A Comprehensive Virulence and Resistance Characteristics of Listeria monocytogenes Isolated from Fish and the Fish Industry Environment
Abstract
:1. Introduction
2. Results
2.1. MLST Analysis
2.2. cgMLST Analysis
2.3. Assessment of Virulence Factor Genotypes across Different Sublineages
2.4. Antimicrobial Resistance and Stress Tolerance Genes
2.5. Comparison of Isolates from Food and A Food-Production Environment with Human L. monocytogenes
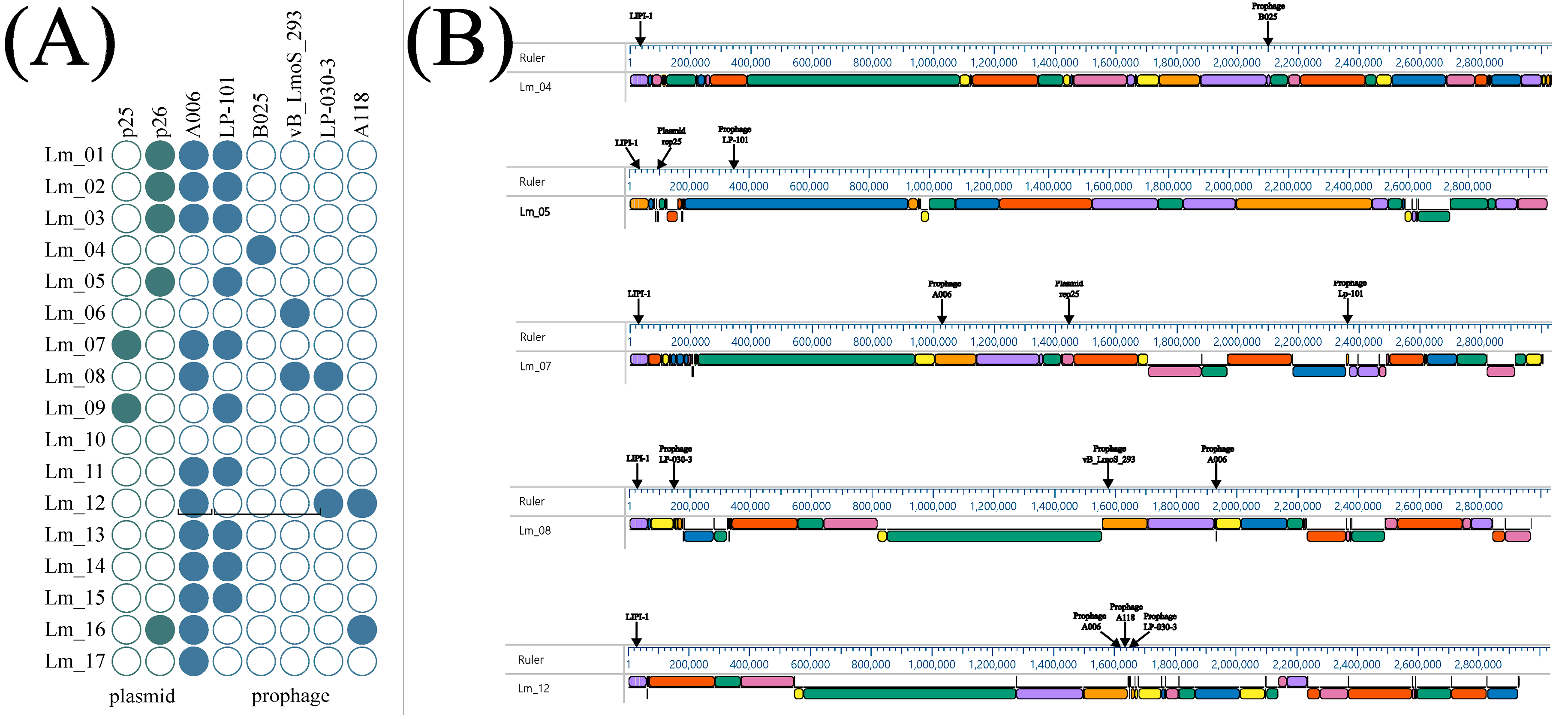
2.6. Detection of Prophage Regions and Plasmid
2.7. Collinearity Analysis
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Library Preparation and Sequencing
4.3. MLST and cgMLST Characterization
4.4. Identification of Virulence-, Antimicrobial-, and Stress-Related Genes
4.5. Prophage and Plasmids Identification
4.6. Collinearity Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tiensuu, T.; Guerreiro, D.N.; Oliveira, A.H.; O’Byrne, C.; Johansson, J. Flick of a switch: Regulatory mechanisms allowing listeria monocytogenes to transition from a saprophyte to a killer. Microbiology 2019, 165, 819–833. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.H.; Park, Y.J.; Kim, M.; Seo, Y.H.; Kim, Y.A.; Choi, J.Y.; Yong, D.; Jeong, S.H.; Lee, K. Increasing Incidence of Listeriosis and Infection-associated Clinical Outcomes. Ann. Lab. Med. 2018, 38, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Bland, R.; Brown, S.R.B.; Waite-Cusic, J.; Kovacevic, J. Probing antimicrobial resistance and sanitizer tolerance themes and their implications for the food industry through the Listeria monocytogenes lens. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1777–1802. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, R.; Halbedel, S.; Lüth, S.; Holzer, A.; Adler, M.; Pietzka, A.; Al Dahouk, S.; Stark, K.; Flieger, A.; Kleta, S.; et al. Invasive listeriosis outbreaks and salmon products: A genomic, epidemiological study. Emerg. Microbes Infect. 2022, 11, 1308–1315. [Google Scholar] [CrossRef]
- Lambertz, S.T.; Nilsson, C.; Brådenmark, A.; Sylvén, S.; Johansson, A.; Jansson, L.M.; Lindblad, M. Prevalence and level of Listeria monocytogenes in ready-to-eat foods in Sweden 2010. Int. J. Food Microbiol. 2012, 160, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Valladares, G.; Danielsson-Tham, M.L.; Tham, W. Implicated Food Products for Listeriosis and Changes in Serovars of Listeria monocytogenes Affecting Humans in Recent Decades. Foodborne Pathog. Dis. 2018, 15, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; Nørrung, B.; et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018, 16, e05134. [Google Scholar] [CrossRef]
- Kurpas, M.; Wieczorek, K.; Osek, J. Ready-to-eat Meat Products As a Source of Listeria Monocytogenes. J. Vet. Res. 2018, 62, 49–55. [Google Scholar] [CrossRef]
- Gillesberg Lassen, S.; Ethelberg, S.; Björkman, J.T.; Jensen, T.; Sørensen, G.; Kvistholm Jensen, A.; Müller, L.; Nielsen, E.M.; Mølbak, K. Two listeria outbreaks caused by smoked fish consumption—Using whole-genome sequencing for outbreak investigations. Clin. Microbiol. Infect. 2016, 22, 620–624. [Google Scholar] [CrossRef]
