Molecular Signature of Biological Aggressiveness in Clear Cell Sarcoma of the Kidney (CCSK)
Abstract
1. Introduction
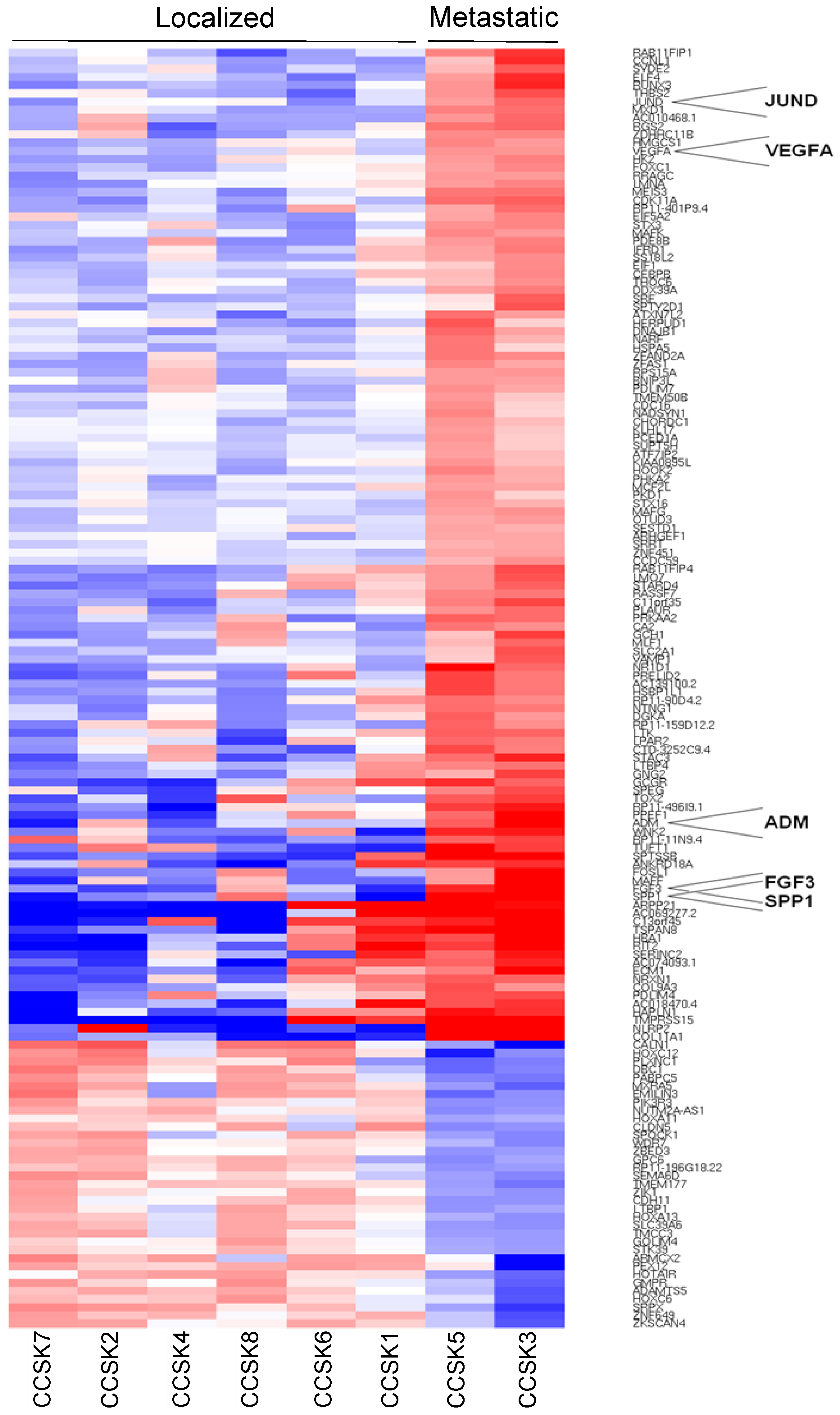
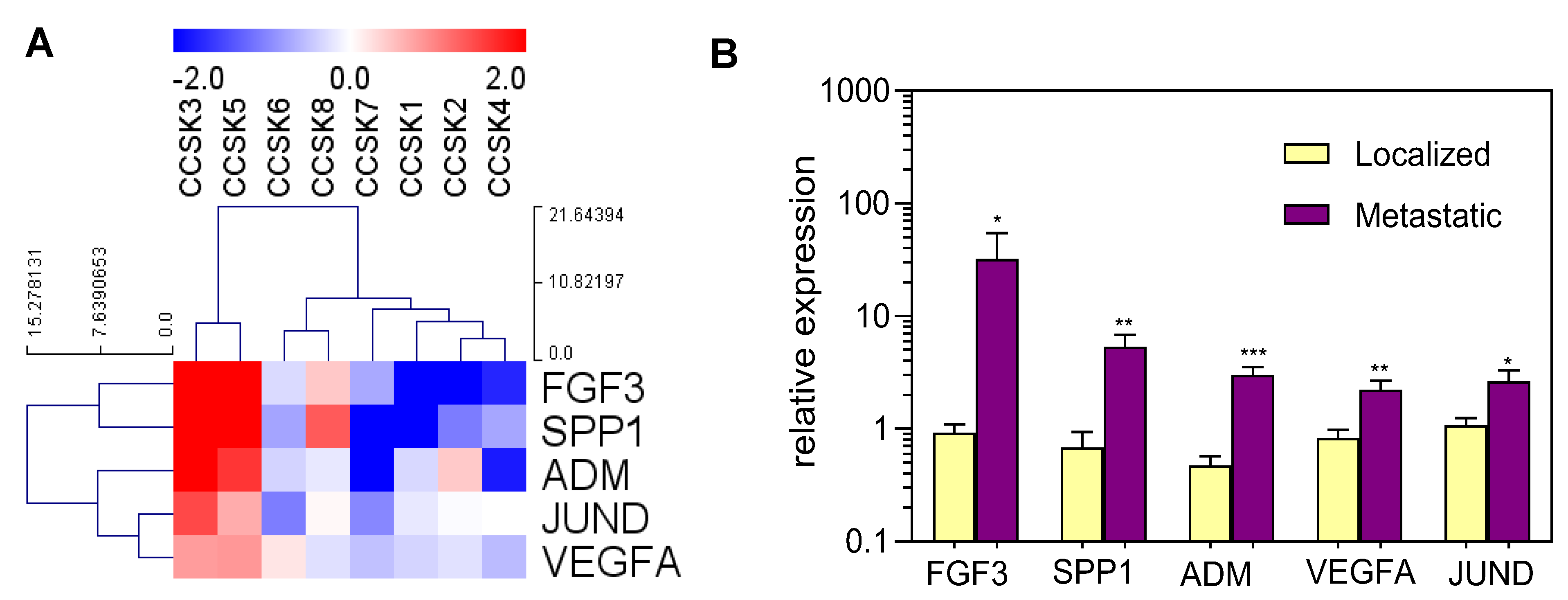
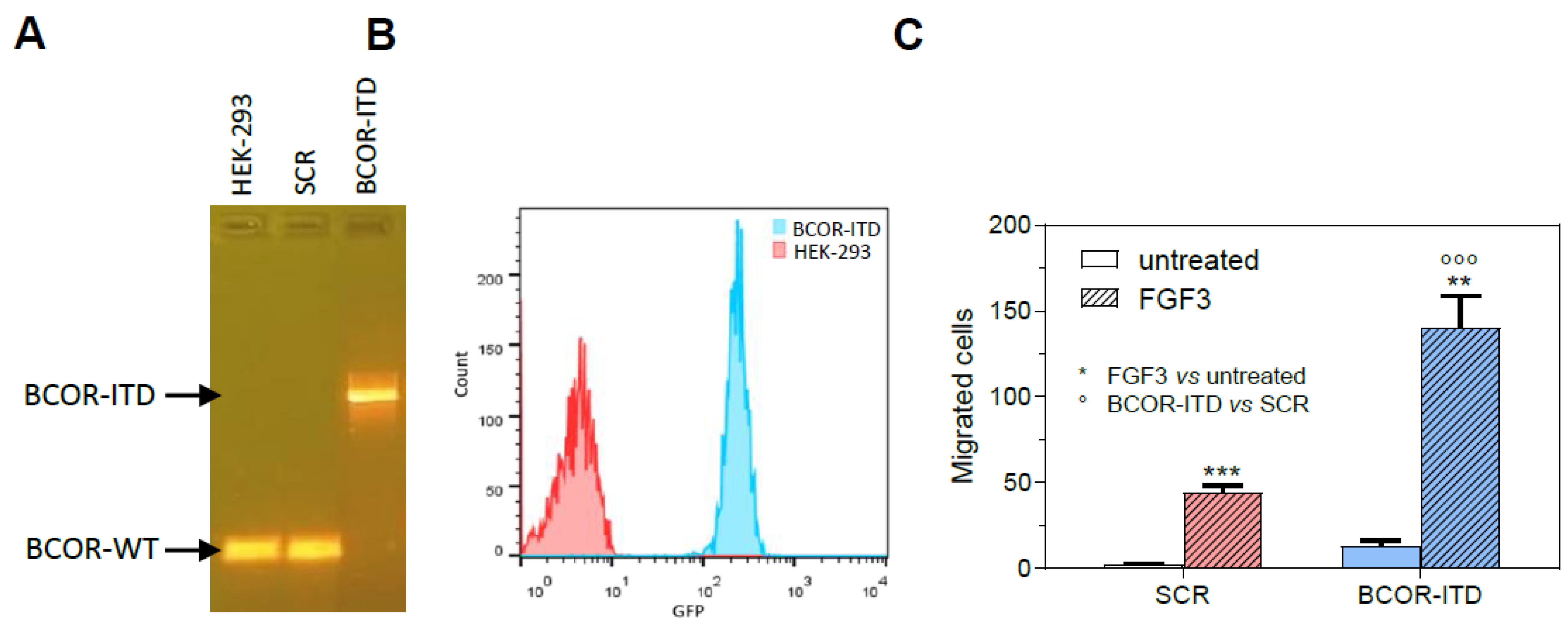
2. Results
3. Discussion
4. Materials and Methods
4.1. Patients and Tumor Samples
4.2. WTS and WES
4.3. Bioinformatic Analysis
4.4. Sanger Sequencing
4.5. Quantitative PCR
4.6. Cell Lines and Gene Editing
4.7. Flow Cytometry
4.8. Migration Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gooskens, S.L.; Furtwängler, R.; Vujanic, G.M.; Dome, J.S.; Graf, N.; van den Heuvel-Eibrink, M.M. Clear cell sarcoma of the kidney: A review. Eur. J. Cancer 2012, 48, 2219–2226. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Vecchiato, L.; Alaggio, R.; Cecchetto, G.; Zorzi, C.; Carli, M. Clear cell sarcoma of the kidney in a newborn. Med. Pediatr. Oncol. 2003, 41, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Newbould, M.J.; Kelsey, A.M. Clear cell sarcoma of the kidney in a 4-month-old infant: A case report. Med. Pediatr. Oncol. 1993, 21, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Honzumi, M.; Itoh, Y.; Umehara, N.; Moriyama, S.; Funada, M. Clear-cell sarcoma of the kidney seen in a 3-day-old newborn. Z Kinderchir. 1983, 38, 422–424. [Google Scholar] [CrossRef]
- van den Heuvel-Eibrink, M.M.; Grundy, P.; Graf, N.; Pritchard-Jones, K.; Bergeron, C.; Patte, C.; van Tinteren, H.; Rey, A.; Langford, C.; Anderson, J.R.; et al. Characteristics and survival of 750 children diagnosed with a renal tumor in the first seven months of life: A collaborative study by the SIOP/GPOH/SFOP, NWTSG, and UKCCSG Wilms’ tumor study groups. Pediatr. Blood Cancer 2008, 50, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Benchekroun, A.; Ghadouane, M.; Zannoud, M.; Alami, M.; Amhajji, R.; Faik, M. Clear cell sarcoma of the kidney in an adult. A case report. Ann. Urol. 2002, 36, 33–35. [Google Scholar] [CrossRef] [PubMed]
