Inhibition of Ribosomal RNA Processing 15 Homolog (RRP15) Suppressed Tumor Growth, Invasion and Epithelial to Mesenchymal Transition (EMT) of Colon Cancer
Abstract
:1. Introduction
2. Results
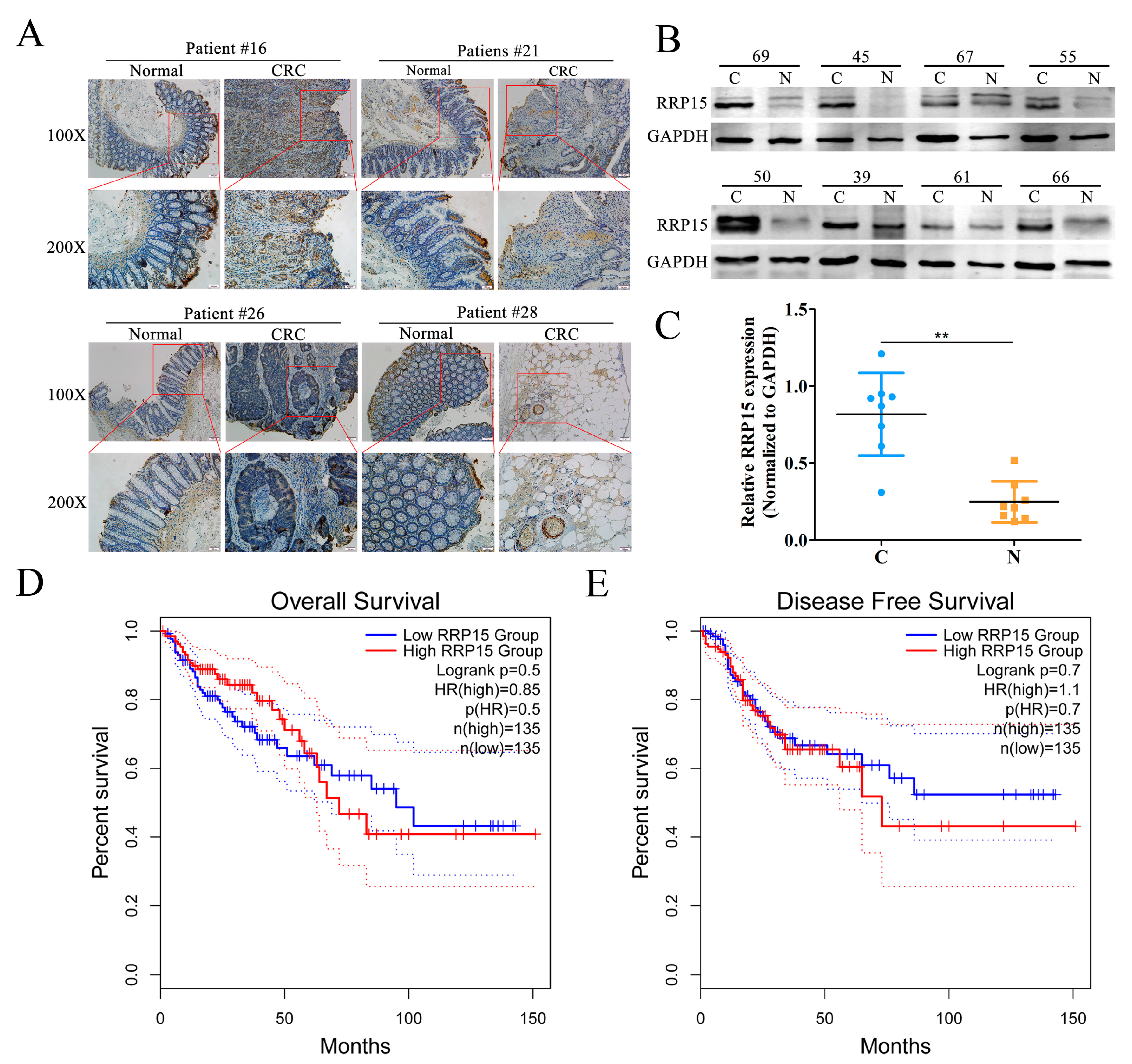
2.1. RRP15 Expression Was Markedly Increased in Human Clinical CC Samples, and Correlated with Poor Prognosis
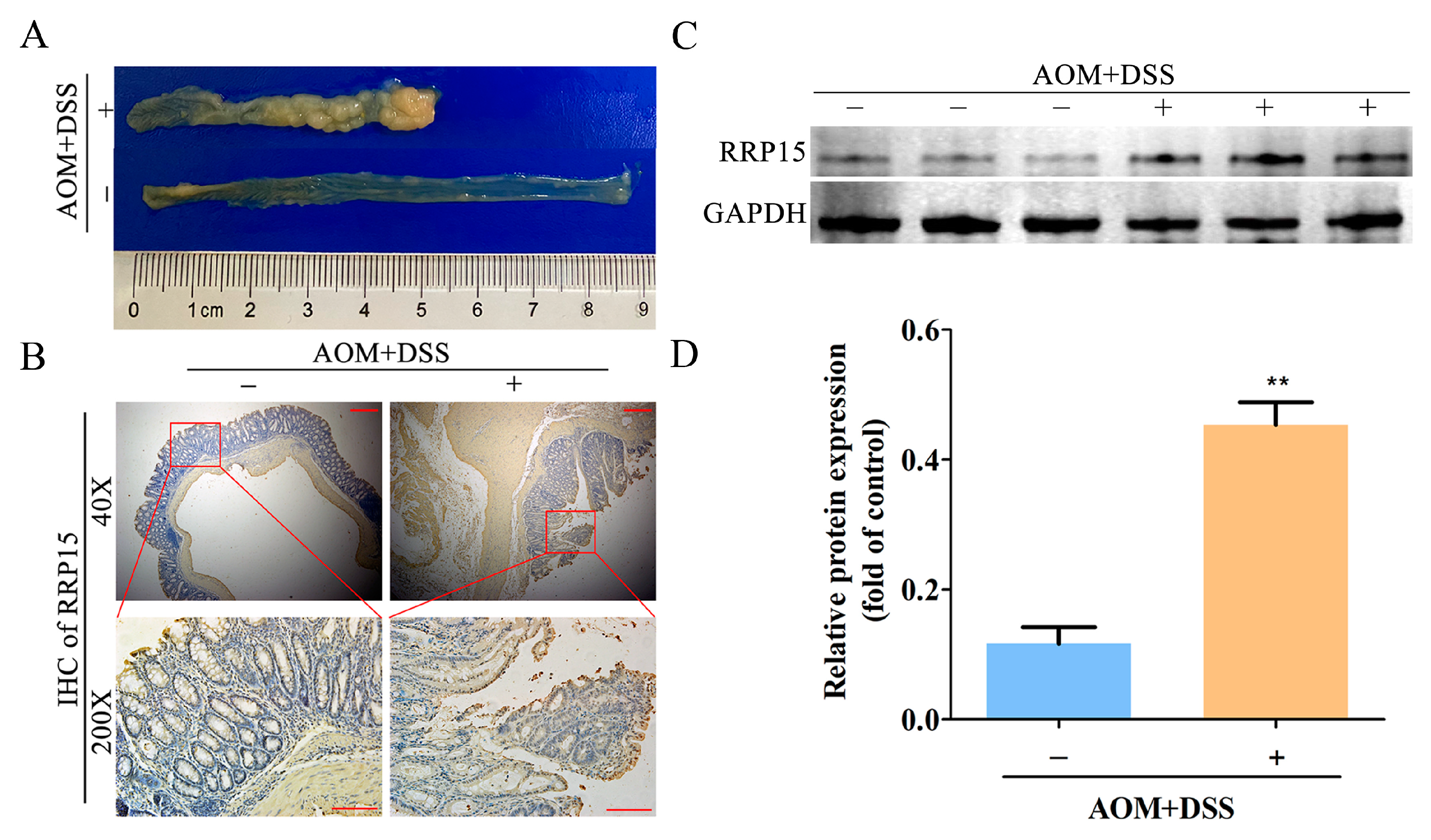
2.2. High RRP15 Expression Identified in Colon Tissues of AOM/DSS-Induced Mice CAC Model
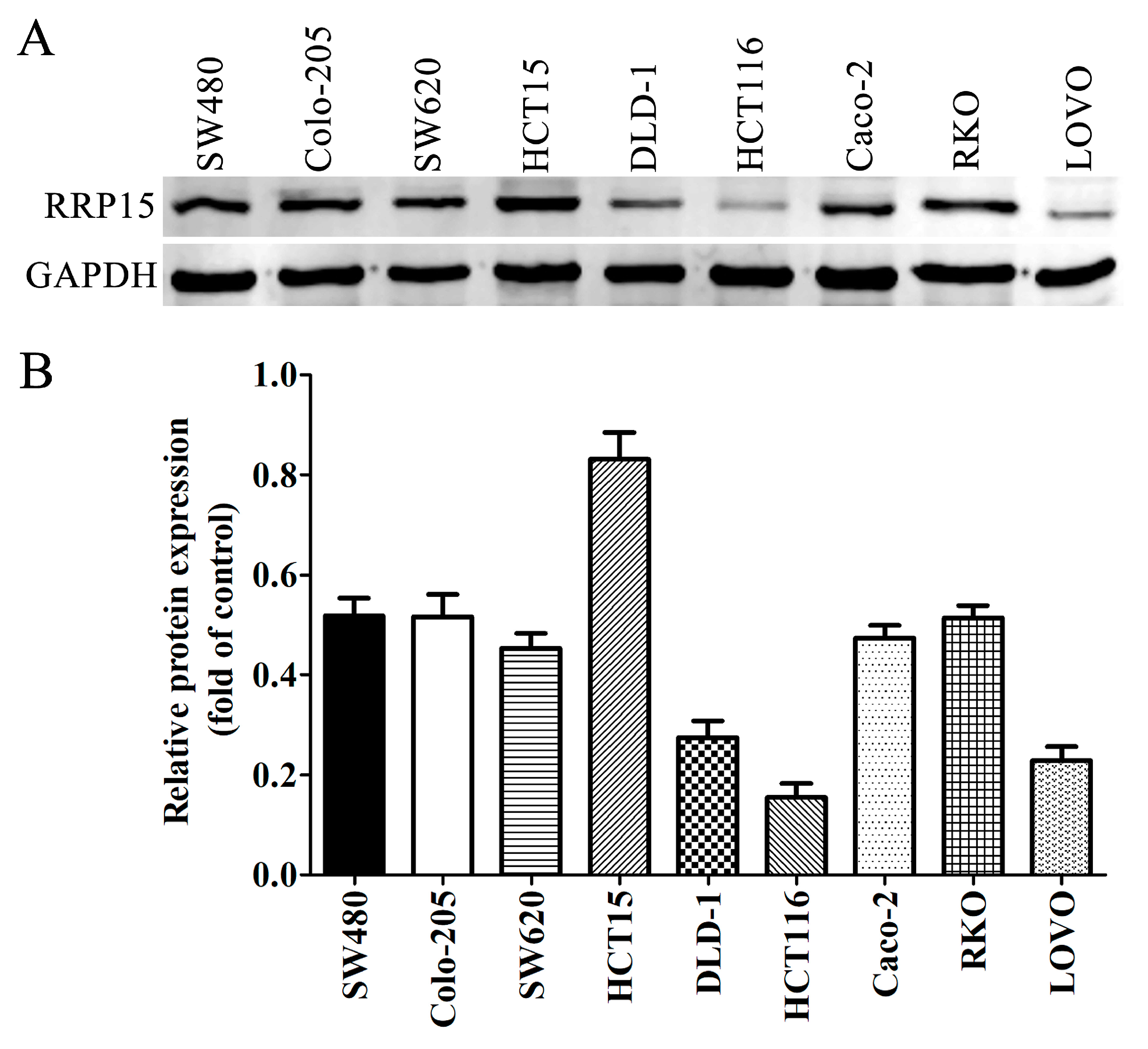
2.3. RRP15 Protein Expression in CC Cell Lines
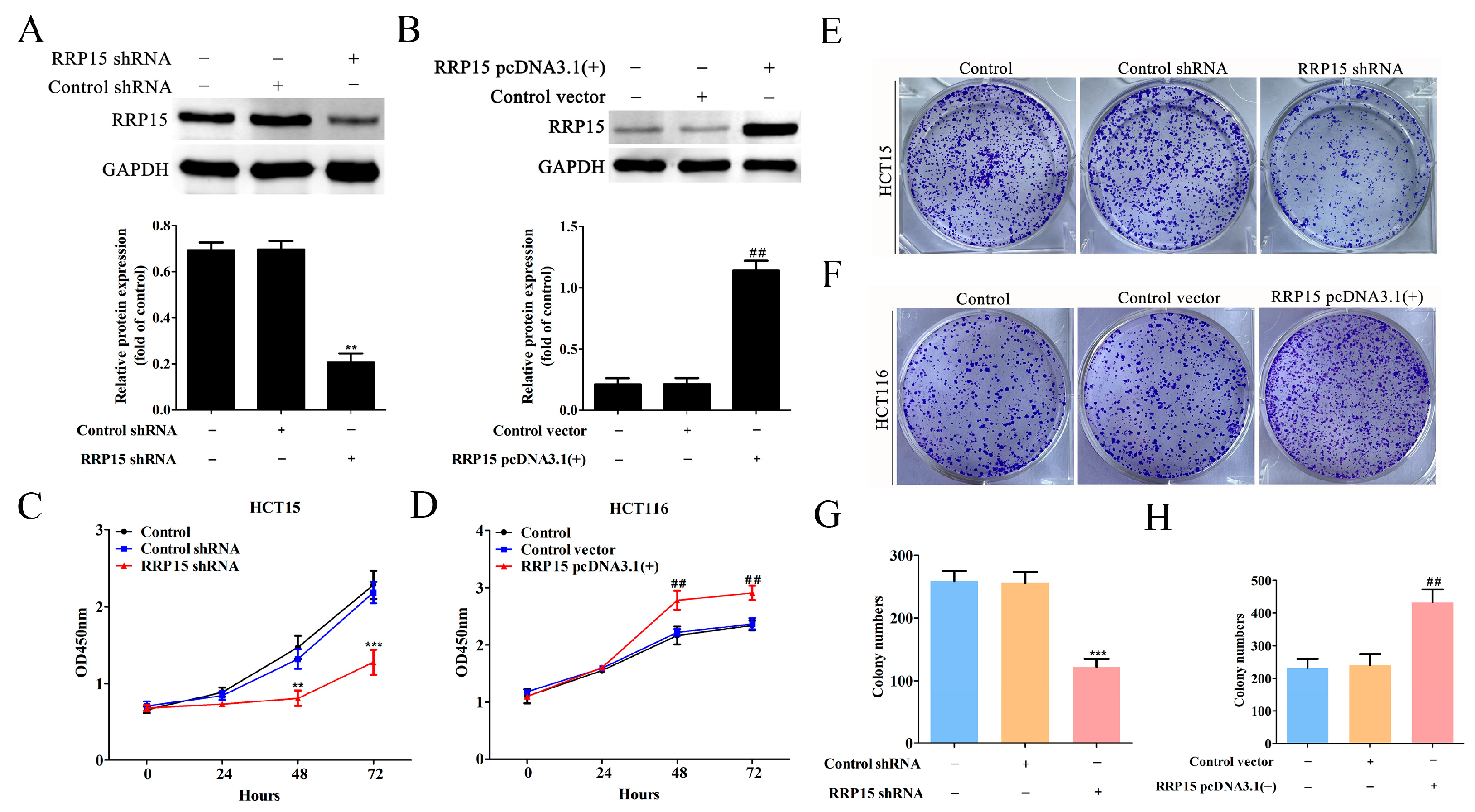
2.4. RRP15 Knockdown Inhibited CC Cell Growth and Colony Formation In Vitro
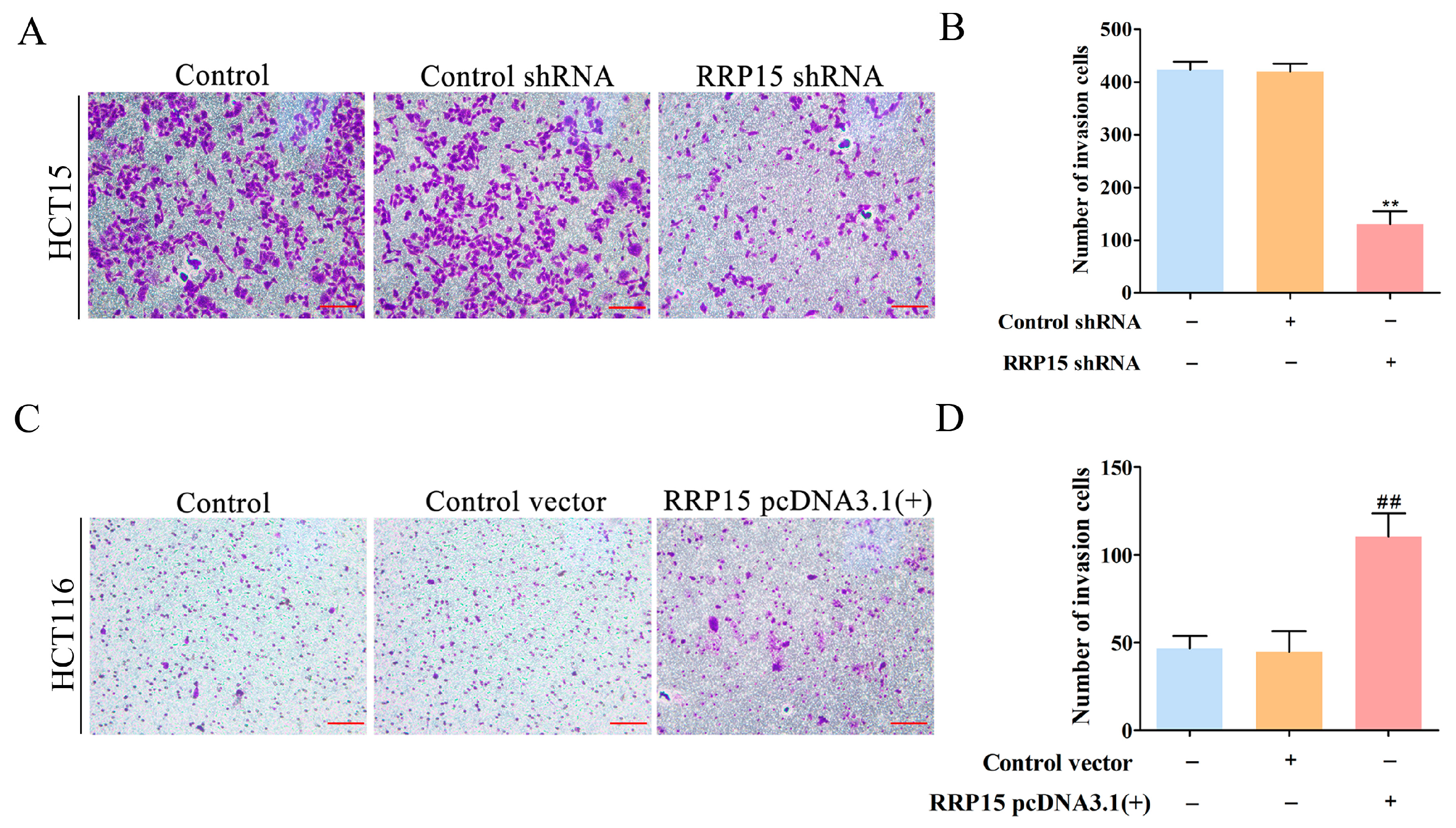
2.5. RRP15 Knockdown Suppressed CC Cells Invasion In Vitro
2.6. RRP15 Knockdown Suppressed CC Cells Growth In Vivo
2.7. RRP15 Expression Positively Correlated with EMT in Human Clinical CC Samples, and RRP15 Knockdown Inhibited EMT Process of CC Cells In Vitro
3. Discussion
4. Materials and Methods
4.1. Clinical Specimens
4.2. Cell Culture
4.3. Plasmid Construction and Transfection
4.4. Cell Count Kit-8 (CCK-8) Assay
4.5. Colony Formation Assay
4.6. Transwell Invasion Assay
4.7. Immunohistochemical Staining (IHC)
4.8. Western Blotting
