Biochemical and Bioinformatic Studies of Mutations of Residues at the Monomer–Monomer Interface of Human Ornithine Aminotransferase Leading to Gyrate Atrophy of Choroid and Retina
Abstract
1. Introduction
2. Results and Discussion
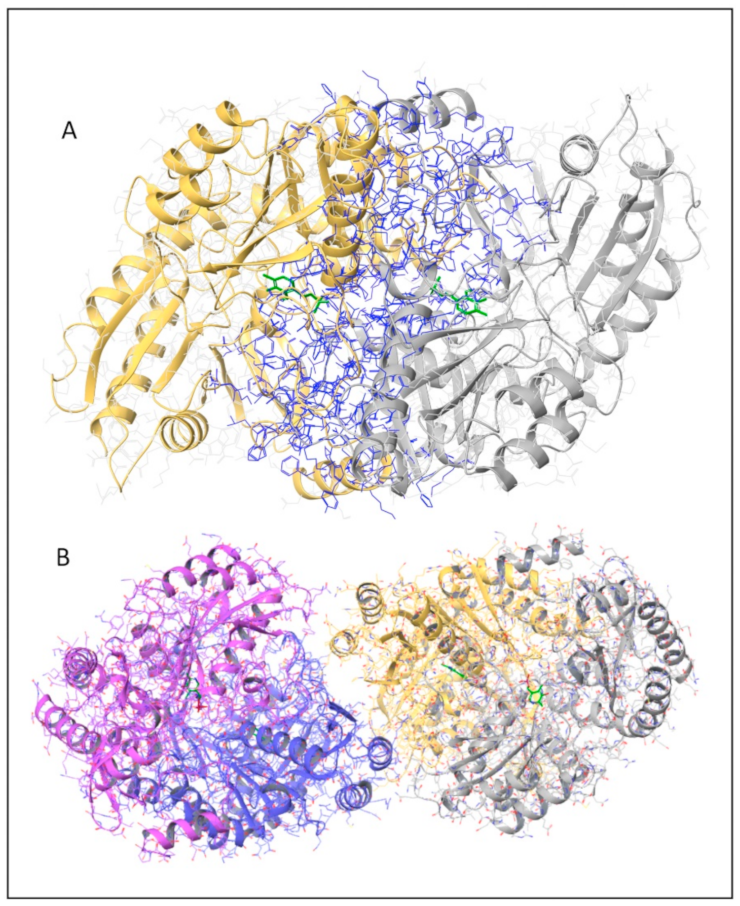
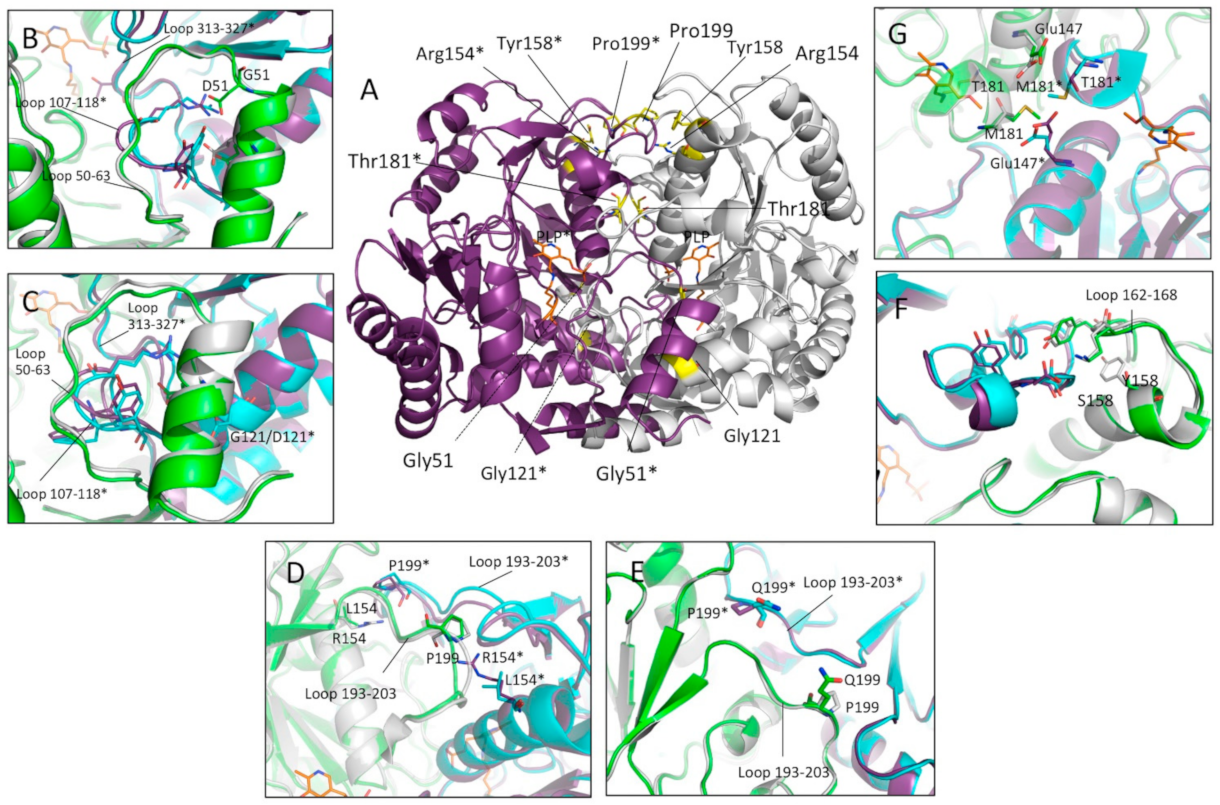
2.1. In Silico Analyses of hOAT Interfaces and of the Mutation Sites
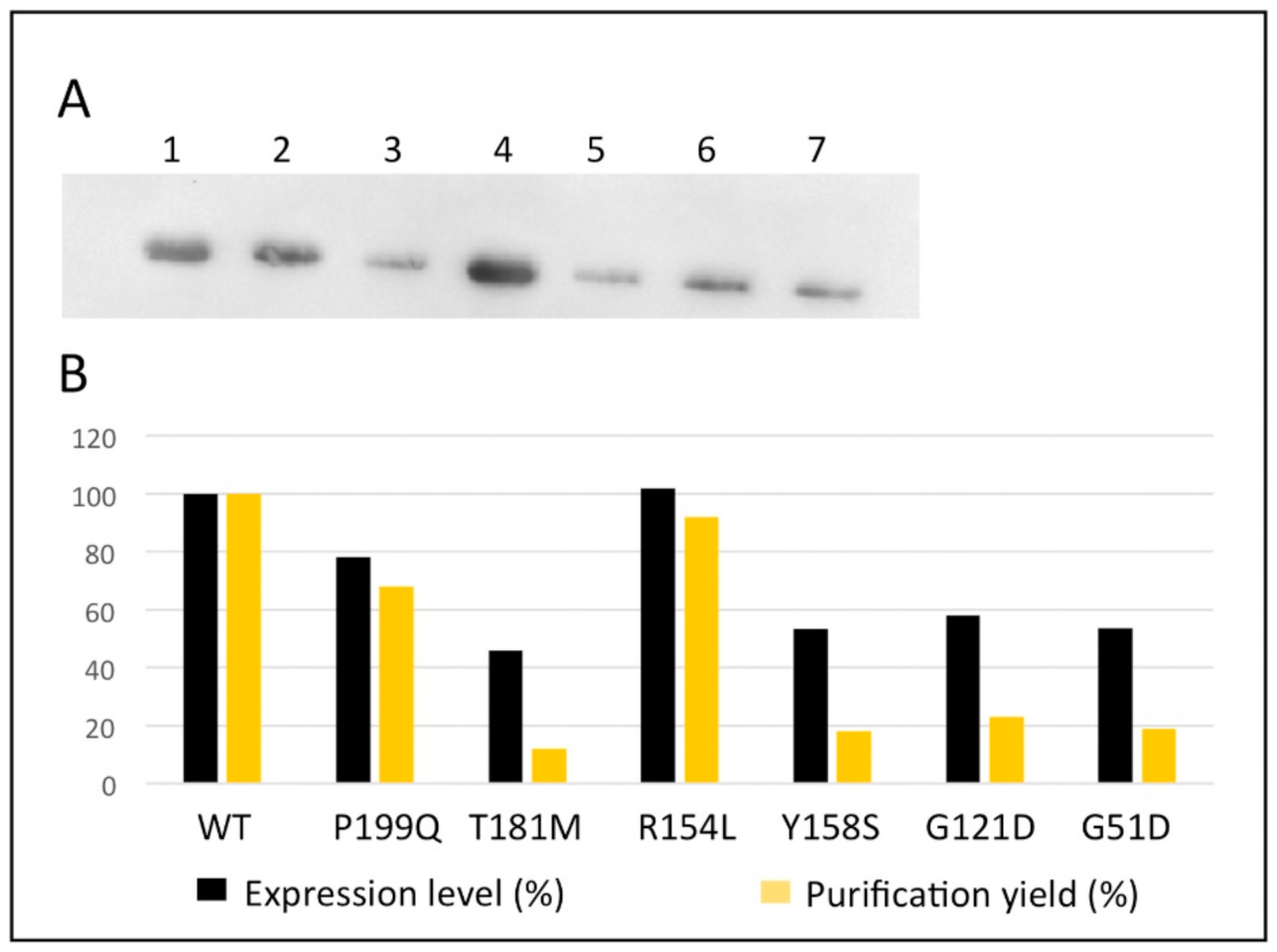
2.2. Expression and Purification of the hOAT Interface Variants
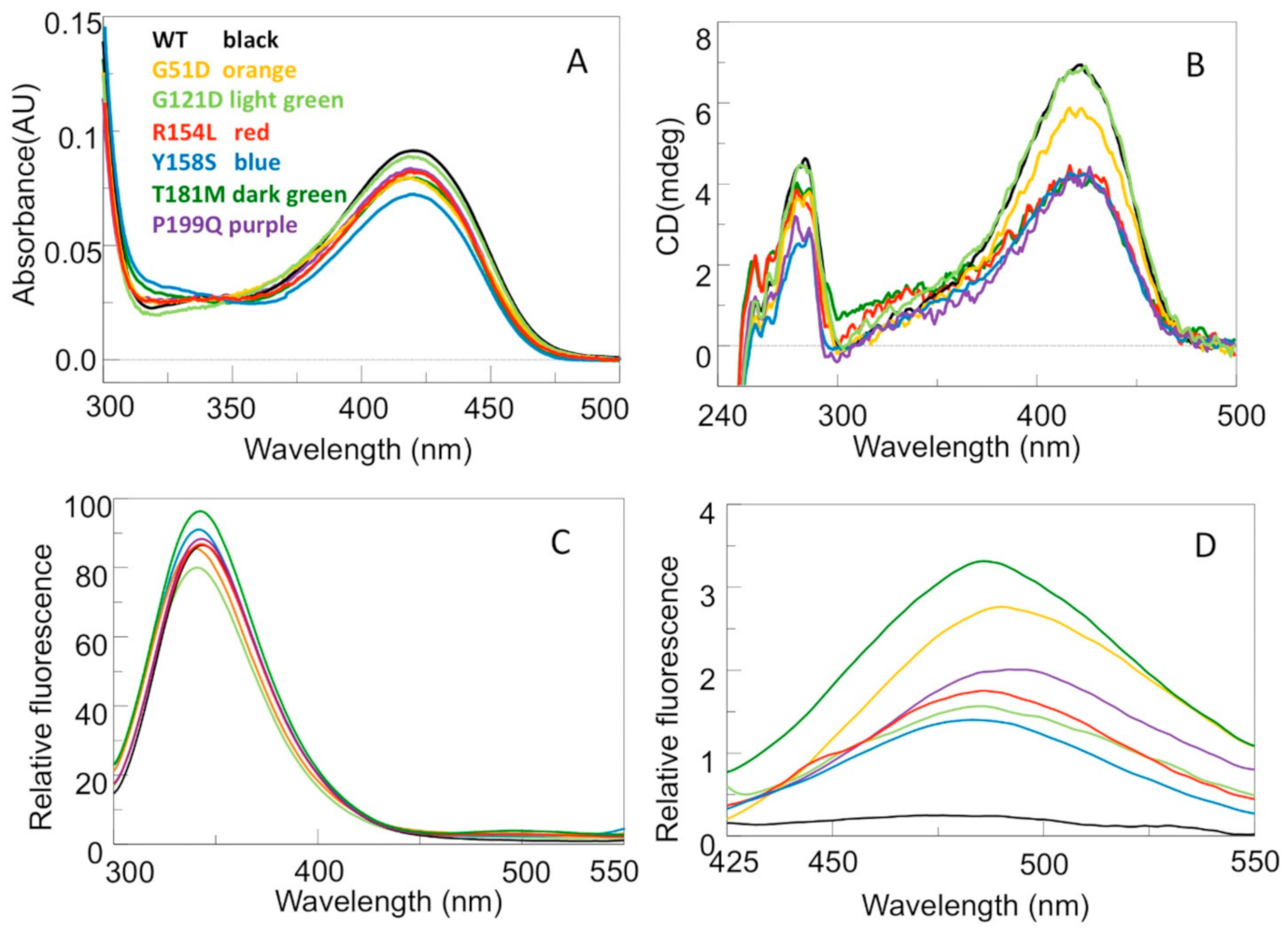
2.3. Impact of the G52D, G121D, R154L, Y158S, T181M and P199Q Mutations on the Spectroscopic Features and Thermal Stability of hOAT
2.4. Impact of the Interface Variants on the Quaternary Structure of hOAT
2.5. Impact of the G52D, G121D, R154L, Y158S, T181M and P199Q Mutations on the Kinetic Features of hOAT
3. Materials and Methods
3.1. Materials
3.2. Computational Studies
3.3. Site Directed Mutagenesis
3.4. Expression and Purification of OAT Variants
3.5. Western-Blot
3.6. Enzyme Activity Assays
3.7. Spectroscopic Measurements and Thermal Stability
3.8. Analytical Size-Exclusion Chromatography (SEC)