- Bhayani, S.B.; Liapis, H.; Kibel, A.S. Adult clear cell sarcoma of the kidney with atrial tumor thrombus. J. Urol. 2001, 165, 896–897. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.K.; Krishnani, N.; Bhandari, M. Clear cell sarcoma of kidney in an adult. Br. J. Urol. 1993, 72, 118. [Google Scholar] [CrossRef] [PubMed]
- Oda, H.; Shiga, J.; Machinami, R. Clear cell sarcoma of kidney. Two cases in adults. Cancer 1993, 71, 2286–2291. [Google Scholar] [CrossRef]
- Argani, P.; Perlman, E.J.; Breslow, N.E.; Browning, N.G.; Green, D.M.; D’Angio, G.J.; Beckwith, J.B. Clear cell sarcoma of the kidney: A review of 351 cases from the National Wilms’ Tumor Study Group Pathology Center. Am. J. Surg. Pathol. 2000, 24, 4–18. [Google Scholar] [CrossRef]
- Lowe, L.H.; Isuani, B.H.; Heller, R.M.; Stein, S.M.; Johnson, J.E.; Navarro, O.M.; Hernanz-Schulman, M. Pediatric renal masses: Wilms’ tumor and beyond. Radiographics 2000, 20, 1585–1603. [Google Scholar] [CrossRef] [PubMed]
- Seibel, N.L.; Li, S.; Breslow, N.E.; Beckwith, J.B.; Green, D.M.; Haase, G.M.; Ritchey, M.L.; Thomas, P.R.; Grundy, P.E.; Finklestein, J.Z.; et al. Effect of duration of treatment on treatment outcome for patients with clear-cell sarcoma of the kidney: A report from the National Wilms’ Tumor Study Group. J. Clin. Oncol. 2004, 22, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Ueno-Yokohata, H.; Okita, H.; Nakasato, K.; Akimoto, S.; Hata, J.; Koshinaga, T.; Fukuzawa, M.; Kiyokawa, N. Consistent in-frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat. Genet. 2015, 47, 861–863. [Google Scholar] [CrossRef] [PubMed]
- Astolfi, A.; Melchionda, F.; Perotti, D.; Fois, M.; Indio, V.; Urbini, M.; Genovese, C.G.; Collini, P.; Salfi, N.; Nantron, M.; et al. Whole transcriptome sequencing identifies BCOR internal tandem duplication as a common feature of clear cell sarcoma of the kidney. Oncotarget 2015, 6, 40934–40939. [Google Scholar] [CrossRef]
- Roy, A.; Kumar, V.; Zorman, B.; Fang, E.; Haines, K.M.; Doddapaneni, H.; Hampton, O.A.; White, S.; Bavle, A.A.; Patel, N.R.; et al. Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat. Commun. 2015, 6, 8891. [Google Scholar] [CrossRef]