4.9. CC Xenografts Mice Model
4.10. Establishment of Azoxymethane (AOM)/Dextran sulphate sodium (DSS)-Induced Mice Colitis-Associated Cancer (CAC) Model
4.11. Gene Correlation Analysis in GEPIA
4.12. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, Z.; Lu, Z.; Li, L.; Ma, M.; Long, F.; Wu, R.; Huang, L.; Chou, J.; Yang, K.; Zhang, Y.; et al. Identification and validation of ferroptosis-related lncRNA signatures as a novel prognostic model for colon cancer. Front. Immunol. 2022, 12, 783362. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, T.; Cai, X.; Dong, J.; Xia, C.; Zhou, Y.; Ding, R.; Yang, R.; Tan, J.; Zhang, L.; et al. Neoadjuvant immunotherapy for MSI-H/dMMR locally advanced colorectal cancer: New strategies and unveiled opportunities. Front. Immunol. 2022, 13, 795972. [Google Scholar] [CrossRef]
- Jaaks, P.; Coker, E.A.; Vis, D.J.; Edwards, O.; Carpenter, E.F.; Leto, S.M.; Dwane, L.; Sassi, F.; Lightfoot, H.; Barthorpe, S.; et al. Effective drug combinations in breast, colon and pancreatic cancer cells. Nature 2022, 603, 166–173. [Google Scholar] [CrossRef]
- Yu, C.; Luo, D.; Yu, J.; Zhang, M.; Zheng, X.; Xu, G.; Wang, J.; Wang, H.; Xu, Y.; Jiang, K.; et al. Genome-wide CRISPR-cas9 knockout screening identifies GRB7 as a driver for MEK inhibitor resistance in KRAS mutant colon cancer. Oncogene 2022, 41, 191–203. [Google Scholar] [CrossRef]