- Lunestad, B.T.; Truong, T.T.T.; Lindstedt, B.A. A multiple-locus variable-number tandem repeat analysis (MLVA) of Listeria monocytogenes isolated from Norwegian salmon-processing factories and from listeriosis patients. Epidemiol. Infect. 2013, 141, 2101–2110. [Google Scholar] [CrossRef]
- Lecuit, M. Listeria monocytogenes, a model in infection biology. Cell Microbiol. 2020, 22, e13186. [Google Scholar] [CrossRef]
- Chen, J.; Luo, X.; Jiang, L.; Jin, P.; Wei, W.; Liu, D.; Fang, W. Molecular characteristics and virulence potential of Listeria monocytogenes isolates from Chinese food systems. Food Microbiol. 2009, 26, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Cossart, P.; Toledo-Arana, A. Listeria monocytogenes, a unique model in infection biology: An overview. Microbes Infect. 2008, 10, 1041–1050. [Google Scholar] [CrossRef]
- Ireton, K.; Mortuza, R.; Gyanwali, G.C.; Gianfelice, A.; Hussain, M. Role of internalin proteins in the pathogenesis of Listeria monocytogenes. Mol. Microbiol. 2021, 116, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Quereda, J.J.; Morón-García, A.; Palacios-Gorba, C.; Dessaux, C.; García-del Portillo, F.; Pucciarelli, M.G.; Ortega, A.D. Pathogenicity and virulence of Listeria monocytogenes: A trip from environmental to medical microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef] [PubMed]
- Quereda, J.J.; Dussurget, O.; Nahori, M.A.; Ghozlane, A.; Volant, S.; Dillies, M.A.; Regnault, B.; Kennedy, S.; Mondot, S.; Villoing, B.; et al. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. USA 2016, 113, 5706–5711. [Google Scholar] [CrossRef] [PubMed]
- Vilchis-Rangel, R.E.; Espinoza-Mellado, M. del R.; Salinas-Jaramillo, I.J.; Martinez-Peña, M.D.; Rodas-Suárez, O.R. Association of Listeria monocytogenes LIPI-1 and LIPI-3 marker llsX with invasiveness. Curr. Microbiol. 2019, 76, 637–643. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308. [Google Scholar] [CrossRef]
- Schiavano, G.F.; Ateba, C.N.; Petruzzelli, A.; Mele, V.; Amagliani, G.; Guidi, F.; De Santi, M.; Pomilio, F.; Blasi, G.; Gattuso, A.; et al. Whole-Genome Sequencing Characterization of Virulence Profiles of Listeria monocytogenes Food and Human Isolates and In Vitro Adhesion/Invasion Assessment. Microorganisms 2022, 10, 62. [Google Scholar] [CrossRef]
- Song, W.; Sun, H.X.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.; Wang, D.; Wang, Y.; Hu, M.; Liu, W.; et al. Prophage Hunter: An integrative hunting tool for active prophages. Nucleic Acids Res. 2019, 47, W74–W80. [Google Scholar] [CrossRef] [Green Version]