- Karlsson, J.; Valind, A.; Gisselsson, D. BCOR internal tandem duplication and YWHAE-NUTM2B/E fusion are mutually exclusive events in clear cell sarcoma of the kidney. Genes Chromosomes Cancer 2016, 55, 120–123. [Google Scholar] [CrossRef]
- Kenny, C.; Bausenwein, S.; Lazaro, A.; Furtwängler, R.; Gooskens, S.L.; van den Heuvel Eibrink, M.; Vokuhl, C.; Leuschner, I.; Graf, N.; Gessler, M.; et al. Mutually exclusive BCOR internal tandem duplications and YWHAE-NUTM2 fusions in clear cell sarcoma of kidney: Not the full story. J Pathol. 2016, 238, 617–620. [Google Scholar] [CrossRef]
- O’Meara, E.; Stack, D.; Lee, C.H.; Garvin, A.J.; Morris, T.; Argani, P.; Han, J.S.; Karlsson, J.; Gisselson, D.; Leuschner, I.; et al. Characterization of the chromosomal translocation t(10;17)(q22;p13) in clear cell sarcoma of kidney. J. Pathol. 2012, 227, 72–80. [Google Scholar] [CrossRef]
- Brownlee, N.A.; Perkins, L.A.; Stewart, W.; Jackle, B.; Pettenati, M.J.; Koty, P.P.; Iskandar, S.S.; Garvin, A.J. Recurring translocation (10;17) and deletion (14q) in clear cell sarcoma of the kidney. Arch. Pathol. Lab. Med. 2007, 131, 446–451. [Google Scholar] [CrossRef]
- Rakheja, D.; Weinberg, A.G.; Tomlinson, G.E.; Partridge, K.; Schneider, N.R. Translocation (10;17)(q22;p13): A recurring translocation in clear cell sarcoma of kidney. Cancer Genet. Cytogenet. 2004, 154, 175–179. [Google Scholar] [CrossRef]
- Huynh, K.D.; Fischle, W.; Verdin, E.; Bardwell, V.J. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000, 14, 1810–1823. [Google Scholar] [CrossRef]
- Wamstad, J.A.; Corcoran, C.M.; Keating, A.M.; Bardwell, V.J. Role of the transcriptional corepressor Bcor in embryonic stem cell differentiation and early embryonic development. PLoS ONE 2008, 3, e2814. [Google Scholar] [CrossRef] [PubMed]
- Astolfi, A.; Fiore, M.; Melchionda, F.; Indio, V.; Bertuccio, S.N.; Pession, A. BCOR involvement in cancer. Epigenomics 2019, 11, 835–855. [Google Scholar] [CrossRef]