- Chen, M.C.; Nhan, D.C.; Hsu, C.H.; Wang, T.F.; Li, C.C.; Ho, T.J.; Mahalakshmi, B.; Chen, M.C.; Yang, L.Y.; Huang, C.Y.; et al. SENP1 participates in Irinotecan resistance in human colon cancer cells. J. Cell. Biochem. 2021, 122, 1277–1294. [Google Scholar] [CrossRef]
- De Marchis, M.L.; Giorgi, A.; Schininà, M.E.; Bozzoni, I.; Fatica, A. Rrp15p, a novel component of pre-ribosomal particles required for 60S ribosome subunit maturation. RNA 2005, 11, 495–502. [Google Scholar] [CrossRef]
- Dong, Z.; Zhu, C.; Zhan, Q.; Jiang, W. The roles of RRP15 in nucleolar formation, ribosome biogenesis and checkpoint control in human cells. Oncotarget 2017, 8, 13240–13252. [Google Scholar] [CrossRef]
- Wu, T.; Ren, M.X.; Chen, G.P.; Jin, Z.M.; Wang, G. Rrp15 affects cell cycle, proliferation, and apoptosis in NIH3T3 cells. FEBS Open Bio 2016, 6, 1085–1092. [Google Scholar] [CrossRef]
- Bhaskar, V.; Desogus, J.; Graff-Meyer, A.; Schenk, A.D.; Cavadini, S.; Chao, J.A. Dynamic association of human Ebp1 with the ribosome. RNA 2021, 27, 411–419. [Google Scholar] [CrossRef]
