A Novel Glycoside Hydrolase DogH Utilizing Soluble Starch to Maltose Improve Osmotic Tolerance in Deinococcus radiodurans
Abstract
:1. Introduction
2. Results
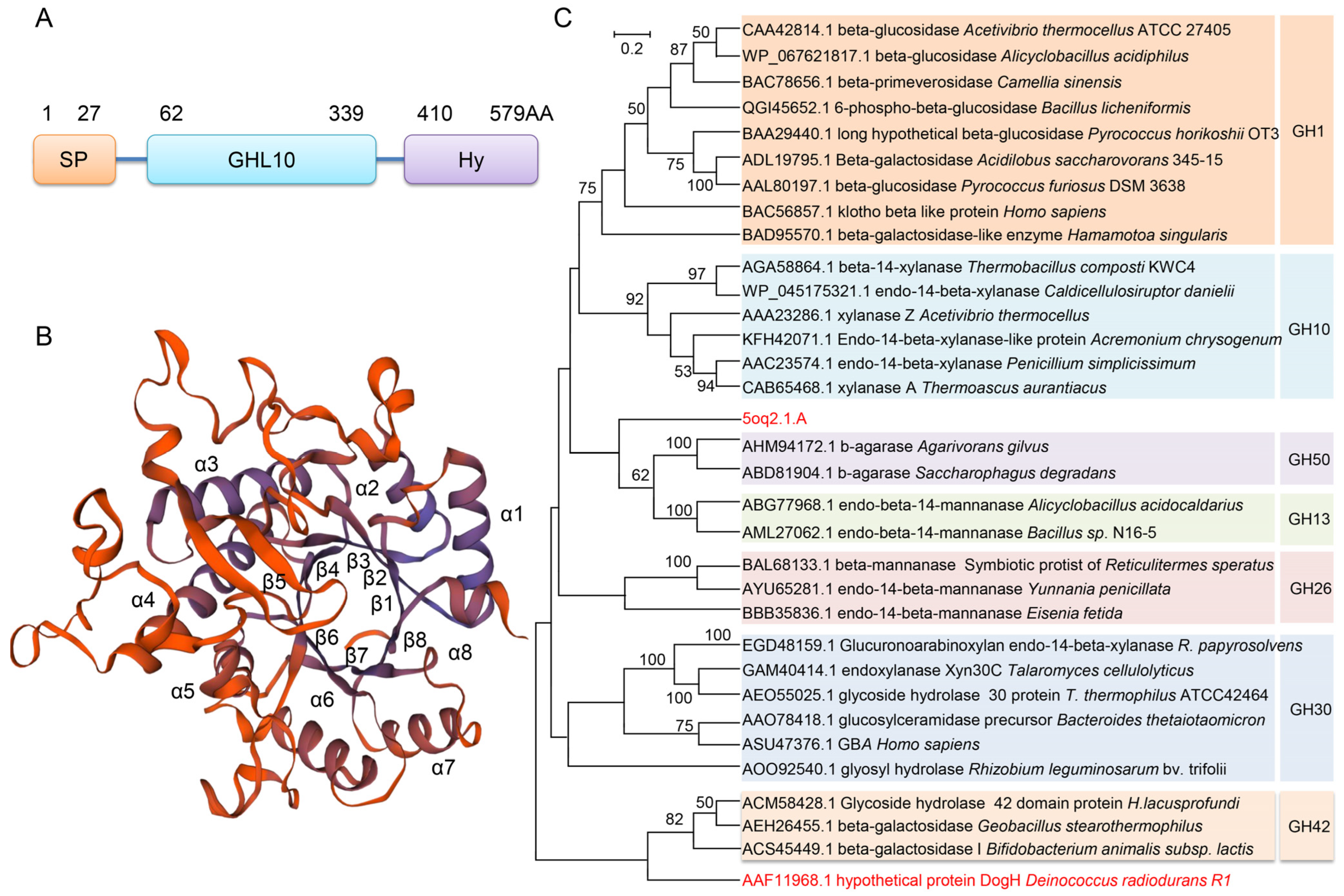
2.1. Domains of DogH and Its Bioinformatics Analysis
2.2. dogH Gene Deletion Reduced Osmotic and Desiccation Stress Tolerance of D. radiodurans
2.3. dogH Gene Deletion Affected the Intracellular Osmoprotectant Content of D. radiodurans
2.4. DogH Glycoside Hydrolase Substrate Specificity and Product Analysis
2.5. DogH Affected Trehalose Content of D. radiodurans
2.6. Different Environments Shape Strains with Different Osmoprotectants
3. Discussion
3.1. The Evolutionary Origin of DogH Glycoside Hydrolase
3.2. Pathways of Trehalose Synthesis in D. radiodurans
3.3. Mechanisms of Osmoregulation in D. radiodurans
4. Materials and Methods
4.1. Strains and Plasmids
4.2. Bioinformatics Analysis
4.3. Abiotic Stress Phenotyping
4.4. Real-Time Fluorescence Quantitative PCR
4.5. Collection and Analysis of Cellular Metabolites
4.6. DogH Expression and Purification
4.7. DogH Glycoside Hydrolase Substrate Profiling
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IUBMB. Enzyme Nomenclature. Recommendations 1992. Supplement: Corrections and additions. Eur. J. Biochem. 1994, 223, 1–5. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucl. Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- van Wyk, N.; Drancourt, M.; Henrissat, B.; Kremer, L. Current perspectives on the families of glycoside hydrolases of Mycobacterium tuberculosis: Their importance and prospects for assigning function to unknowns. Glycobiology 2017, 27, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Krämer, R.; Morbach, S. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Mol. Microbiol. 2003, 49, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- Reina-Bueno, M.; Argandoña, M.; Salvador, M.; Rodríguez-Moya, J.; Iglesias-Guerra, F.; Csonka, L.N.; Nieto, J.J.; Vargas, C. Role of Trehalose in Salinity and Temperature Tolerance in the Model Halophilic Bacterium Chromohalobacter salexigens. PLoS ONE 2012, 7, e33587. [Google Scholar] [CrossRef]
- Stam, M.R.; Danchin, E.G.J.; Rancurel, C.; Coutinho, P.M.; Henrissat, B. Dividing the large glycoside hydrolase family 13 into subfamilies: Towards improved functional annotations of -amylase-related proteins. Protein Eng. Des. Sel. 2006, 19, 555–562. [Google Scholar] [CrossRef]
- MacElroy, R.D. Some comments on the evolution of extremophiles. Biosystems 1974, 6, 74–75. [Google Scholar] [CrossRef]
- Banasik, M.; Stanisławska-Sachadyn, A.; Hildebrandt, E.; Sachadyn, P. In vitro affinity of Deinococcus radiodurans MutS towards mismatched DNA exceeds that of its orthologues from Escherichia coli and Thermus thermophilus. J. Biotechnol. 2017, 252, 55–64. [Google Scholar] [CrossRef]
- Blasius, M.; Sommer, S.; Hübscher, U. Deinococcus radiodurans: What belongs to the survival kit? Crit. Rev. Biochem. Mol. Biol. 2008, 43, 221–238. [Google Scholar] [CrossRef]