- van den Boom, V.; Maat, H.; Geugien, M.; Rodríguez López, A.; Sotoca, A.M.; Jaques, J.; Brouwers-Vos, A.Z.; Fusetti, F.; Groen, R.W.; Yuan, H.; et al. Non-canonical PRC1.1 Targets Active Genes Independent of H3K27me3 and Is Essential for Leukemogenesis. Cell Rep. 2016, 14, 332–346. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; O’Loghlen, A. PRC1 complex diversity: Where is it taking us? Trends Cell Biol. 2014, 24, 632–641. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF homologs; CBX proteins; and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef]
- Schwartz, Y.B.; Pirrotta, V. A new world of Polycombs: Unexpected partnerships and emerging functions. Nat. Rev. Genet. 2013, 14, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Yamaza, T.; Lee, J.S.; Yu, J.; Wang, S.; Fan, G.; Shi, S.; Wang, C.Y. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat. Cell Biol. 2009, 11, 1002–1009. [Google Scholar] [CrossRef]
- Karlsson, J.; Valind, A.; Jansson, C.; O’Sullivan, M.J.; Holmquist Mengelbier, L.; Gisselsson, D. Aberrant epigenetic regulation in clear cell sarcoma of the kidney featuring distinct DNA hypermethylation and EZH2 overexpression. Oncotarget 2016, 7, 11127–11136. [Google Scholar] [CrossRef]
- Karlsson, J.; Holmquist Mengelbier, L.; Ciornei, C.D.; Naranjo, A.; O’Sullivan, M.J.; Gisselsson, D. Clear cell sarcoma of the kidney demonstrates an embryonic signature indicative of a primitive nephrogenic origin. Genes Chromosomes Cancer 2014, 53, 381–391. [Google Scholar] [CrossRef]
- Cutcliffe, C.; Kersey, D.; Huang, C.C.; Zeng, Y.; Walterhouse, D.; Perlman, E.J.; Renal Tumor Committee of the Children’s Oncology Group. Clear cell sarcoma of the kidney: Up-regulation of neural markers with activation of the sonic hedgehog and Akt pathways. Clin. Cancer Res. 2005, 11, 7986–7994. [Google Scholar] [CrossRef] [PubMed]
- Gooskens, S.L.; Gadd, S.; Guidry Auvil, J.M.; Gerhard, D.S.; Khan, J.; Patidar, R.; Meerzaman, D.; Chen, Q.R.; Hsu, C.H.; Yan, C.; et al. TCF21 hypermethylation in genetically quiescent clear cell sarcoma of the kidney. Oncotarget 2015, 6, 15828–15841. [Google Scholar] [CrossRef]