- Ferguson, B.; Handoko, H.Y.; Mukhopadhyay, P.; Chitsazan, A.; Balmer, L.; Morahan, G.; Walker, G.J. Different genetic mechanisms mediate spontaneous versus UVR-induced malignant melanoma. ELife 2019, 8, e42424. [Google Scholar] [CrossRef]
- Zhao, D.; Qian, L.; Zhuang, D.; Wang, L.; Cao, Y.; Zhou, F.; Zhang, S.; Liu, Y.; Liang, Y.; Zhang, W.; et al. Inhibition of ribosomal RNA processing 15 Homolog (RRP15), which is overexpressed in hepatocellular carcinoma, suppresses tumour growth via induction of senescence and apoptosis. Cancer Lett. 2021, 519, 315–327. [Google Scholar] [CrossRef]
- Tian, M.; Wang, X.; Sun, J.; Lin, W.; Chen, L.; Liu, S.; Wu, X.; Shi, L.; Xu, P.; Cai, X. IRF3 prevents colorectal tumorigenesis via inhibiting the nuclear translocation of β-catenin. Nat. Commun. 2020, 11, 5762. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Wu, F.; Ying, K.; Xu, Y.; Shan, L.; Lv, Y.; Gao, X.; Xu, D.; Lu, J.; Xie, B. Therapeutic effects of dihydroartemisinin in multiple stages of colitis-associated colorectal cancer. Theranostics 2021, 11, 6225–6239. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ding, K.; Wang, Y.; Yan, T.; Xu, Y.; Deng, Z.; Lin, W.; Zhang, L.; Zhu, W.; Zhao, R.; et al. Wumei Pill ameliorates AOM/DSS-induced colitis-associated colon cancer through inhibition of inflammation and oxidative stress by regulating S-Adenosylhomocysteine hydrolase- (AHCY-) mediated hedgehog signaling in mice. Oxid. Med. Cell. Longev. 2022, 2022, 4061713. [Google Scholar] [CrossRef]