- Slade, D.; Radman, M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef] [Green Version]
- White, O.; Eisen, J.A.; Heidelberg, J.F.; Hickey, E.K.; Peterson, J.D.; Dodson, R.J.; Haft, D.H.; Gwinn, M.L.; Nelson, W.C.; Richardson, D.L.; et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 1999, 286, 1571–1577. [Google Scholar] [CrossRef]
- Ruhal, R.; Kataria, R.; Choudhury, B. Trends in bacterial trehalose metabolism and significant nodes of metabolic pathway in the direction of trehalose accumulation. Microb. Biotechnol. 2013, 6, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wei, X.; Liu, G.-L.; Hu, Z.; Chi, Z.-M.; Chi, Z. Glycerol, trehalose and vacuoles had relations to pullulan synthesis and osmotic tolerance by the whole genome duplicated strain Aureobasidium melanogenum TN3-1 isolated from natural honey. Int. J. Biol. Macromol. 2020, 165, 131–140. [Google Scholar] [CrossRef]
- Chi, Z.; Liu, J.; Ji, J.; Meng, Z. Enhanced conversion of soluble starch to trehalose by a mutant of Saccharomycopsis fibuligera sdu. J. Biotechnol. 2003, 102, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lu, G.; Huang, X.; Guo, H.; Su, X.; Han, L.; Zhang, Y.; Qi, Z.; Xiao, Y.; Cheng, H. Overexpression of the caryophyllene synthase gene GhTPS1 in cotton negatively affects multiple pests while attracting parasitoids. Pest Manag. Sci. 2020, 76, 1722–1730. [Google Scholar] [CrossRef]
- Gao, L.; Zhou, Z.; Chen, X.; Zhang, W.; Lin, M.; Chen, M. Comparative Proteomics Analysis Reveals New Features of the Oxidative Stress Response in the Polyextremophilic Bacterium Deinococcus radiodurans. Microorganisms 2020, 8, 451. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, W.J.; Kirby, J.M.; Roberts, A.K.; Shone, C.C.; Acharya, K.R. The molecular structure of the glycoside hydrolase domain of Cwp19 from Clostridium difficile. FEBS J. 2017, 284, 4343–4357. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Giovannoni, S.J.; Woese, C.R. The Deinococcus-Thermus phylum and the effect of rRNA composition on phylogenetic tree construction. Syst. Appl. Microbiol. 1989, 11, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Omelchenko, M.V.; Wolf, Y.I.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Daly, M.J.; Koonin, E.V.; Makarova, K.S. Comparative genomics of Thermus thermophilus and Deinococcus radiodurans: Divergent routes of adaptation to thermophily and radiation resistance. BMC Evol. Biol. 2005, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Aravind, L.; Wolf, Y.I.; Tatusov, R.L.; Minton, K.W.; Koonin, E.V.; Daly, M.J. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 2001, 65, 44–79. [Google Scholar] [CrossRef] [Green Version]