- Huang, C.C.; Cutcliffe, C.; Coffin, C.; Sorensen, P.H.; Beckwith, J.B.; Perlman, E.J.; Renal Tumor Committee of the Children’s Oncology Group. Classification of malignant pediatric renal tumors by gene expression. Pediatr. Blood Cancer 2006, 46, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Z.; Zhou, Z.; Ren, Y. Revealing the role of VEGFA in clear cell sarcoma of the kidney by protein-protein interaction network and significant pathway analysis. Oncol. Lett. 2016, 11, 953–958. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, C.H.; Ou, W.B.; Mariño-Enriquez, A.; Zhu, M.; Mayeda, M.; Wang, Y.; Guo, X.; Brunner, A.L.; Amant, F.; French, C.A.; et al. 14-3-3 fusion oncogenes in high-grade endometrial stromal sarcoma. Proc. Natl. Acad. Sci. USA 2012, 109, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Little, S.E.; Bax, D.A.; Rodriguez-Pinilla, M.; Natrajan, R.; Messahel, B.; Pritchard-Jones, K.; Vujanic, G.M.; Reis-Filho, J.S.; Jones, C. Multifaceted dysregulation of the epidermal growth factor receptor pathway in clear cell sarcoma of the kidney. Clin. Cancer Res. 2007, 13, 4360–4364. [Google Scholar] [CrossRef]
- Mirkovic, J.; Calicchio, M.; Fletcher, C.D.; Perez-Atayde, A.R. Diffuse and strong cyclin D1 immunoreactivity in clear cell sarcoma of the kidney. Histopathology 2015, 67, 306–312. [Google Scholar] [CrossRef]
- Jet Aw, S.; Hong Kuick, C.; Hwee Yong, M.; Wen Quan Lian, D.; Wang, S.; Liang Loh, A.H.; Ling, S.; Lian Peh, G.; Yen Soh, S.; Pheng Loh, A.H.; et al. Novel Karyotypes and Cyclin D1 Immunoreactivity in Clear Cell Sarcoma of the Kidney. Pediatr. Dev. Pathol. 2015, 18, 297–304. [Google Scholar]
- Ueno, H.; Okita, H.; Akimoto, S.; Kobayashi, K.; Nakabayashi, K.; Hata, K.; Fujimoto, J.; Hata, J.; Fukuzawa, M.; Kiyokawa, N. DNA methylation profile distinguishes clear cell sarcoma of the kidney from other pediatric renal tumors. PLoS ONE 2013, 8, e62233. [Google Scholar] [CrossRef][Green Version]
- Boo, Y.J.; Fisher, J.C.; Haley, M.J.; Cowles, R.A.; Kandel, J.J.; Yamashiro, D.J. Vascular characterization of clear cell sarcoma of the kidney in a child: A case report and review. J. Pediatr. Surg. 2009, 44, 2031–2036. [Google Scholar] [CrossRef]