- Lin, T.A.; Lin, W.S.; Chou, Y.C.; Nagabhushanam, K.; Ho, C.T.; Pan, M.H. Oxyresveratrol inhibits human colon cancer cell migration through regulating epithelial-mesenchymal transition and microRNA. Food Funct. 2021, 12, 9658–9668. [Google Scholar] [CrossRef]
- Liang, F.; Lv, Y.; Qiao, X.; Zhang, S.; Shen, S.; Wang, C.; Xu, G.; Zou, X.; Wang, L.; Zhang, B. Cinchonine-induced cell death in pancreatic cancer cells by downregulating RRP15. Cell Biol. Int. 2023. [Google Scholar] [CrossRef]
- Lungulescu, C.; Ghimpau, V.; Gheonea, D.I.; Sur, D.; Lungulescu, C.V. The Role of neuropilin-2 in the epithelial to mesenchymal transition of colorectal cancer: A systematic review. Biomedicines 2022, 10, 172. [Google Scholar] [CrossRef]
- Mullins, R.D.Z.; Pal, A.; Barrett, T.F.; Heft Neal, M.E.; Puram, S.V. Epithelial-mesenchymal plasticity in tumor immune evasion. Cancer Res. 2022, 82, 2329–2343. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhang, Y.; Dai, H.; Han, B. Epithelial-mesenchymal transition-mediated tumor therapeutic resistance. Molecules 2022, 27, 4750. [Google Scholar] [CrossRef]
