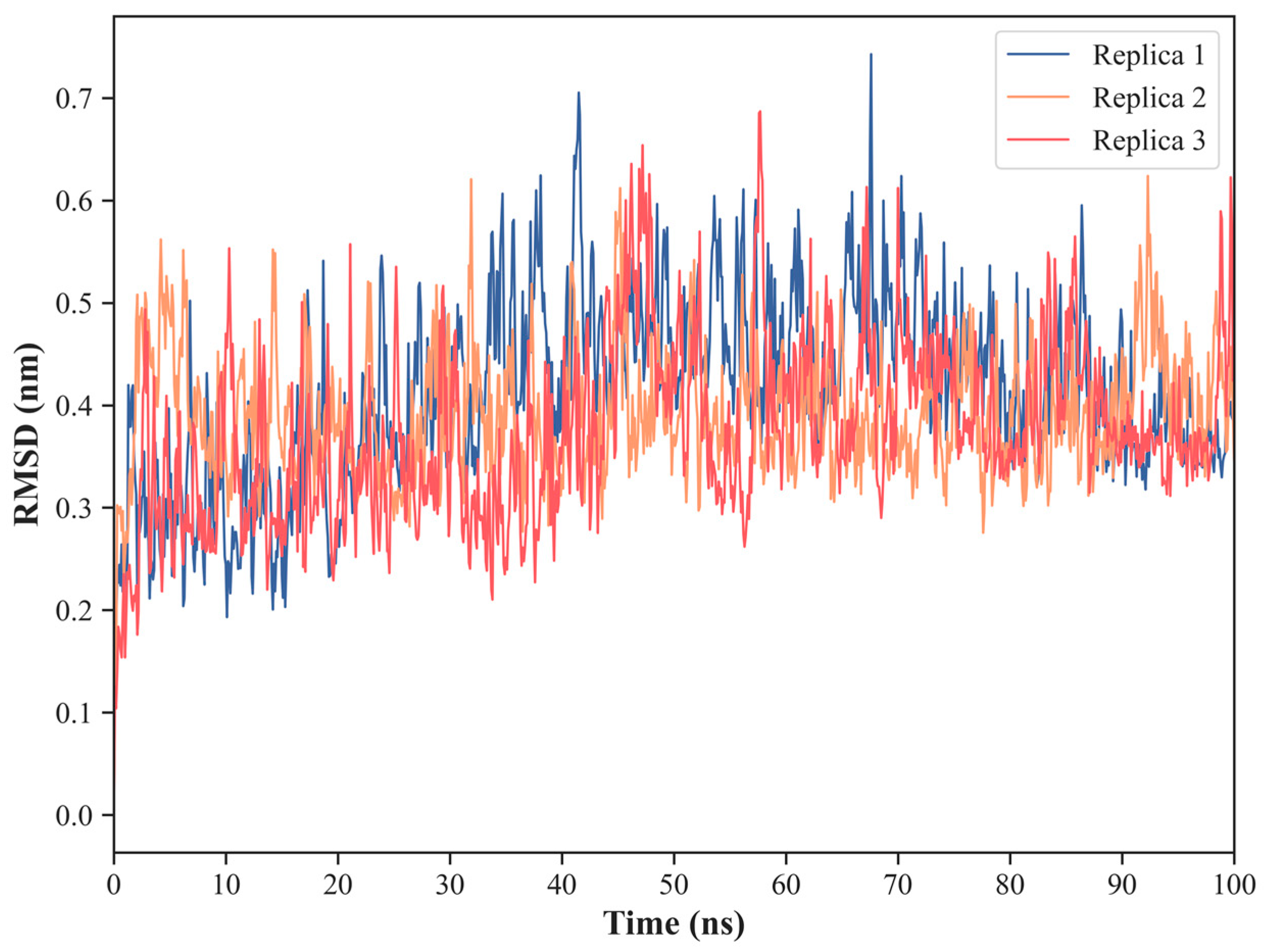
2.1. NS1 Structure’s Dynamics
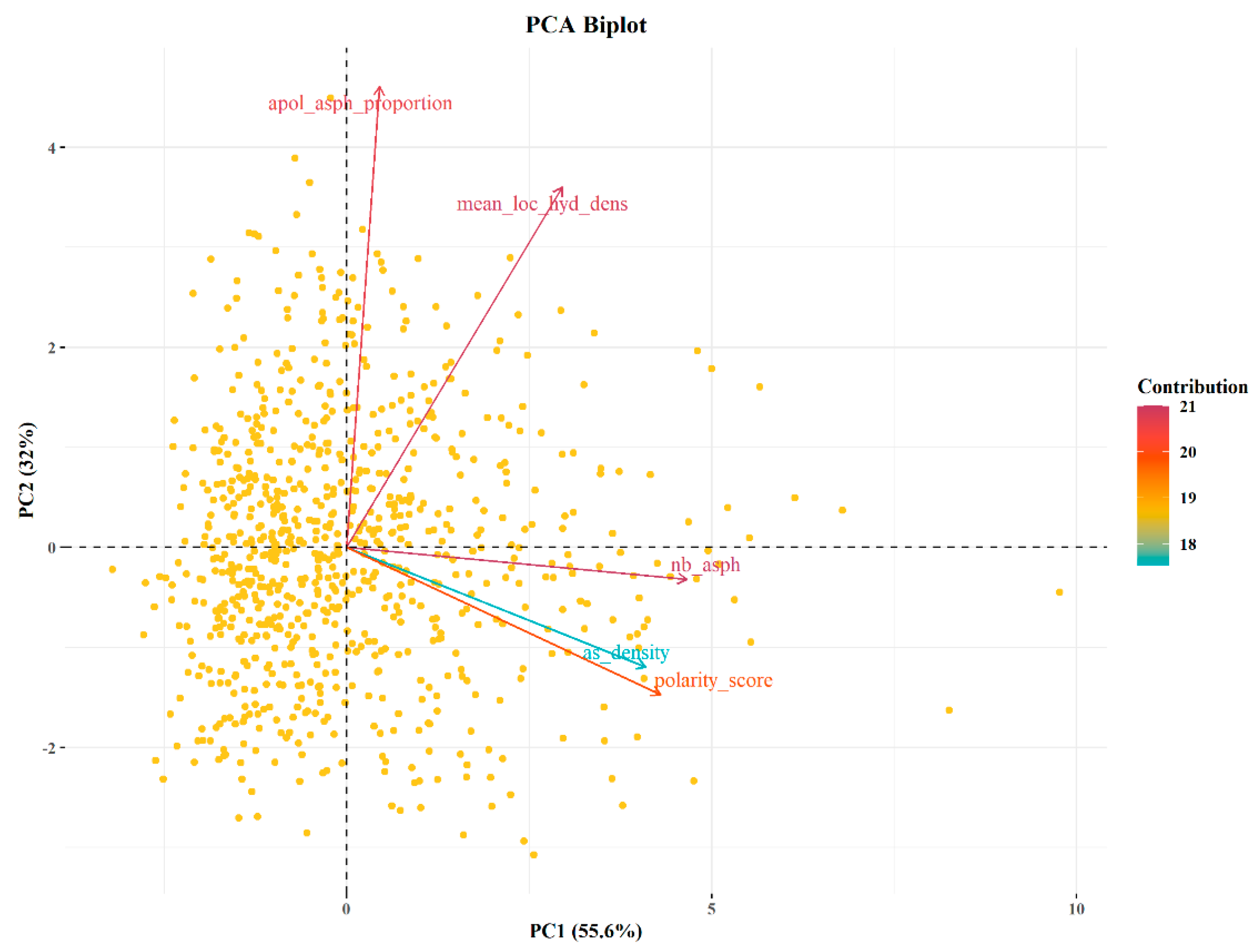
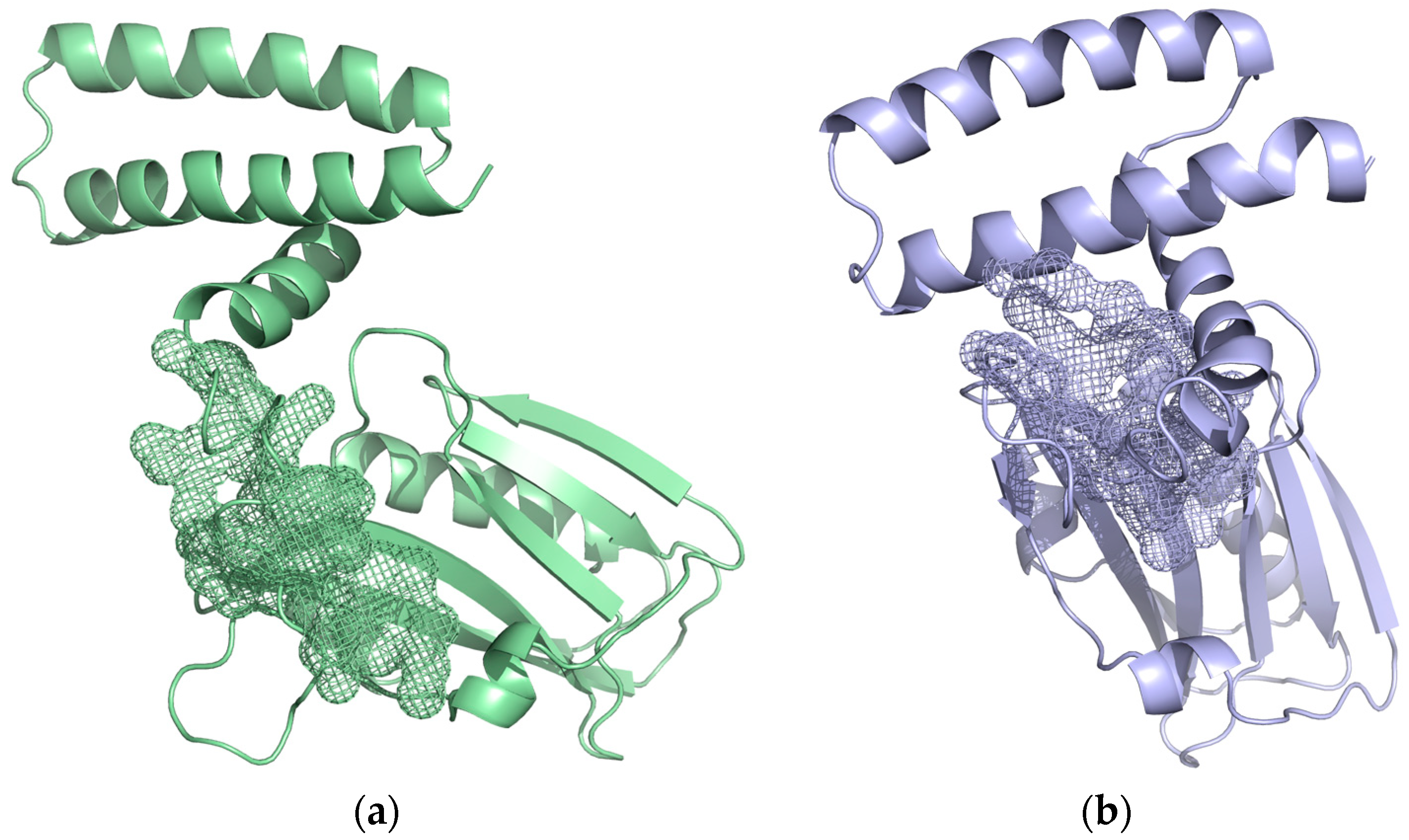
2.2. Pocket Identification and Analysis
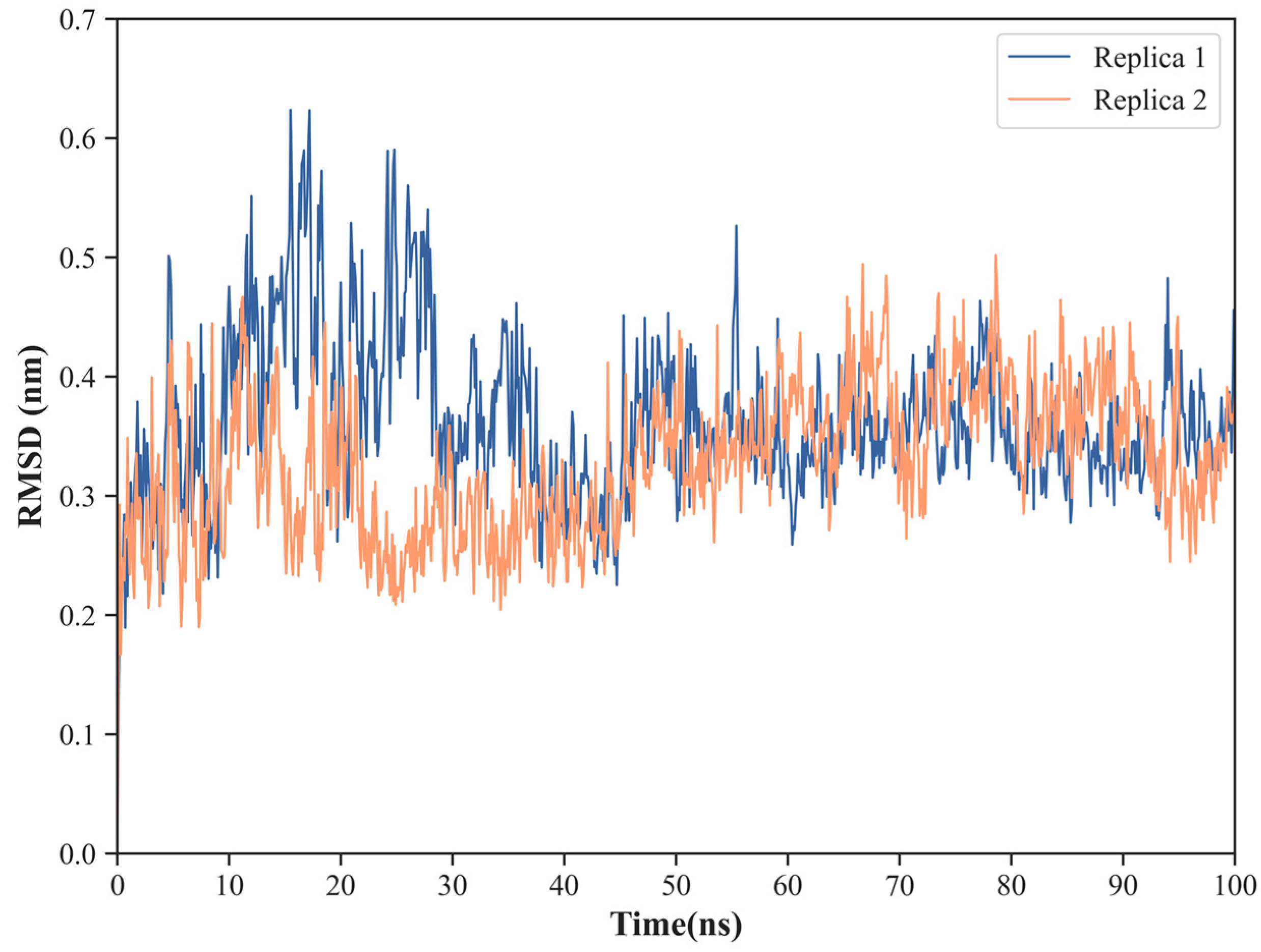
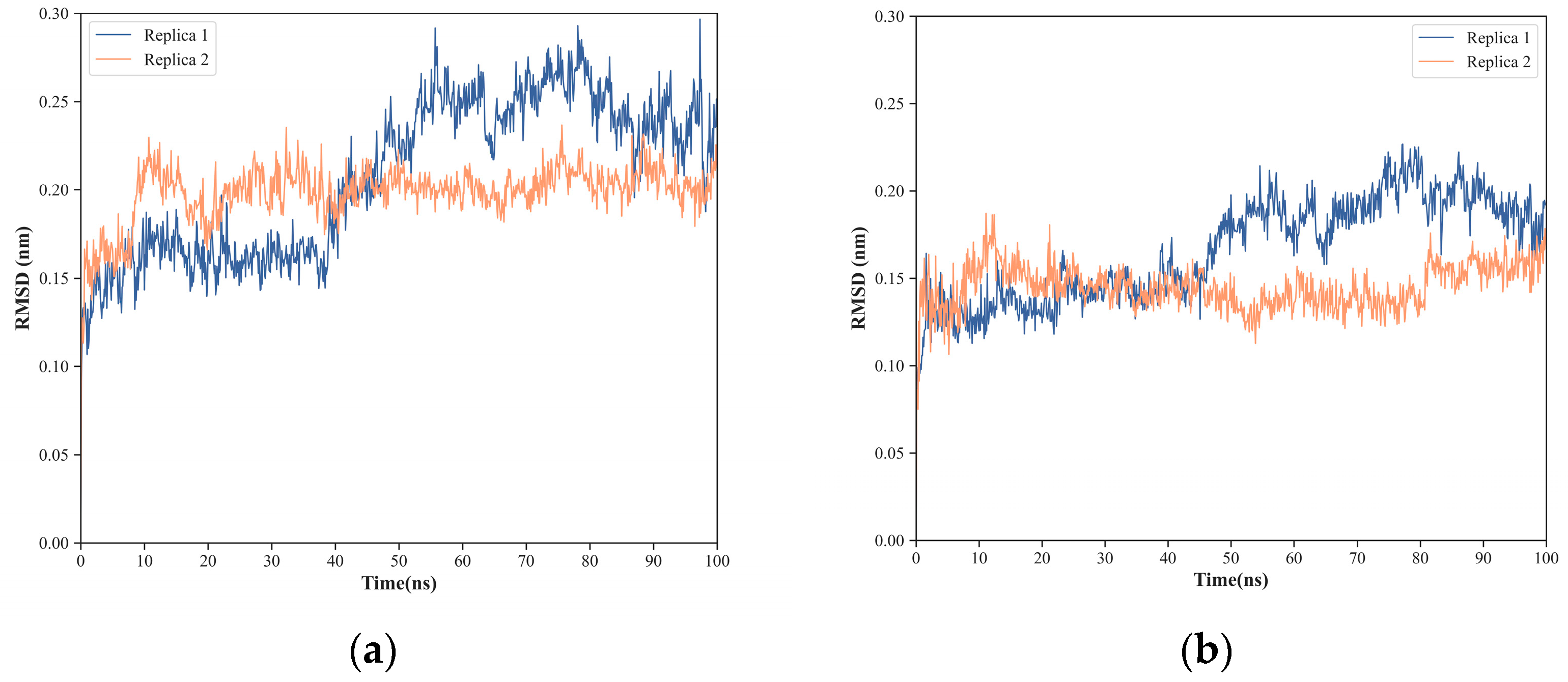
2.3. Characterization of Interactions between Influenza NS1 and Human PI3K or TRIM25
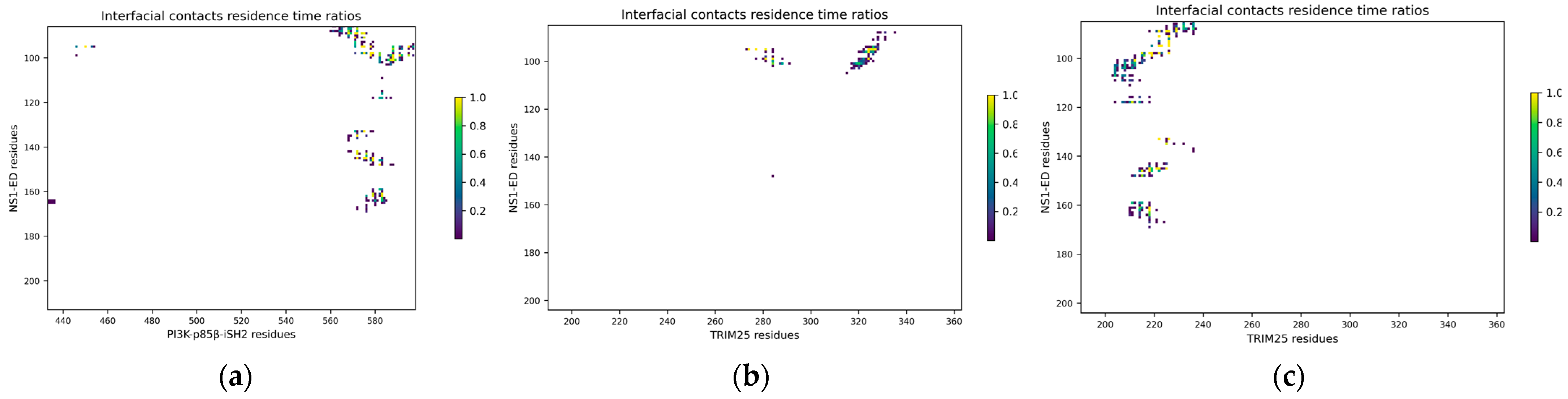
2.4. Binding of NS1 to PI3K and TRIM25 Is Mostly Driven by the Same Six Hotspot Residues
3. Materials and Methods
3.1. NS1 Three-Dimensional Protein Structures
3.2. Molecular Dynamics (MD) Protocol and Analysis