- Jordan, I.K.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Gene conversions in genes encoding outer-membrane proteins in H. pylori and C. pneumoniae. Trends Genet. 2001, 17, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Gury, J.; Barthelmebs, L.; Tran, N.P.; Diviès, C.; Cavin, J.-F. Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl. Environ. Microbiol. 2004, 70, 2146–2153. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Omelchenko, M.V.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Lapidus, A.; Copeland, A.; Kim, E.; Land, M.; et al. Deinococcus geothermalis: The pool of extreme radiation resistance genes shrinks. PLoS ONE 2007, 2, e955. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Chen, M.; Zhang, W.; Lu, W.; Wang, J.; Yang, M.; Zhao, P.; Tang, R.; Li, X.; Hao, Y.; et al. Genome sequence and transcriptome analysis of the radioresistant bacterium Deinococcus gobiensis: Insights into the extreme environmental adaptations. PLoS ONE 2012, 7, e34458. [Google Scholar] [CrossRef] [PubMed]
- Murawska, M.; Ladurner, A.G. Bromodomain AAA+ ATPases get into shape. Nucleus 2020, 11, 32–34. [Google Scholar] [CrossRef]
- Ogura, T.; Wilkinson, A.J. AAA+ superfamily ATPases: Common structure--diverse function. Genes Cells 2001, 6, 575–597. [Google Scholar] [CrossRef]
- Pollet, A.; Delcour, J.A.; Courtin, C.M. Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families. Crit. Rev. Biotechnol. 2010, 30, 176–191. [Google Scholar] [CrossRef]
- Kirsch, F.; Klähn, S.; Hagemann, M. Salt-Regulated Accumulation of the Compatible Solutes Sucrose and Glucosylglycerol in Cyanobacteria and Its Biotechnological Potential. Front. Microbiol. 2019, 10, 2139. [Google Scholar] [CrossRef]
- Empadinhas, N.; da Costa, M.S. Diversity and biosynthesis of compatible solutes in hyper/thermophiles. Int. Microbiol. 2006, 9, 199–206. [Google Scholar]
- Ash, C. Trehalose confers superpowers. Science 2017, 358, 1398–1399. [Google Scholar] [CrossRef]
- Argüelles, J.C. Physiological roles of trehalose in bacteria and yeasts: A comparative analysis. Arch. Microbiol. 2000, 174, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Primavesi, L.F.; Jhurreea, D.; Zhang, Y. Trehalose Metabolism and Signaling. Annu. Rev. Plant Biol. 2008, 59, 417–441. [Google Scholar] [CrossRef] [PubMed]
- Stracke, C.; Meyer, B.H.; Hagemann, A.; Jo, E.; Lee, A.; Albers, S.-V.; Cha, J.; Bräsen, C.; Siebers, B. Salt Stress Response of Sulfolobus acidocaldarius Involves Complex Trehalose Metabolism Utilizing a Novel Trehalose-6-Phosphate Synthase (TPS)/Trehalose-6-Phosphate Phosphatase (TPP) Pathway. Appl. Environ. Microbiol. 2020, 86, e01565-20. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, J.; Wei, D.; Wang, Y.; Chen, X.; Xing, L.; Li, M. Isolation and Identification of a Thermophilic Strain Producing Trehalose Synthase from Geothermal Water in China. Biosci. Biotechnol. Biochem. 2008, 72, 2019–2024. [Google Scholar] [CrossRef]
- De Smet, K.A.L.; Weston, A.; Brown, I.N.; Young, D.B.; Robertson, B.D. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 2000, 146 Pt 1, 199–208. [Google Scholar] [CrossRef]
- Maruta, K.; Mitsuzumi, H.; Nakada, T.; Kubota, M.; Chaen, H.; Fukuda, S.; Sugimoto, T.; Kurimoto, M. Cloning and sequencing of a cluster of genes encoding novel enzymes of trehalose biosynthesis from thermophilic archaebacterium Sulfolobus acidocaldarius. Biochim. Biophys. Acta 1996, 1291, 177–181. [Google Scholar] [CrossRef]