3.9. HPLC Analysis of the Coenzymes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Inana, G.; Totsuka, S.; Redmond, M.; Dougherty, T.; Nagle, J.; Shiono, T.; Ohura, T.; Kominami, E.; Katunuma, N. Molecular cloning of human ornithine aminotransferase mRNA. Proc. Natl. Acad. Sci. USA 1986, 83, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Simmaco, M.; John, R.A.; Barra, D.; Bossa, F. The primary structure of ornithine aminotransferase. Identification of active-site sequence and site of post-translational proteolysis. FEBS Lett. 1986, 199, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Eliot, A.C.; Kirsch, J.F. Pyridoxal phosphate enzymes: Mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 2004, 73, 383–415. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.W.; Hennig, M.; Hohenester, E.; Jansonius, J.N.; Schirmer, T. Crystal structure of human recombinant ornithine aminotransferase. J. Mol. Biol. 1998, 277, 81–102. [Google Scholar] [CrossRef] [PubMed]
- Montioli, R.; Zamparelli, C.; Borri Voltattorni, C.; Cellini, B. Oligomeric State and Thermal Stability of Apo- and Holo-Human Ornithine delta-Aminotransferase. Protein J. 2017, 36, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Takki, K.K.; Milton, R.C. The natural history of gyrate atrophy of the choroid and retina. Ophthalmology 1981, 88, 292–301. [Google Scholar] [CrossRef]
- Montioli, R.; Bellezza, I.; Desbats, M.A.; Borri Voltattorni, C.; Salviati, L.; Cellini, B. Deficit of human ornithine aminotransferase in gyrate atrophy: Molecular, cellular, and clinical aspects. Biochim. Biophys. Acta Proteins Proteom. 2021, 1869, 140555. [Google Scholar] [CrossRef]
- Montioli, R.; Desbats, M.A.; Grottelli, S.; Doimo, M.; Bellezza, I.; Borri Voltattorni, C.; Salviati, L.; Cellini, B. Molecular and cellular basis of ornithine delta-aminotransferase deficiency caused by the V332M mutation associated with gyrate atrophy of the choroid and retina. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3629–3638. [Google Scholar] [CrossRef]
- Montioli, R.; Paiardini, A.; Giardina, G.; Zanzoni, S.; Cutruzzola, F.; Cellini, B.; Borri Voltattorni, C. R180T variant of delta-ornithine aminotransferase associated with gyrate atrophy: Biochemical, computational, X-ray and NMR studies provide insight into its catalytic features. FEBS J. 2019, 286, 2787–2798. [Google Scholar] [CrossRef]