3.3. Molecular Dynamics Simulations of the NS1-PI3K and NS1-TRIM25 Complexes
3.4. Druggable Pockets Identification and Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of Global Seasonal Influenza-Associated Respiratory Mortality: A Modelling Study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef]
- World Health Organization Global Influenza Strategy 2019–2030; Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in Publication (CIP) Data; World Health Organization: Geneve, Switzerland, 2019.
- Macias, A.E.; McElhaney, J.E.; Chaves, S.S.; Nealon, J.; Nunes, M.C.; Samson, S.I.; Seet, B.T.; Weinke, T.; Yu, H. The Disease Burden of Influenza beyond Respiratory Illness. Vaccine 2021, 39, A6–A14. [Google Scholar] [CrossRef]
- Putri, W.C.W.S.; Muscatello, D.J.; Stockwell, M.S.; Newall, A.T. Economic Burden of Seasonal Influenza in the United States. Vaccine 2018, 36, 3960–3966. [Google Scholar] [CrossRef] [PubMed]
- Preaud, E.; Durand, L.; Macabeo, B.; Farkas, N.; Sloesen, B.; Palache, A.; Shupo, F.; Samson, S.I. Annual Public Health and Economic Benefits of Seasonal Influenza Vaccination: A European Estimate. BMC Public Health 2014, 14, 813. [Google Scholar] [CrossRef] [PubMed]
- Influenza (Seasonal). Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 8 March 2022).
- Bisen, P.S.; Raghuvanshi, R. Emerging Epidemics: Management and Control, 1st ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2013; p. 422. [Google Scholar]
- Fauci, A.S. Harrison’s Principles of Internal Medicine, 17th ed.; McGraw-Hill Medical: New York, NY, USA, 2008; ISBN 978-0-07-159991-7/978-0-07-147692-8/978-0-07-146633-2/978-0-07-147691-1. [Google Scholar]
- CDC Seasonal Flu vs. Pandemic Flu Infographic. Centers for Disease Control and Prevention. 2018. Available online: https://www.cdc.gov/flu/resource-center/freeresources/graphics/seasonal-vs-pandemic-flu-infographic.htm (accessed on 11 October 2021).