- Pan, Z.; Cai, J.; Lin, J.; Zhou, H.; Peng, J.; Liang, J.; Xia, L.; Yin, Q.; Zou, B.; Zheng, J.; et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol. Cancer 2020, 19, 71. [Google Scholar] [CrossRef]
- Gupta, R.; Bhatt, L.K.; Johnston, T.P.; Prabhavalkar, K.S. Colon cancer stem cells: Potential target for the treatment of colorectal cancer. Cancer Biol. Ther. 2019, 20, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Han, X.; Wei, B.; Fang, J.; Wei, H. RSPO2 enriches LGR5(+) spheroid colon cancer stem cells and promotes its metastasis by epithelial-mesenchymal transition. Am. J. Transl. Res. 2016, 8, 354–364. [Google Scholar] [PubMed]
- Acikgoz, E.; Tatar, C.; Oktem, G. Triptolide inhibits CD133+/CD44+ colon cancer stem cell growth and migration through triggering apoptosis and represses epithelial-mesenchymal transition via downregulating expressions of snail, slug, and twist. J. Cell. Biochem. 2020, 121, 3313–3324. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Long, C.; Wang, F.; Lou, B.; Yuan, M.; Duan, F.; Yang, Y.; Li, J.; Qian, X.; Zeng, J.; et al. METTL3 attenuates proliferative vitreoretinopathy and epithelial-mesenchymal transition of retinal pigment epithelial cells via wnt/β-catenin pathway. J. Cell. Mol. Med. 2021, 25, 4220–4234. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ko, D.; Yoon, J.; Lee, Y.; Kim, S. TMEM52B suppression promotes cancer cell survival and invasion through modulating E-cadherin stability and EGFR activity. J. Exp. Clin. Cancer Res. 2021, 40, 58. [Google Scholar] [CrossRef] [PubMed]
- Mathur, P.S.; Gierut, J.J.; Guzman, G.; Xie, H.; Xicola, R.M.; Llor, X.; Chastkofsky, M.I.; Perekatt, A.O.; Tyner, A.L. Kinase-dependent and -independent roles for PTK6 in colon cancer. Mol. Cancer Res. 2016, 14, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Liu, J.; Zhao, L.; Zhang, P.; Wang, W.; Shou, D.; Ji, J.; Li, C.; Chai, K.; Dong, Y. Inoscavin A, a pyrone compound isolated from a Sanghuangporus vaninii extract, inhibits colon cancer cell growth and induces cell apoptosis via the hedgehog signaling pathway. Phytomedicine 2022, 96, 153852. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.H.; Zhou, H.; Cooper, L.; Huang, J.L.; Zhu, S.B.; Zhao, X.X.; Ding, H.; Pan, Y.L.; Rong, L. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front. Immunol. 2019, 10, 6. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, Z.; Xu, Y.; Cai, Y.; Lin, W.; Zhang, L.; Jiang, A.; Zhou, Y.; Zhao, R.; Zhao, H.; Liu, Z.; et al. Inhibition of Ribosomal RNA Processing 15 Homolog (RRP15) Suppressed Tumor Growth, Invasion and Epithelial to Mesenchymal Transition (EMT) of Colon Cancer. Int. J. Mol. Sci. 2023, 24, 3528. https://doi.org/10.3390/ijms24043528