- Montioli, R.; Sgaravizzi, G.; Desbats, M.A.; Grottelli, S.; Voltattorni, C.B.; Salviati, L.; Cellini, B. Molecular and Cellular Studies Reveal Folding Defects of Human Ornithine Aminotransferase Variants Associated with Gyrate Atrophy of the Choroid and Retina. Front. Mol. Biosci. 2021, 8, 695205. [Google Scholar] [CrossRef]
- Brody, L.C.; Mitchell, G.A.; Obie, C.; Michaud, J.; Steel, G.; Fontaine, G.; Robert, M.F.; Sipila, I.; Kaiser-Kupfer, M.; Valle, D. Ornithine delta-aminotransferase mutations in gyrate atrophy. Allelic heterogeneity and functional consequences. J. Biol. Chem. 1992, 267, 3302–3307. [Google Scholar] [CrossRef]
- Sergouniotis, P.I.; Davidson, A.E.; Lenassi, E.; Devery, S.R.; Moore, A.T.; Webster, A.R. Retinal structure, function, and molecular pathologic features in gyrate atrophy. Ophthalmology 2012, 119, 596–605. [Google Scholar] [CrossRef]
- Cui, X.; Jauregui, R.; Park, K.S.; Tsang, S.H. Multimodal characterization of a novel mutation causing vitamin B6-responsive gyrate atrophy. Ophthalmic Genet. 2018, 39, 512–516. [Google Scholar] [CrossRef]