- Wannet, W.J.; Op den Camp, H.J.; Wisselink, H.W.; van der Drift, C.; Van Griensven, L.J.; Vogels, G.D. Purification and characterization of trehalose phosphorylase from the commercial mushroom Agaricus bisporus. Biochim. Biophys. Acta 1998, 1425, 177–188. [Google Scholar] [CrossRef]
- Qu, Q.; Lee, S.-J.; Boos, W. TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archaeon Thermococcus litoralis. J. Biol. Chem. 2004, 279, 47890–47897. [Google Scholar] [CrossRef]
- Ryu, S.-I.; Park, C.-S.; Cha, J.; Woo, E.-J.; Lee, S.-B. A novel trehalose-synthesizing glycosyltransferase from Pyrococcus horikoshii: Molecular cloning and characterization. Biochem. Biophys. Res. Commun. 2005, 329, 429–436. [Google Scholar] [CrossRef]
- Silva, Z.; Alarico, S.; Nobre, A.; Horlacher, R.; Marugg, J.; Boos, W.; Mingote, A.I.; da Costa, M.S. Osmotic Adaptation of Thermus thermophilus RQ-1: Lesson from a Mutant Deficient in Synthesis of Trehalose. J. Bacteriol. 2003, 185, 5943–5952. [Google Scholar] [CrossRef]
- Krisko, A.; Radman, M. Biology of Extreme Radiation Resistance: The Way of Deinococcus radiodurans. Cold Spring Harb. Perspect. Biol. 2013, 5, a012765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, N.; Li, J.; Shin, H.-D.; Du, G.; Chen, J.; Liu, L. Microbial response to environmental stresses: From fundamental mechanisms to practical applications. Appl. Microbiol. Biotechnol. 2017, 101, 3991–4008. [Google Scholar] [CrossRef] [PubMed]
- Mocali, S.; Chiellini, C.; Fabiani, A.; Decuzzi, S.; de Pascale, D.; Parrilli, E.; Tutino, M.L.; Perrin, E.; Bosi, E.; Fondi, M.; et al. Ecology of cold environments: New insights of bacterial metabolic adaptation through an integrated genomic-phenomic approach. Sci. Rep. 2017, 7, 839. [Google Scholar] [CrossRef] [PubMed]
- Im, S.; Joe, M.; Kim, D.; Park, D.-H.; Lim, S. Transcriptome analysis of salt-stressed Deinococcus radiodurans and characterization of salt-sensitive mutants. Res. Microbiol. 2013, 164, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Dani, P.; Ujaoney, A.K.; Apte, S.K.; Basu, B. Regulation of potassium dependent ATPase (kdp) operon of Deinococcus radiodurans. PLoS ONE 2017, 12, e0188998. [Google Scholar] [CrossRef]
- Yu, J.; Li, T.; Dai, S.; Weng, Y.; Li, J.; Li, Q.; Xu, H.; Hua, Y.; Tian, B. A tamB homolog is involved in maintenance of cell envelope integrity and stress resistance of Deinococcus radiodurans. Sci. Rep. 2017, 7, 45929. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, C.; Wang, Z.; Lin, M.; Wang, J.; Wu, M. Pleiotropic roles of late embryogenesis abundant proteins of Deinococcus radiodurans against oxidation and desiccation. Comput. Struct. Biotechnol. J. 2021, 19, 3407–3415. [Google Scholar] [CrossRef]