- Heinitz, M.L.; Johnson, J.M. The incidence of Listeria spp., Salmonella spp., and Clostridium botulinum in smoked fish and shellfish. J. Food Prot. 1998, 61, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.K.; Siitonen, A.; Heiskanen, P.; Haajanen, H.; Björkroth, K.J.; Korkeala, H.J. Molecular Epidemiology of an Outbreak of Febrile Gastroenteritis Caused by Listeria monocytogenes in Cold-Smoked Rainbow Trout. J. Clin. Microbiol. 1999, 37, 2358. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.; Zaiser, A.; Leitner, R.; Quijada, N.M.; Pracser, N.; Pietzka, A.; Ruppitsch, W.; Schmitz-Esser, S.; Wagner, M.; Rychli, K. Virulence characterization and comparative genomics of Listeria monocytogenes sequence type 155 strains. BMC Genomics 2020, 21, 847. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ji, Q.; Li, S.; Liu, M. Prevalence and Genetic Diversity of Listeria monocytogenes Isolated From Retail Pork in Wuhan, China. Front. Microbiol. 2021, 12, 459. [Google Scholar] [CrossRef]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef]
- Hurley, D.; Luque-Sastre, L.; Parker, C.T.; Huynh, S.; Eshwar, A.K.; Nguyen, S.V.; Andrews, N.; Moura, A.; Fox, E.M.; Jordan, K.; et al. Whole-Genome Sequencing-Based Characterization of 100 Listeria monocytogenes Isolates Collected from Food Processing Environments over a Four-Year Period. mSphere 2019, 4, e00252-19. [Google Scholar] [CrossRef] [PubMed]
- Thomassen, G.M.B.; Krych, L.; Knøchel, S.; Mehli, L. ON-rep-seq as a rapid and cost-effective alternative to whole-genome sequencing for species-level identification and strain-level discrimination of Listeria monocytogenes contamination in a salmon processing plant. Microbiologyopen 2021, 10, e1246. [Google Scholar] [CrossRef]
- Wieczorek, K.; Bomba, A.; Osek, J. Whole-Genome Sequencing-Based Characterization of Listeria monocytogenes from Fish and Fish Production Environments in Poland. Int. J. Mol. Sci. 2020, 21, 9419. [Google Scholar] [CrossRef]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef]
- Gram, L. Potential Hazards in Cold-Smoked Fish: Listeria monocytogenes. J. Food Sci. 2001, 66, S1072–S1081. [Google Scholar] [CrossRef]
- Rahman, S.; Das, A.K. A subtractive proteomics and immunoinformatics approach towards designing a potential multi-epitope vaccine against pathogenic Listeria monocytogenes. Microb. Pathog. 2022, 172, 105782. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Bernal, G.; Müller-Altrock, S.; González-Zorn, B.; Scortti, M.; Herrmann, P.; Monzó, H.J.; Lacharme, L.; Kreft, J.; Vázquez-Boland, J.A. A spontaneous genomic deletion in Listeria ivanovii identifies LIPI-2, a species-specific pathogenicity island encoding sphingomyelinase and numerous internalins. Mol. Microbiol. 2006, 59, 415–432. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yao, H.; Doijad, S.; Kong, S.; Shen, Y.; Cai, X.; Tan, W.; Wang, Y.; Feng, Y.; Ling, Z.; et al. A hybrid sub-lineage of Listeria monocytogenes comprising hypervirulent isolates. Nat. Commun. 2019, 10, 4283. [Google Scholar] [CrossRef]
- Cotter, P.D.; Draper, L.A.; Lawton, E.M.; Daly, K.M.; Groeger, D.S.; Casey, P.G.; Ross, R.P.; Hill, C. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008, 4, e1000144. [Google Scholar] [CrossRef]