- Mashima, Y.; Shiono, T.; Tamai, M.; Inana, G. Heterogeneity and uniqueness of ornithine aminotransferase mutations found in Japanese gyrate atrophy patients. Curr. Eye Res. 1996, 15, 792–796. [Google Scholar] [CrossRef]
- Kaufman, D.L.; Ramesh, V.; McClatchey, A.I.; Menkes, J.H.; Tobin, A.J. Detection of point mutations associated with genetic diseases by an exon scanning technique. Genomics 1990, 8, 656–663. [Google Scholar] [CrossRef]
- Cellini, B.; Bertoldi, M.; Montioli, R.; Laurents, D.V.; Paiardini, A.; Voltattorni, C.B. Dimerization and folding processes of Treponema denticola cystalysin: The role of pyridoxal 5′-phosphate. Biochemistry 2006, 45, 14140–14154. [Google Scholar] [CrossRef]
- Cellini, B.; Lorenzetto, A.; Montioli, R.; Oppici, E.; Voltattorni, C.B. Human liver peroxisomal alanine:glyoxylate aminotransferase: Different stability under chemical stress of the major allele, the minor allele, and its pathogenic G170R variant. Biochimie 2010, 92, 1801–1811. [Google Scholar] [CrossRef]
- Xiong, P.; Zhang, C.; Zheng, W.; Zhang, Y. BindProfX: Assessing Mutation-Induced Binding Affinity Change by Protein Interface Profiles with Pseudo-Counts. J. Mol. Biol. 2017, 429, 426–434. [Google Scholar] [CrossRef]
- Manning, L.R.; Jenkins, W.T.; Hess, J.R.; Vandegriff, K.; Winslow, R.M.; Manning, J.M. Subunit dissociations in natural and recombinant hemoglobins. Protein Sci. 1996, 5, 775–781. [Google Scholar] [CrossRef]
- Cellini, B.; Montioli, R.; Oppici, E.; Astegno, A.; Voltattorni, C.B. The chaperone role of the pyridoxal 5′-phosphate and its implications for rare diseases involving B6-dependent enzymes. Clin. Biochem. 2014, 47, 158–165. [Google Scholar] [CrossRef]
- Hayasaka, S.; Saito, T.; Nakajima, H.; Takaku, Y.; Shiono, T.; Mizuno, K.; Ohmura, K.; Tada, K. Gyrate atrophy with hyperornithinaemia: Different types of responsiveness to vitamin B6. Br. J. Ophthalmol. 1981, 65, 478–483. [Google Scholar] [CrossRef] [PubMed]
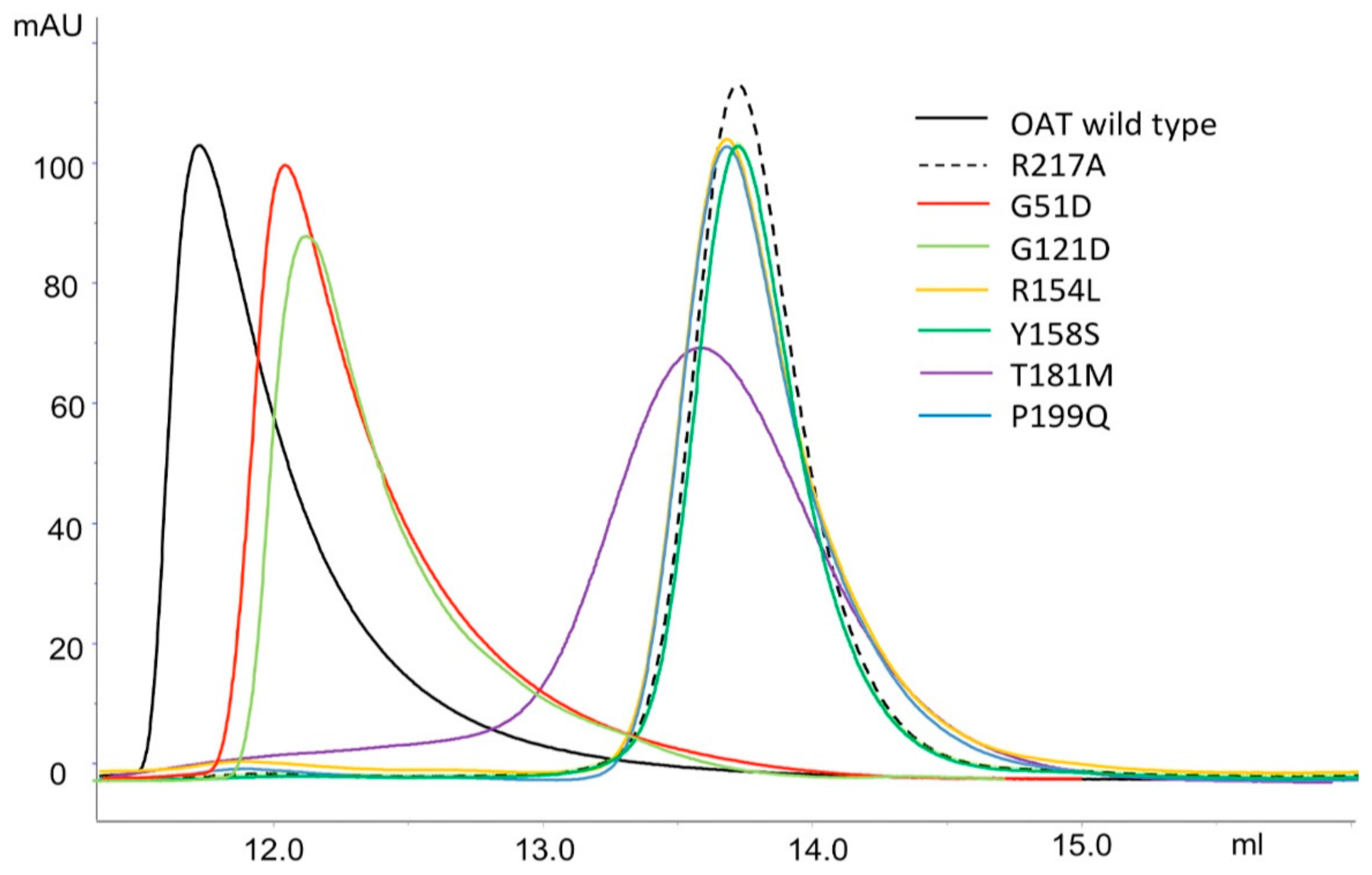
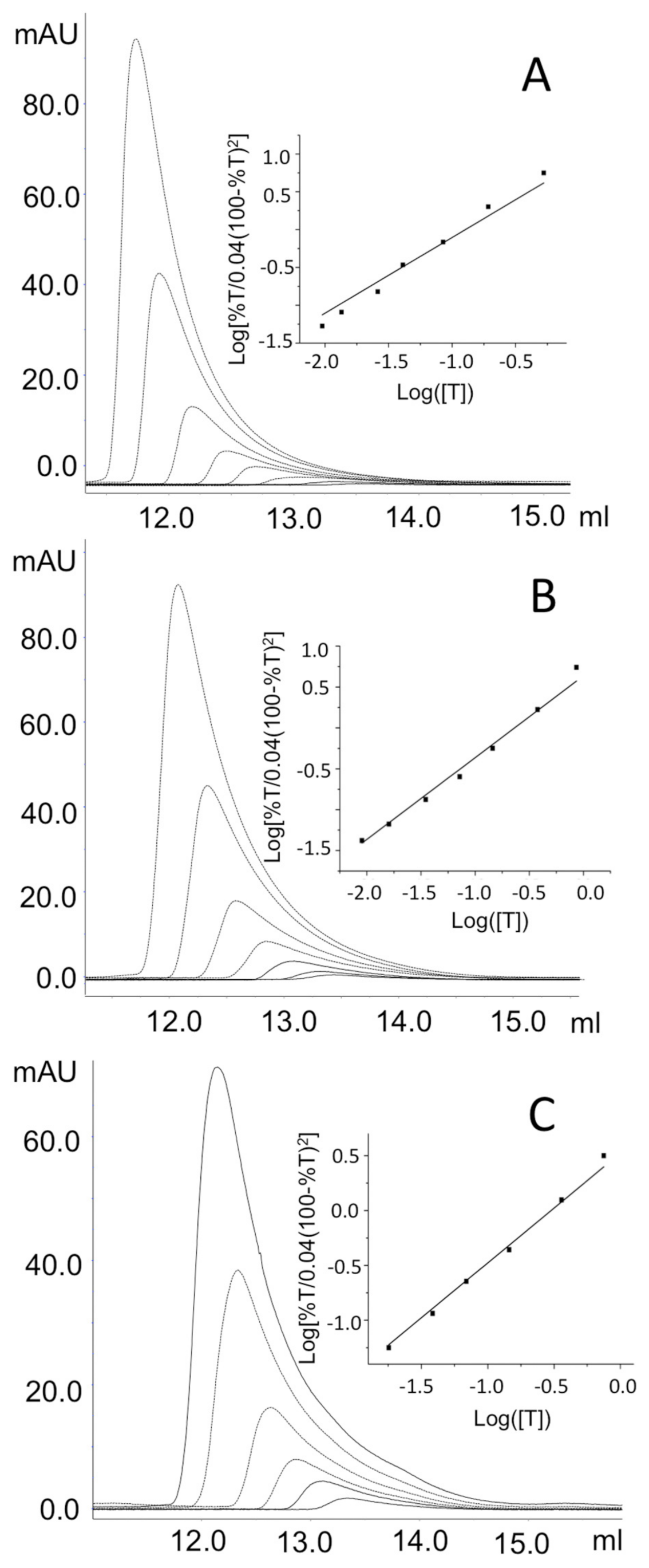
| Enzymatic Species | ΔΔG (kcal/mol) | ΔTm (°C) | KDtet-dim (µM) |
|---|---|---|---|
| OAT wild-type | 0 | 0 | 0.13 ± 0.01 |
| G51D | 1.95 | −0.5 | 0.24 ± 0.01 |