- Rosário-Ferreira, N.; Preto, A.J.; Melo, R.; Moreira, I.S.; Brito, R.M.M. The Central Role of Non-Structural Protein 1 (NS1) in Influenza Biology and Infection. Int. J. Mol. Sci. 2020, 21, 1511. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, R.; Amicizia, D.; Lai, P.L.; Bragazzi, N.L.; Panatto, D. Compounds with Anti-Influenza Activity: Present and Future of Strategies for the Optimal Treatment and Management of Influenza. Part I: Influenza Life-Cycle and Currently Available Drugs. J. Prev. Med. Hyg. 2014, 55, 69–85. [Google Scholar]
- Carrat, F.; Flahault, A. Influenza Vaccine: The Challenge of Antigenic Drift. Vaccine 2007, 25, 6852–6862. [Google Scholar] [CrossRef] [PubMed]
- Paules, C.; Subbarao, K. Influenza. Lancet 2017, 390, 697–708. [Google Scholar] [CrossRef]
- Mubareka, S.; Palese, P. Influenza Virus. In Influenza Vaccines for the Future; Springer Basel: Basel, Switzerland, 2011; pp. 3–26. ISBN 978-3-0346-0278-5, 978-3-0346-0279-2. [Google Scholar]
- Hussain, M.; Galvin, H.; Haw, T.Y.; Nutsford, A.; Husain, M. Drug Resistance in Influenza A Virus:The Epidemiology and Management. Infect. Drug Resist. 2017, 10, 121–134. [Google Scholar] [CrossRef]
- Engel, D.A. The Influenza Virus NS1 Protein as a Therapeutic Target. Antivir. Res. 2013, 99, 409–416. [Google Scholar] [CrossRef]
- García-Sastre, A.; Egorov, A.; Matassov, D.; Brandt, S.; Levy, D.E.; Durbin, J.E.; Palese, P.; Muster, T. Influenza A Virus Lacking the NS1 Gene Replicates in Interferon-Deficient Systems. Virology 1998, 252, 324–330. [Google Scholar] [CrossRef]
- Talon, J.; Salvatore, M.; O’Neill, R.E.; Nakaya, Y.; Zheng, H.; Muster, T.; García-Sastre, A.; Palese, P. Influenza A and B Viruses Expressing Altered NS1 Proteins: A Vaccine Approach. Proc. Natl. Acad. Sci. USA 2000, 97, 4309–4314. [Google Scholar] [CrossRef] [PubMed]
- Koliopoulos, M.G.; Lethier, M.; van der Veen, A.G.; Haubrich, K.; Hennig, J.; Kowalinski, E.; Stevens, R.V.; Martin, S.R.; Reis e Sousa, C.; Cusack, S.; et al. Molecular Mechanism of Influenza A NS1-Mediated TRIM25 Recognition and Inhibition. Nat. Commun. 2018, 9, 1820. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-H.; Zhao, B.; Shi, J.; Savage, N.; Shen, Q.; Byrnes, J.; Yang, L.; Hwang, W.; Li, P. Molecular Recognition of a Host Protein by NS1 of Pandemic and Seasonal Influenza A Viruses. Proc. Natl. Acad. Sci. USA 2020, 117, 6550–6558. [Google Scholar] [CrossRef]
- Chang, F.; Lee, J.T.; Navolanic, P.M.; Steelman, L.S.; Shelton, J.G.; Blalock, W.L.; Franklin, R.A.; McCubrey, J.A. Involvement of PI3K/Akt Pathway in Cell Cycle Progression, Apoptosis, and Neoplastic Transformation: A Target for Cancer Chemotherapy. Leukemia 2003, 17, 590–603. [Google Scholar] [CrossRef]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the Phosphoinositide 3-Kinase Pathway in Cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Zhirnov, O.P.; Klenk, H.-D. Control of Apoptosis in Influenza Virus-Infected Cells by up-Regulation of Akt and P53 Signaling. Apoptosis 2007, 12, 1419–1432. [Google Scholar] [CrossRef]
- Shin, Y.-K.; Li, Y.; Liu, Q.; Anderson, D.H.; Babiuk, L.A.; Zhou, Y. SH3 Binding Motif 1 in Influenza A Virus NS1 Protein Is Essential for PI3K/Akt Signaling Pathway Activation. J. Virol. 2007, 81, 12730–12739. [Google Scholar] [CrossRef]
- Ehrhardt, C.; Wolff, T.; Pleschka, S.; Planz, O.; Beermann, W.; Bode, J.G.; Schmolke, M.; Ludwig, S. Influenza A Virus NS1 Protein Activates the PI3K/Akt Pathway To Mediate Antiapoptotic Signaling Responses. J. Virol. 2007, 81, 3058–3067. [Google Scholar] [CrossRef]
- Ayllon, J.; García-Sastre, A.; Hale, B.G. Influenza A Viruses and PI3K: Are There Time, Place and Manner Restrictions? Virulence 2012, 3, 411–414. [Google Scholar] [CrossRef]
- Gallacher, M.; Brown, S.G.; Hale, B.G.; Fearns, R.; Olver, R.E.; Randall, R.E.; Wilson, S.M. Cation Currents in Human Airway Epithelial Cells Induced by Infection with Influenza A Virus. J. Physiol. 2009, 587, 3159–3173. [Google Scholar] [CrossRef]
- Versteeg, G.A.; Rajsbaum, R.; Sánchez-Aparicio, M.T.; Maestre, A.M.; Valdiviezo, J.; Shi, M.; Inn, K.-S.; Fernandez-Sesma, A.; Jung, J.; García-Sastre, A. The E3-Ligase TRIM Family of Proteins Regulates Signaling Pathways Triggered by Innate Immune Pattern-Recognition Receptors. Immunity 2013, 38, 384–398. [Google Scholar] [CrossRef] [PubMed]