- Wong, M.K.; Ng, C.C.Y.; Kuick, C.H.; Aw, S.J.; Rajasegaran, V.; Lim, J.Q.; Sudhanshi, J.; Loh, E.; Yin, M.; Ma, J.; et al. Clear cell sarcomas of the kidney are characterised by BCOR gene abnormalities; including exon 15 internal tandem duplications and BCOR-CCNB3 gene fusion. Histopathology 2018, 72, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.; Dave, B.K.; Desai, S.B. Clear cell sarcoma of the kidney--a study of seven cases over a period of three years. Indian J. Pathol. Microbiol. 2007, 50, 270–273. [Google Scholar]
- Choi, Y.J.; Jung, W.H.; Jung, S.H.; Park, C. Clear cell sarcoma of the kidney--immunohistochemical study and flow cytometric DNA analysis of 7 cases. Yonsei Med. J. 1994, 35, 336–343. [Google Scholar] [CrossRef]
- Bonadio, J.; Perlman, E.J. Immunohistochemical analysis of 61 clear cell sarcomas of the kidney for a panel including NGFR and CD99. Mod. Pathol. 2008, 21, 218. [Google Scholar]
- Chen, S.; Li, M.; Li, R.; Cao, J.; Wu, Q.; Zhou, T.; Cai, Z.; Li, N. Clear cell sarcoma of the kidney in children: A clinopathologic analysis of three cases. Int. J. Clin. Exp. Pathol. 2020, 13, 771–777. [Google Scholar]
- Uddin, N.; Minhas, K.; Abdul-Ghafar, J.; Ahmed, A.; Ahmad, Z. Expression of cyclin D1 in clear cell sarcoma of kidney. Is it useful in differentiating it from its histological mimics? Diagn. Pathol. 2019, 14, 13. [Google Scholar] [CrossRef]
- Furtwängler, R.; Gooskens, S.L.; van Tinteren, H.; de Kraker, J.; Schleiermacher, G.; Bergeron, C.; de Camargo, B.; Acha, T.; Godzinski, J.; Sandstedt, B.; et al. Clear cell sarcomas of the kidney registered on International Society of Pediatric Oncology (SIOP) 93-01 and SIOP 2001 protocols: A report of the SIOP Renal Tumour Study Group. Eur. J. Cancer 2013, 49, 3497–3506. [Google Scholar] [CrossRef] [PubMed]