| G121D | 3.83 | −1.3 | 0.30 ± 0.01 |
| R154L | 8.68 | −12 | >>10 |
| Y158S | 2.07 | −12 | >>10 |
| T181M | 11.52 | −8.9 | >10 |
| P199Q | 10.82 | −10.5 | >>10 |
| Enzyme | Substrate | Cosubstrate | kcat (s−1) | KM(L-Orn) (mM) | KM(α-KG) (mM) | kcat/KM (mM−1s−1) |
|---|---|---|---|---|---|---|
| OAT WT [5] | L-Orn | α-KG | 34.9 ± 0.6 | 6.5 ± 0.4 | 5.4 ± 0.3 | |
| α-KG | L-Orn | 35.7 ± 0.7 | 3.9 ± 0.5 | 9.1 ± 1.2 | ||
| T181M | L-Orn | α-KG | 6.8 ± 0.3 | 91 ± 10 | 0.075 ± 0.009 | |
| α-KG | L-Orn | 7.2 ± 0.4 | 14.7 ± 2.8 | 0.49 ± 0.10 | ||
| P199Q | L-Orn | α-KG | 0.25 ± 0.01 | 18.8 ± 1.9 | 1.3 × 10−3 ± 2 × 10−4 | |
| α-KG | L-Orn | 0.39 ± 0.03 | 87.6 ± 0.5 | 4.4 × 10−4 ± 3 × 10−5 | ||
| Y158S | L-Orn | α-KG | 1.55 ± 0.13 | 3.4 ± 0.7 | 0.45 ± 0.10 | |
| α-KG | L-Orn | 2.19 ± 0.14 | 31.0 ± 6.4 | 0.07 ± 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Floriani, F.; Borri Voltattorni, C.; Cellini, B.; Montioli, R. Biochemical and Bioinformatic Studies of Mutations of Residues at the Monomer–Monomer Interface of Human Ornithine Aminotransferase Leading to Gyrate Atrophy of Choroid and Retina. Int. J. Mol. Sci. 2023, 24, 3369. https://doi.org/10.3390/ijms24043369
Floriani F, Borri Voltattorni C, Cellini B, Montioli R. Biochemical and Bioinformatic Studies of Mutations of Residues at the Monomer–Monomer Interface of Human Ornithine Aminotransferase Leading to Gyrate Atrophy of Choroid and Retina. International Journal of Molecular Sciences. 2023; 24(4):3369. https://doi.org/10.3390/ijms24043369
Chicago/Turabian StyleFloriani, Fulvio, Carla Borri Voltattorni, Barbara Cellini, and Riccardo Montioli. 2023. "Biochemical and Bioinformatic Studies of Mutations of Residues at the Monomer–Monomer Interface of Human Ornithine Aminotransferase Leading to Gyrate Atrophy of Choroid and Retina" International Journal of Molecular Sciences 24, no. 4: 3369. https://doi.org/10.3390/ijms24043369
APA StyleFloriani, F., Borri Voltattorni, C., Cellini, B., & Montioli, R. (2023). Biochemical and Bioinformatic Studies of Mutations of Residues at the Monomer–Monomer Interface of Human Ornithine Aminotransferase Leading to Gyrate Atrophy of Choroid and Retina. International Journal of Molecular Sciences, 24(4), 3369. https://doi.org/10.3390/ijms24043369