- Martín-Vicente, M.; Medrano, L.M.; Resino, S.; García-Sastre, A.; Martínez, I. TRIM25 in the Regulation of the Antiviral Innate Immunity. Front. Immunol. 2017, 8, 1187. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Peisley, A.; Tetrault, D.; Li, Z.; Egelman, E.H.; Magor, K.E.; Walz, T.; Penczek, P.A.; Hur, S. Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I. Mol. Cell 2014, 55, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; He, X.; Zheng, H.; Huang, L.J.; Hou, F.; Yu, Z.; de la Cruz, M.J.; Borkowski, B.; Zhang, X.; Chen, Z.J.; et al. Structural Basis for the Prion-like MAVS Filaments in Antiviral Innate Immunity. eLife 2014, 3, e01489. [Google Scholar] [CrossRef]
- Atkinson, S.C.; Heaton, S.M.; Audsley, M.D.; Kleifeld, O.; Borg, N.A. TRIM25 and DEAD-Box RNA Helicase DDX3X Cooperate to Regulate RIG-I-Mediated Antiviral Immunity. Int. J. Mol. Sci. 2021, 22, 9094. [Google Scholar] [CrossRef]
- Prada-Gracia, D.; Huerta-Yépez, S.; Moreno-Vargas, L.M. Application of Computational Methods for Anticancer Drug Discovery, Design, and Optimization. Bol. Med. Hosp. Infant. Mex. 2016, 73, 411–423. [Google Scholar] [CrossRef]
- Yasmin, T.; Ali, M.T.; Haque, S.; Hossain, M. Interaction of Quercetin of Onion with Axon Guidance Protein Receptor, NRP-1 Plays Important Role in Cancer Treatment: An In Silico Approach. Interdiscip. Sci. Comput. Life Sci. 2017, 9, 184–191. [Google Scholar] [CrossRef]
- Moorthy, N.S.H.N.; Sousa, S.F.; Ramos, M.J.; Fernandes, P.A. Molecular Dynamic Simulations and Structure-Based Pharmacophore Development for Farnesyltransferase Inhibitors Discovery. J. Enzym. Inhib. Med. Chem. 2016, 31, 1428–1442. [Google Scholar] [CrossRef]
- Milani, M.; Donalisio, M.; Bonotto, R.M.; Schneider, E.; Arduino, I.; Boni, F.; Lembo, D.; Marcello, A.; Mastrangelo, E. Combined in Silico and in Vitro Approaches Identified the Antipsychotic Drug Lurasidone and the Antiviral Drug Elbasvir as SARS-CoV2 and HCoV-OC43 Inhibitors. Antivir. Res. 2021, 189, 105055. [Google Scholar] [CrossRef]
- Macalino, S.J.Y.; Basith, S.; Clavio, N.A.B.; Chang, H.; Kang, S.; Choi, S. Evolution of In Silico Strategies for Protein-Protein Interaction Drug Discovery. Molecules 2018, 23, 1963. [Google Scholar] [CrossRef] [PubMed]
- Wehrhan, L.; Hillisch, A.; Mundt, S.; Tersteegen, A.; Meier, K. Druggability Assessment for Selected Serine Proteases in a Pharmaceutical Industry Setting. ChemMedChem 2020, 15, 2010–2018. [Google Scholar] [CrossRef] [PubMed]
- Salo-Ahen, O.M.H.; Alanko, I.; Bhadane, R.; Bonvin, A.M.J.J.; Honorato, R.V.; Hossain, S.; Juffer, A.H.; Kabedev, A.; Lahtela-Kakkonen, M.; Larsen, A.S.; et al. Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development. Processes 2020, 9, 71. [Google Scholar] [CrossRef]
- Scannell, J.W.; Blanckley, A.; Boldon, H.; Warrington, B. Diagnosing the Decline in Pharmaceutical R&D Efficiency. Nat. Rev. Drug Discov. 2012, 11, 191–200. [Google Scholar] [CrossRef]
- Hingorani, A.D.; Kuan, V.; Finan, C.; Kruger, F.A.; Gaulton, A.; Chopade, S.; Sofat, R.; MacAllister, R.J.; Overington, J.P.; Hemingway, H.; et al. Improving the Odds of Drug Development Success through Human Genomics: Modelling Study. Sci. Rep. 2019, 9, 18911. [Google Scholar] [CrossRef]
- Hay, M.; Thomas, D.W.; Craighead, J.L.; Economides, C.; Rosenthal, J. Clinical Development Success Rates for Investigational Drugs. Nat. Biotechnol. 2014, 32, 40–51. [Google Scholar] [CrossRef]
- Le Guilloux, V.; Schmidtke, P.; Tuffery, P. Fpocket: An Open Source Platform for Ligand Pocket Detection. BMC Bioinform. 2009, 10, 168. [Google Scholar] [CrossRef]
- Mitra, S.; Kumar, D.; Hu, L.; Sankaran, B.; Moosa, M.M.; Rice, A.P.; Ferreon, J.C.; Ferreon, A.C.M.; Prasad, B.V.V. Influenza A Virus Protein NS1 Exhibits Strain-Independent Conformational Plasticity. J. Virol. 2019, 93, e00917-19. [Google Scholar] [CrossRef]
- Carrillo, B.; Choi, J.-M.; Bornholdt, Z.A.; Sankaran, B.; Rice, A.P.; Prasad, B.V.V. The Influenza A Virus Protein NS1 Displays Structural Polymorphism. J. Virol. 2014, 88, 4113–4122. [Google Scholar] [CrossRef]
- Fan, S.; Macken, C.A.; Li, C.; Ozawa, M.; Goto, H.; Iswahyudi, N.F.N.; Nidom, C.A.; Chen, H.; Neumann, G.; Kawaoka, Y. Synergistic Effect of the PDZ and P85β-Binding Domains of the NS1 Protein on Virulence of an Avian H5N1 Influenza A Virus. J. Virol. 2013, 87, 4861–4871. [Google Scholar] [CrossRef]
- Hale, B.G.; Kerry, P.S.; Jackson, D.; Precious, B.L.; Gray, A.; Killip, M.J.; Randall, R.E.; Russell, R.J. Structural Insights into Phosphoinositide 3-Kinase Activation by the Influenza A Virus NS1 Protein. Proc. Natl. Acad. Sci. USA 2010, 107, 1954–1959. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Anderson, D.H.; Liu, Q.; Zhou, Y. Mechanism of Influenza A Virus NS1 Protein Interaction with the P85β, but Not the P85α, Subunit of Phosphatidylinositol 3-Kinase (PI3K) and Up-Regulation of PI3K Activity. J. Biol. Chem. 2008, 283, 23397–23409. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Azeim, S.; Chermak, E.; Vangone, A.; Oliva, R.; Cavallo, L. MDcons: Intermolecular Contact Maps as a Tool to Analyze the Interface of Protein Complexes from Molecular Dynamics Trajectories. BMC Bioinform. 2014, 15, S1. [Google Scholar] [CrossRef] [PubMed]