- Gooskens, S.L.; Furtwängler, R.; Spreafico, F.; van Tinteren, H.; de Kraker, J.; Vujanic, G.M.; Leuschner, I.; Coulomb-L’Herminé, A.; Godzinski, J.; Schleiermacher, G.; et al. Treatment and outcome of patients with relapsed clear cell sarcoma of the kidney: A combined SIOP and AIEOP study. Br. J. Cancer 2014, 111, 227–233. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Wang, Z.; Gearhart, M.D.; Lee, Y.W.; Kumar, I.; Ramazanov, B.; Zhang, Y.; Hernandez, C.; Lu, A.Y.; Neuenkirchen, N.; Deng, J.; et al. A Non-canonical BCOR-PRC1.1 Complex Represses Differentiation Programs in Human ESCs. Cell Stem Cell 2018, 22, 235–251.e9. [Google Scholar] [CrossRef] [PubMed]
- Boulard, M.; Edwards, J.R.; Bestor, T.H. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat. Genet. 2015, 47, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 2012, 1, e00205. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Johansen, J.V.; Helin, K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell 2013, 49, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulos, I.; Thorsen, J.; Gorunova, L.; Haugom, L.; Bjerkehagen, B.; Davidson, B.; Heim, S.; Micci, F. Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22-translocation. Genes Chromosomes Cancer 2013, 52, 610–618. [Google Scholar] [PubMed]
- Hoang, L.N.; Aneja, A.; Conlon, N.; Delair, D.F.; Middha, S.; Benayed, R.; Hensley, M.L.; Park, K.J.; Hollmann, T.J.; Hameed, M.R.; et al. Novel High-grade Endometrial Stromal Sarcoma: A Morphologic Mimicker of Myxoid Leiomyosarcoma. Am. J. Surg. Pathol. 2017, 41, 12–24. [Google Scholar] [CrossRef]
- Rajasekhar, V.K.; Vemuri, M.C. Regulatory Networks in Stem Cells, 1st ed.; Humana Totowa: Totowa, NJ, USA, 2009; pp. 285–290. [Google Scholar]
- Goel, H.L.; Mercurio, A.M. VEGF targets the tumour cell. Nat. Rev. Cancer 2013, 13, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Claesson-Welsh, L.; Welsh, M. VEGFA and tumour angiogenesis. J. Intern. Med. 2013, 273, 114–127. [Google Scholar] [CrossRef]
- Matsumoto, K.; Ema, M. Roles of VEGF-A signalling in development, regeneration, and tumours. J. Biochem. 2014, 156, 1–10. [Google Scholar] [CrossRef]
- Icer, M.A.; Gezmen-Karadag, M. The multiple functions and mechanisms of osteopontin. Clin. Biochem. 2018, 59, 17–24. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Q.; Alam, A.; Cui, J.; Suen, K.C.; Soo, A.P.; Eguchi, S.; Gu, J.; Ma, D. The role of osteopontin in the progression of solid organ tumour. Cell Death Dis. 2018, 9, 356. [Google Scholar] [CrossRef]