- Alsheikh, M.K.; Heyen, B.J.; Randall, S.K. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J. Biol. Chem. 2003, 278, 40882–40889. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, J.; Liu, X.; Liu, Y.; Guo, C.; Zhang, L.; Han, J.; Wu, X.; Xue, D.; Gomaa, A.E.; et al. DrwH, a novel WHy domain-containing hydrophobic LEA5C protein from Deinococcus radiodurans, protects enzymatic activity under oxidative stress. Sci. Rep. 2017, 7, 9281. [Google Scholar] [CrossRef]
- Awile, O.; Krisko, A.; Sbalzarini, I.F.; Zagrovic, B. Intrinsically disordered regions may lower the hydration free energy in proteins: A case study of nudix hydrolase in the bacterium Deinococcus radiodurans. PLoS Comput. Biol. 2010, 6, e1000854. [Google Scholar] [CrossRef]
- Oren, A. Microbial life at high salt concentrations: Phylogenetic and metabolic diversity. Saline Syst. 2008, 4, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alarico, S.; Empadinhas, N.; Simões, C.; Silva, Z.; Henne, A.; Mingote, A.; Santos, H.; da Costa, M.S. Distribution of genes for synthesis of trehalose and Mannosylglycerate in Thermus spp. and direct correlation of these genes with halotolerance. Appl. Environ. Microbiol. 2005, 71, 2460–2466. [Google Scholar] [CrossRef] [PubMed]
- Santos, H.; da Costa, M.S. Compatible solutes of organisms that live in hot saline environments. Environ. Microbiol. 2002, 4, 501–509. [Google Scholar] [CrossRef]
- Agapov, A.A.; Kulbachinskiy, A.V. Mechanisms of stress resistance and gene regulation in the radioresistant bacterium Deinococcus radiodurans. Biochem. Mosc. 2015, 80, 1201–1216. [Google Scholar] [CrossRef]
- Wang, L.; Xu, G.; Chen, H.; Zhao, Y.; Xu, N.; Tian, B.; Hua, Y. DrRRA: A novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Mol. Microbiol. 2008, 67, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Wang, L.; Chen, H.; Lu, H.; Ying, N.; Tian, B.; Hua, Y. RecO is essential for DNA damage repair in Deinococcus radiodurans. J. Bacteriol. 2008, 190, 2624–2628. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Hong, Y.; Guo, M.; Wang, J. ENJ algorithm can construct triple phylogenetic trees. Mol. Ther. Nucleic Acids 2021, 23, 286–293. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Guo, L.; Zhao, M.; Tang, Y.; Han, J.; Gui, Y.; Ge, J.; Jiang, S.; Dai, Q.; Zhang, W.; Lin, M.; et al. Modular Assembly of Ordered Hydrophilic Proteins Improve Salinity Tolerance in Escherichia coli. Int. J. Mol. Sci. 2021, 22, 4482. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, M.; Lv, M.; Lin, M.; Wang, J.; Zuo, K. A Novel Small RNA, DsrO, in Deinococcus radiodurans Promotes Methionine Sulfoxide Reductase (msrA) Expression for Oxidative Stress Adaptation. Appl. Environ. Microbiol. 2022, 88, e0003822. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Fredriksen, L.; Stokke, R.; Jensen, M.S.; Westereng, B.; Jameson, J.-K.; Steen, I.H.; Eijsink, V.G.H. Discovery of a Thermostable GH10 Xylanase with Broad Substrate Specificity from the Arctic Mid-Ocean Ridge Vent System. Appl. Environ. Microbiol. 2019, 85, e02970-18. [Google Scholar] [CrossRef] [Green Version]








| Gene ID | Description | Fold Change |
|---|---|---|
| dogH(dr2412) | Hypothetical protein | 2.8 |