Deng Z, Xu Y, Cai Y, Lin W, Zhang L, Jiang A, Zhou Y, Zhao R, Zhao H, Liu Z, et al. Inhibition of Ribosomal RNA Processing 15 Homolog (RRP15) Suppressed Tumor Growth, Invasion and Epithelial to Mesenchymal Transition (EMT) of Colon Cancer. International Journal of Molecular Sciences. 2023; 24(4):3528. https://doi.org/10.3390/ijms24043528
Chicago/Turabian StyleDeng, Zirong, Yun Xu, Yuchen Cai, Weiling Lin, Libei Zhang, Aoqing Jiang, Yuhang Zhou, Rui Zhao, Heyan Zhao, Zhaoguo Liu, and et al. 2023. "Inhibition of Ribosomal RNA Processing 15 Homolog (RRP15) Suppressed Tumor Growth, Invasion and Epithelial to Mesenchymal Transition (EMT) of Colon Cancer" International Journal of Molecular Sciences 24, no. 4: 3528. https://doi.org/10.3390/ijms24043528
APA StyleDeng, Z., Xu, Y., Cai, Y., Lin, W., Zhang, L., Jiang, A., Zhou, Y., Zhao, R., Zhao, H., Liu, Z., & Yan, T. (2023). Inhibition of Ribosomal RNA Processing 15 Homolog (RRP15) Suppressed Tumor Growth, Invasion and Epithelial to Mesenchymal Transition (EMT) of Colon Cancer. International Journal of Molecular Sciences, 24(4), 3528. https://doi.org/10.3390/ijms24043528