- Guidi, F.; Chiaverini, A.; Repetto, A.; Lorenzetti, C.; Centorotola, G.; Bazzucchi, V.; Palombo, B.; Gattuso, A.; Pomilio, F.; Blasi, G. Hyper-Virulent Listeria monocytogenes Strains Associated With Respiratory Infections in Central Italy. Front. Cell Infect. Microbiol. 2021, 11, 765540. [Google Scholar] [CrossRef]
- Palaiodimou, L.; Fanning, S.; Fox, E.M. Genomic insights into persistence of Listeria species in the food processing environment. J. Appl. Microbiol. 2021, 131, 2082–2094. [Google Scholar] [CrossRef] [PubMed]
- Olier, M.; Pierre, F.; Rousseaux, S.; Lemaitre, J.P.; Rousset, A.; Piveteau, P.; Guzzo, J. Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 2003, 71, 1217–1224. [Google Scholar] [CrossRef]
- Van Stelten, A.; Nightingale, K.K. Development and implementation of a multiplex single-nucleotide polymorphism genotyping assay for detection of virulence-attenuating mutations in the Listeria monocytogenes virulence-associated gene inlA. Appl. Environ. Microbiol. 2008, 74, 7365–7375. [Google Scholar] [CrossRef]
- Li, L.; Olsen, R.H.; Ye, L.; Wang, W.; Shi, L.; Yan, H.; Meng, H. Characterization of Antimicrobial Resistance of Listeria monocytogenes Strains Isolated from a Pork Processing Plant and Its Respective Meat Markets in Southern China. Foodborne Pathog. Dis. 2016, 13, 262–268. [Google Scholar] [CrossRef]
- Wilson, A.; Gray, J.; Scott Chandry, P.; Fox, E.M. Phenotypic and Genotypic Analysis of Antimicrobial Resistance among Listeria monocytogenes Isolated from Australian Food Production Chains. Genes 2018, 9, 80. [Google Scholar] [CrossRef] [Green Version]
- Mafuna, T.; Matle, I.; Magwedere, K.; Pierneef, R.E.; Reva, O.N. Whole Genome-Based Characterization of Listeria monocytogenes Isolates Recovered From the Food Chain in South Africa. Front. Microbiol. 2021, 12, 669287. [Google Scholar] [CrossRef] [PubMed]
- Kurpas, M.; Osek, J.; Moura, A.; Leclercq, A.; Lecuit, M.; Wieczorek, K. Genomic Characterization of Listeria monocytogenes Isolated From Ready-to-Eat Meat and Meat Processing Environments in Poland. Front. Microbiol. 2020, 11, 1412. [Google Scholar] [CrossRef] [PubMed]
- Schar, D.; Klein, E.Y.; Laxminarayan, R.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in aquaculture. Sci. Reports 2020, 10, 21878. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist. Updat. 2010, 13, 151–171. [Google Scholar] [CrossRef]
- Sacha, P.; Jaworowska, J.; Ojdana, D.; Wieczorek, P.; Czaban, S.; Tryniszewska, E. Occurrence of the aacA4 gene among multidrug resistant strains of Pseudomonas aeruginosa isolated from bronchial secretions obtained from the Intensive Therapy Unit at University Hospital in Bialystok, Poland. Folia Histochem. Cytobiol. 2012, 50, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Hingston, P.; Brenner, T.; Hansen, L.T.; Wang, S. Comparative Analysis of Listeria monocytogenes Plasmids and Expression Levels of Plasmid-Encoded Genes during Growth under Salt and Acid Stress Conditions. Toxins 2019, 11, 426. [Google Scholar] [CrossRef]