- Méndez, R.; Leplae, R.; De Maria, L.; Wodak, S.J. Assessment of Blind Predictions of Protein-Protein Interactions: Current Status of Docking Methods. Proteins Struct. Funct. Bioinform. 2003, 52, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Hale, B.G.; Jackson, D.; Chen, Y.H.; Lamb, R.A.; Randall, R.E. Influenza A Virus NS1 Protein Binds P85β and Activates Phosphatidylinositol-3-Kinase Signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 14194–14199. [Google Scholar] [CrossRef]
- Ylösmäki, L.; Schmotz, C.; Ylösmäki, E.; Saksela, K. Reorganization of the Host Cell Crk(L)–PI3 Kinase Signaling Complex by the Influenza A Virus NS1 Protein. Virology 2015, 484, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Dubrow, A.; Kim, I.; Topo, E.; Cho, J.-H. Understanding the Binding Transition State After the Conformational Selection Step: The Second Half of the Molecular Recognition Process Between NS1 of the 1918 Influenza Virus and Host P85β. Front. Mol. Biosci. 2021, 8, 716477. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Yang, Z.; Lasker, K.; Schneidman-Duhovny, D.; Webb, B.; Huang, C.C.; Pettersen, E.F.; Goddard, T.D.; Meng, E.C.; Sali, A.; Ferrin, T.E. UCSF Chimera, MODELLER, and IMP: An Integrated Modeling System. J. Struct. Biol. 2012, 179, 269–278. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved Side-Chain Torsion Potentials for the Amber ff99SB Protein Force Field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 14101. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, H.C.S.; Hu, Z. Using PyMOL as a Platform for Computational Drug Design. WIREs Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Hajduk, P.J.; Huth, J.R.; Tse, C. Predicting Protein Druggability. Drug Discov. Today 2005, 10, 1675–1682. [Google Scholar] [CrossRef]





| NS1 Residues | PI3K Residues |
|---|---|
| Ser 87 | His 450 |
| Arg 88 | Met 568 |
| Tyr 89 | Arg 571 |
| Thr 91 | Lys 572 |
| Met 93 | Arg 574 |
| Leu 95 | Asp 575 |
| Glu 96 | Gln 576 |
| Glu 97 | Leu 578 |
| Met 98 | Val 579 |
| Ser 99 | Trp 580 |
| Arg 100 | Thr 582 |
| Asp 101 | Gln 583 |
| Trp 102 | Lys 584 |
| Cys 116 | Ala 586 |
| Arg 118 | Arg 587 |
| Asn 133 | Gln 588 |
| Phe 134 | Lys 589 |
| Ser 135 | Ile 591 |
| Asn 133 | Asn 592 |
| Phe 134 | Leu 595 |
| Ser 135 | |
| Glu 142 | |
| Thr 143 | |
| Leu 144 | |
| Ile 145 | |
| Leu 146 | |
| Arg 148 | |
| Glu 159 | |
| Ser 161 | |
| Pro 162 | |
| Leu 163 | |
| Pro 164 | |
| Ser 165 |
| NS1 Residues | TRIM25 |
|---|---|
| Ser 87 | Val 208 (chain N) |
| Arg 88 | Ser 211 (chain N) |
| Tyr 89 | Gln 212 (chain N) |
| Leu 90 | Asn 214 (chain N) |
| Thr 91 | Gly 215 (chain N) |
| Met 93 | Arg 218 (chain N) |
| Thr 94 | Ala 219 (chain N) |
| Leu 95 | Asp 221 (chain N) |
| Glu 96 | Asp 222 (chain N) |
| Met 98 | Val 223 (chain N) |
| Ser 99 | Asn 225 (chain N) |
| Glu 101 | Arg 226 (chain N) |
| Arg 118 | Asp 229 (chain N) |
| Asn 133 | Met 232 (chain N) |
| Ser 135 | Arg 236 (chain N) |
| Ile 145 | Phe 274 (chain I) |
| Leu 146 | Ile 277 (chain I) |
| Arg 148 | Leu 281 (chain I) |
| Glu 159 | Val 322 (chain I) |
| Ser 161 | Tyr 323 (chain I) |
| Pro 162 | Ile 324 (chain I) |
| Pro 164 | Pro 325 (chain I) |
| Glu 326 (chain I) | |
| Val 327 (chain I) |
| NS1 Residues | PI3K Residues | Residence Time Ratio |
|---|---|---|
| Tyr 89 | Met 568 | 1.00 |
| Tyr 89 | Lys 572 | 1.00 |
| Tyr 89 | Asp 575 | 1.00 |
| Tyr 89 | Arg 571 | 1.00 |
| Ser 135 | Lys 572 | 1.00 |
| Ser 99 | Leu 578 | 1.00 |
| Ser 99 | Ile 591 | 1.00 |
| Met 98 | Leu 578 | 1.00 |
| Met 98 | Asp 575 | 1.00 |
| Leu 146 | Val 579 | 1.00 |
| Leu 95 | Leu 595 | 1.00 |
| Leu 95 | Leu 578 | 1.00 |
| Leu 95 | His 450 | 1.00 |
| Leu 95 | Arg 574 | 1.00 |
| Ile 145 | Val 579 | 1.00 |
| Ile 145 | Lys 572 | 1.00 |
| Ile 145 | Gln 576 | 1.00 |
| Ile 145 | Asp 575 | 1.00 |
| Asn 133 | Asp 575 | 1.00 |
| Thr 91 | Arg 571 | 0.99 |
| Asp 101 | Gln 588 | 0.99 |
| Ser 99 | Gln 588 | 0.98 |
| Met 98 | Val 579 | 0.98 |
| Leu 95 | Asp 575 | 0.98 |
| Arg 148 | Thr 582 | 0.98 |
| Ser 161 | Gln 583 | 0.97 |
| Pro 162 | Gln 583 | 0.97 |
| Arg 100 | Gln 588 | 0.97 |
| Glu 142 | Lys 572 | 0.96 |
| Thr 143 | Lys 572 | 0.95 |
| Pro 162 | Val 579 | 0.95 |
| Tyr 89 | Arg 574 | 0.94 |
| Asp 101 | Arg 587 | 0.93 |
| Thr 91 | Asp 575 | 0.92 |
| Ser 161 | Val 579 | 0.92 |
| Ser 87 | Lys 572 | 0.92 |
| Thr 143 | Gln 576 | 0.90 |