| glgX(dr0264) | 1,4-alpha-glucan branching enzyme | 9.3 |
| glgB(dr1848) | 1,4-alpha-glucan branching enzyme | 10.2 |
| treY(dr0463) | Maltooligosyl trehalose synthase | 5.2 |
| treZ(dr0464) | Trehalose trehalohydrolase | 3.5 |
| treS(dr0933) | Trehalose synthase | 5.7 |
| dr1141 | Pullulanase/amylase | 3.9 |
| Carbohydrate | Monosaccharides and Bonds Present |
|---|---|
| Cellobiose | D-glucose-β-1,4-D-glucose |
| a-cyclodextrin | Cyclo-[-D-glucose-α-1,4-]6 |
| Lactose | D-galactose-β-1,4-D-glucose |
| Maltose | D-glucose-α-1,4-D-glucose |
| Melibiose | D-galactose-α-1,6-D-glucose |
| Pullulan | [-D-glucose-α-1,4-D-glucose-a-1,4-D-glucose-α-1,6-]n |
| Starch | [-D-glucose-α-1,4-]n…D-glucose-α-1,6-D-glucose… |
| Sucrose | D-glucose-α-β-1,2-D-fructose |
| Trehalose | D-glucose-α-α-1,1-D-glucose |
| Plasmid/Strain | Description | Source |
|---|---|---|
| pRADZ3 | Shuttle vector for E. coli and D. radiodurans, Chlr (D. radiodurans), Ampr (E. coli) | Laboratory stock |
| pKatAPH3 | To amplify the kanamycin resistance gene | Laboratory stock |
| D. radiodurans R1 | Wild type, served as the strain for generation of the mutants | Laboratory stock |
| ∆dogH mutant | D. radiodurans with genomic deletion of dogH gene dogH mutant with pRADZ3 shuttle plasmid introduced into its genome | This study |
| ∆dogH-pRADZ3 | Complementation of the dogH deletion in D. radiodurans, transformation of dogH mutant with pRADZ3 plasmid | This study |
| ∆dogH-com | expressing D. radiodurans dogH gene | This study |
| WT-dogH-over | Overexpressing D. radiodurans dogH gene | This study |
| E. coli DH5a | The strain expressing the shuttle plasmid pRADZ3 | CW Biotech |
| pET28a(+) | KanR oripBR322 lacIq T7p | Novagen |
| E. coli BL21 | F- ompT hsdSB(rB- mB-) gal dcm(DE3) | TransGen Biotech |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gui, Y.; Lin, M.; Yan, Y.; Jiang, S.; Zhou, Z.; Wang, J. A Novel Glycoside Hydrolase DogH Utilizing Soluble Starch to Maltose Improve Osmotic Tolerance in Deinococcus radiodurans. Int. J. Mol. Sci. 2023, 24, 3437. https://doi.org/10.3390/ijms24043437
Gui Y, Lin M, Yan Y, Jiang S, Zhou Z, Wang J. A Novel Glycoside Hydrolase DogH Utilizing Soluble Starch to Maltose Improve Osmotic Tolerance in Deinococcus radiodurans. International Journal of Molecular Sciences. 2023; 24(4):3437. https://doi.org/10.3390/ijms24043437
Chicago/Turabian StyleGui, Yuan, Min Lin, Yongliang Yan, Shijie Jiang, Zhengfu Zhou, and Jin Wang. 2023. "A Novel Glycoside Hydrolase DogH Utilizing Soluble Starch to Maltose Improve Osmotic Tolerance in Deinococcus radiodurans" International Journal of Molecular Sciences 24, no. 4: 3437. https://doi.org/10.3390/ijms24043437
APA StyleGui, Y., Lin, M., Yan, Y., Jiang, S., Zhou, Z., & Wang, J. (2023). A Novel Glycoside Hydrolase DogH Utilizing Soluble Starch to Maltose Improve Osmotic Tolerance in Deinococcus radiodurans. International Journal of Molecular Sciences, 24(4), 3437. https://doi.org/10.3390/ijms24043437