- Harvey, J.; Gilmour, A. Characterization of recurrent and sporadic Listeria monocytogenes isolates from raw milk and nondairy foods by pulsed-field gel electrophoresis, monocin typing, plasmid profiling, and cadmium and antibiotic resistance determination. Appl. Environ. Microbiol. 2001, 67, 840–847. [Google Scholar] [CrossRef]
- Canchaya, C.; Giubellini, V.; Ventura, M.; De Los Reyes-Gavilán, C.G.; Margolles, A. Mosaic-Like Sequences Containing Transposon, Phage, and Plasmid Elements among Listeria monocytogenes Plasmids. Appl. Environ. Microbiol. 2010, 76, 4851. [Google Scholar] [CrossRef]
- Elhanafi, D.; Utta, V.; Kathariou, S. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998-1999 outbreak. Appl. Environ. Microbiol. 2010, 76, 8231–8238. [Google Scholar] [CrossRef]
- Katharios-Lanwermeyer, S.; Rakic-Martinez, M.; Elhanafi, D.; Ratani, S.; Tiedje, J.M.; Kathariou, S. Coselection of cadmium and benzalkonium chloride resistance in conjugative transfers from nonpathogenic Listeria spp. to other Listeriae. Appl. Environ. Microbiol. 2012, 78, 7549–7556. [Google Scholar] [CrossRef] [Green Version]
- Ryu, J.; Park, S.H.; Yeom, Y.S.; Shrivastav, A.; Lee, S.H.; Kim, Y.R.; Kim, H.Y. Simultaneous detection of Listeria species isolated from meat processed foods using multiplex PCR. Food Control 2013, 32, 659–664. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Marín, O.; González, B.; Poupin, M.J. From Microbial Dynamics to Functionality in the Rhizosphere: A Systematic Review of the Opportunities With Synthetic Microbial Communities. Front. Plant Sci. 2021, 12, 843. [Google Scholar] [CrossRef] [PubMed]
- Shayanthan, A.; Ordoñez, P.A.C.; Oresnik, I.J. The Role of Synthetic Microbial Communities (SynCom) in Sustainable Agriculture. Front. Agron. 2022, 4, 58. [Google Scholar] [CrossRef]
- Felício, M.T.S.; Hogg, T.; Gibbs, P.; Teixeira, P.; Wiedmann, M. Recurrent and sporadic Listeria monocytogenes contamination in alheiras represents considerable diversity, including virulence-attenuated isolates. Appl. Environ. Microbiol. 2007, 73, 3887–3895. [Google Scholar] [CrossRef]
- Gelbíčová, T.; Koláčková, I.; Pantůček, R.; Karpíšková, R. A novel mutation leading to a premature stop codon in inlA of Listeria monocytogenes isolated from neonatal listeriosis. New Microbiol. 2015, 38, 293–296. [Google Scholar] [PubMed]
- Handa-Miya, S.; Kimura, B.; Takahashi, H.; Sato, M.; Ishikawa, T.; Igarashi, K.; Fujii, T. Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int. J. Food Microbiol. 2007, 117, 312–318. [Google Scholar] [CrossRef]
- Jonquières, R.; Bierne, H.; Mengaud, J.; Cossart, P. The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 1998, 66, 3420–3422. [Google Scholar] [CrossRef]
- Nightingale, K.K.; Windham, K.; Martin, K.E.; Yeung, M.; Wiedmann, M. Select Listeria monocytogenes subtypes commonly found in foods carry disctinct nonsense mutations in inlA. Appl. Environ. Microbiol. 2005, 71, 8764–8772. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [Green Version]