| Ser 99 | Leu 595 | 0.90 |
| Glu 96 | Gln 588 | 0.87 |
| Ser 99 | Thr 582 | 0.86 |
| Arg 88 | Met 568 | 0.85 |
| Met 98 | Thr 582 | 0.84 |
| Asp 101 | Lys 589 | 0.82 |
| Leu 163 | Gln 583 | 0.78 |
| Tyr 89 | Gln 569 | 0.74 |
| Leu 95 | Gln 588 | 0.73 |
| Ser 99 | Asn 592 | 0.72 |
| Asp 101 | Ala 586 | 0.72 |
| NS1 Residues | TRIM25 Residues | Residence Time Ratio |
|---|---|---|
| Tyr 89 | Asp 229 | 1.00 |
| Tyr 89 | Asp 222 | 1.00 |
| Tyr 89 | Asn 225 | 1.00 |
| Tyr 89 | Arg 226 | 1.00 |
| Thr 94 | Arg 226 | 1.00 |
| Thr 91 | Asp 222 | 1.00 |
| Thr 91 | Arg 226 | 1.00 |
| Ser 161 | Arg 218 | 1.00 |
| Ser 99 | Ala 219 | 1.00 |
| Pro 162 | Arg 218 | 1.00 |
| Met 98 | Asp 222 | 1.00 |
| Met 98 | Arg 226 | 1.00 |
| Met 98 | Arg 218 | 1.00 |
| Met 98 | Ala 219 | 1.00 |
| Met 93 | Arg 226 | 1.00 |
| Leu 146 | Arg 218 | 1.00 |
| Leu 95 | Val 223 | 1.00 |
| Leu 95 | Asp 222 | 1.00 |
| Leu 95 | Arg 226 | 1.00 |
| Ile 145 | Asp 222 | 1.00 |
| Ile 145 | Asp 221 | 1.00 |
| Ile 145 | Asn 225 | 1.00 |
| Ile 145 | Arg 218 | 1.00 |
| Glu 101 | Gln 212 | 1.00 |
| Asn 133 | Asp 222 | 1.00 |
| Asn 133 | Arg 226 | 1.00 |
| Leu 146 | Gly 215 | 0.95 |
| Ser 135 | Asn 225 | 0.94 |
| Arg 88 | Asp 229 | 0.94 |
| Met 98 | Gly 215 | 0.93 |
| Arg 118 | Ser 211 | 0.93 |
| Arg 148 | Gly 215 | 0.87 |
| Leu 163 | Arg 218 | 0.84 |
| Ser 99 | Ala 216 | 0.83 |
| Ser 161 | Asn 214 | 0.82 |
| Pro 164 | Arg 218 | 0.81 |
| Arg 88 | Arg 236 | 0.75 |
| Ser 87 | Met 232 | 0.74 |
| Ile 145 | Ala 219 | 0.74 |
| Leu 146 | Ala 219 | 0.71 |
| NS1 Residues | TRIM25 Residues | Residence Time Ratio |
|---|---|---|
| Ser 99 | Leu 281 | 1.00 |
| Ser 99 | Ile 324 | 1.00 |
| Leu 95 | Pro 325 | 1.00 |
| Leu 95 | Phe 274 | 1.00 |
| Leu 95 | Ile 324 | 1.00 |
| Leu 95 | Ile 277 | 1.00 |
| Glu 96 | Ile 324 | 1.00 |
| Leu 95 | Val 327 | 0.99 |
| Ser 99 | Lys 284 | 0.97 |
| Leu 95 | Leu 281 | 0.97 |
| Leu 95 | Glu 326 | 0.94 |
| Glu 101 | Lys 284 | 0.83 |
| Glu 101 | Lys 320 | 0.78 |
| Residues | Number of Pairwise Residue–Residue Interactions |
|---|---|
| Y89 | 6 |
| L95 | 6 |
| S99 | 6 |
| M98 | 4 |
| D101 | 4 |
| I145 | 4 |
| T91 | 2 |
| T143 | 2 |
| S161 | 2 |
| P162 | 2 |
| S87 | 1 |
| R88 | 1 |
| E96 | 1 |
| R100 | 1 |
| N133 | 1 |
| S135 | 1 |
| E142 | 1 |
| L146 | 1 |
| R148 | 1 |
| L163 | 1 |
| Residues | Number of Pairwise Residue–Residue Interactions |
|---|---|
| L95 | 10 |
| M98 | 5 |
| S99 | 5 |
| I145 | 5 |
| Y89 | 4 |
| E101 | 3 |
| L146 | 3 |
| R88 | 2 |
| T91 | 2 |
| N133 | 2 |
| S161 | 2 |
| S87 | 1 |
| M93 | 1 |
| T94 | 1 |
| E96 | 1 |
| R118 | 1 |
| S135 | 1 |
| R148 | 1 |
| P162 | 1 |
| L163 | 1 |
| P164 | 1 |
| Residue NS1-ED | Residue PI3K-p85β | Residence Time (%) |
|---|---|---|
| P162 | Q583 | 26.97 |
| Y89 | D575 | 76.22 |
| N133 | D575 | 4.7 |
| T91 | D575 | 0.3 |
| S99 | Q588 | 1.9 |
| S87 | K572 | 14.29 |
| D101 | Q588 | 42.66 |
| D101 | Q588 | 0.3 |
| D101 | Q588 | 4.6 |
| R118 | Q583 | 3.6 |
| D101 | R587 | 20.78 |
| P164 | W580 | 0.3 |
| Y89 | R571 | 0.2 |
| S165 | K584 | 0.2 |
| S161 | Q583 | 1.3 |
| S165 | S434 | 0.1 |
| T143 | K572 | 0.9 |
| S99 | N592 | 0.3 |
| S161 | Q583 | 9.19 |
| E96 | Q588 | 42.46 |
| S99 | Q588 | 22.78 |
| R88 | N561 | 1.4 |
| T91 | R571 | 0.4 |
| S87 | Q569 | 0.1 |
| A86 | Q569 | 0.1 |
| A86 | K572 | 0.1 |
| W102 | R587 | 0.1 |
| E159 | Q583 | 0.3 |
| M98 | T582 | 0.9 |
| E142 | K572 | 6.59 |
| S135 | K572 | 1.5 |
| Y89 | M568 | 2 |
| M93 | R571 | 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cunha, A.E.S.; Loureiro, R.J.S.; Simões, C.J.V.; Brito, R.M.M. Unveiling New Druggable Pockets in Influenza Non-Structural Protein 1: NS1–Host Interactions as Antiviral Targets for Flu. Int. J. Mol. Sci. 2023, 24, 2977. https://doi.org/10.3390/ijms24032977
Cunha AES, Loureiro RJS, Simões CJV, Brito RMM. Unveiling New Druggable Pockets in Influenza Non-Structural Protein 1: NS1–Host Interactions as Antiviral Targets for Flu. International Journal of Molecular Sciences. 2023; 24(3):2977. https://doi.org/10.3390/ijms24032977
Chicago/Turabian StyleCunha, Andreia E. S., Rui J. S. Loureiro, Carlos J. V. Simões, and Rui M. M. Brito. 2023. "Unveiling New Druggable Pockets in Influenza Non-Structural Protein 1: NS1–Host Interactions as Antiviral Targets for Flu" International Journal of Molecular Sciences 24, no. 3: 2977. https://doi.org/10.3390/ijms24032977
APA StyleCunha, A. E. S., Loureiro, R. J. S., Simões, C. J. V., & Brito, R. M. M. (2023). Unveiling New Druggable Pockets in Influenza Non-Structural Protein 1: NS1–Host Interactions as Antiviral Targets for Flu. International Journal of Molecular Sciences, 24(3), 2977. https://doi.org/10.3390/ijms24032977