- Rousseaux, S.; Olier, M.; Lemaître, J.P.; Piveteau, P.; Guzzo, J. Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 2004, 70, 2180–2185. [Google Scholar] [CrossRef] [PubMed]
- Van Stelten, A.; Simpson, J.M.; Ward, T.J.; Nightingale, K.K. Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Appl. Environ. Microbiol. 2010, 76, 2783–2790. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| No. | Source | Year of Isolation | Clonal Complex | Sublineage | cgMLST Type | Serogroup |
|---|---|---|---|---|---|---|
| Lm_1 | smoked salmon | 2018 | CC8 | SL8 | CT1151 | IIa |
| Lm_2 | smoked salmon | 2018 | CC8 | SL8 | CT1151 | IIa |
| Lm_3 | smoked salmon | 2018 | CC121 | SL121 | CT909 | IIa |
| Lm_4 | food processing premises | 2018 | CC101 | SL101 | CT11711 | IIa |
| Lm_5 | smoked salmon | 2018 | CC121 | SL121 | CT909 | IIa |
| Lm_6 | food processing premises | 2018 | CC59 | SL59 | CT2283 | IIb |
| Lm_7 | raw salmon | 2018 | CC193 | SL193 | CT11716 | IIa |
| Lm_8 | raw salmon | 2018 | CC6 | SL6 | CT5641 | IVb |
| Lm_9 | smoked salmon | 2018 | CC31 | SL31 | CT7227 | IIa |
| Lm_10 | food processing premises | 2018 | CC77 | SL77 | CT11718 | IIb |
| Lm_11 | smoked salmon | 2018 | CC8 | SL8 | CT1151 | IIa |
| Lm_12 | food processing premises | 2018 | CC37 | SL37 | CT11717 | IIa |
| Lm_13 | food processing premises | 2018 | CC6 | SL6 | CT443 | IVb |
| Lm_14 | raw salmon | 2018 | CC8 | SL8 | CT7220 | IIa |
| Lm_15 | raw salmon | 2018 | CC9 | SL9 | CT1824 | IIc |
| Lm_16 | smoked salmon | 2018 | CC121 | SL121 | CT893 | IIa |
| Lm_17 | smoked salmon | 2018 | CC87 | SL87 | CT58 | IIb |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakrzewski, A.J.; Kurpas, M.; Zadernowska, A.; Chajęcka-Wierzchowska, W.; Fraqueza, M.J. A Comprehensive Virulence and Resistance Characteristics of Listeria monocytogenes Isolated from Fish and the Fish Industry Environment. Int. J. Mol. Sci. 2023, 24, 3581. https://doi.org/10.3390/ijms24043581
Zakrzewski AJ, Kurpas M, Zadernowska A, Chajęcka-Wierzchowska W, Fraqueza MJ. A Comprehensive Virulence and Resistance Characteristics of Listeria monocytogenes Isolated from Fish and the Fish Industry Environment. International Journal of Molecular Sciences. 2023; 24(4):3581. https://doi.org/10.3390/ijms24043581
Chicago/Turabian StyleZakrzewski, Arkadiusz Józef, Monika Kurpas, Anna Zadernowska, Wioleta Chajęcka-Wierzchowska, and Maria João Fraqueza. 2023. "A Comprehensive Virulence and Resistance Characteristics of Listeria monocytogenes Isolated from Fish and the Fish Industry Environment" International Journal of Molecular Sciences 24, no. 4: 3581. https://doi.org/10.3390/ijms24043581
APA StyleZakrzewski, A. J., Kurpas, M., Zadernowska, A., Chajęcka-Wierzchowska, W., & Fraqueza, M. J. (2023). A Comprehensive Virulence and Resistance Characteristics of Listeria monocytogenes Isolated from Fish and the Fish Industry Environment. International Journal of Molecular Sciences, 24(4), 3581. https://doi.org/10.3390/ijms24043581