- Bandopadhyay, M.; Bulbule, A.; Butti, R.; Chakraborty, G.; Ghorpade, P.; Ghosh, P.; Gorain, M.; Kale, S.; Kumar, D.; Kumar, S.; et al. Osteopontin as a therapeutic target for cancer. Expert Opin. Ther. Targets 2014, 18, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.F.; Lett, G.S.; Haubein, N.C. Osteopontin is a marker for cancer aggressiveness and patient survival. Br. J. Cancer 2010, 103, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Ramchandani, D.; Weber, G.F. Interactions between osteopontin and vascular endothelial growth factor: Implications for skeletal disorders. Bone 2015, 81, 7–15. [Google Scholar] [CrossRef]
- Dai, X.; Ma, W.; Jha, R.K.; He, X. Adrenomedullin and its expression in cancers and bone. A literature review. Front. Biosci. 2010, 2, 1073–1080. [Google Scholar]
- Deville, J.L.; Salas, S.; Figarella-Branger, D.; Ouafik, L.; Daniel, L. Adrenomedullin as a therapeutic target in angiogenesis. Expert Opin. Ther. Targets 2010, 14, 1059–1072. [Google Scholar] [CrossRef] [PubMed]
- Larráyoz, I.M.; Martínez-Herrero, S.; García-Sanmartín, J.; Ochoa-Callejero, L.; Martínez, A. Adrenomedullin and tumour microenvironment. J. Transl. Med. 2014, 12, 339. [Google Scholar] [CrossRef] [PubMed]
- Cheung, B.M.; Tang, F. Adrenomedullin: Exciting new horizons. Recent Pat. Endocr. Metab. Immune Drug Discov. 2012, 6, 4–17. [Google Scholar] [CrossRef]
- Schönauer, R.; Els-Heindl, S.; Beck-Sickinger, A.G. Adrenomedullin–new perspectives of a potent peptide hormone. J. Pept. Sci. 2017, 23, 472–485. [Google Scholar] [CrossRef]
- Dickson, C.; Smith, R.; Brookes, S.; Peters, G. Tumorigenesis by mouse mammary tumor virus: Proviral activation of a cellular gene in the common integration region int-2. Cell 1984, 37, 529–536. [Google Scholar] [CrossRef]
- Ornitz, D.M.; Itoh, N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev. Dev. Biol. 2015, 4, 215–266. [Google Scholar] [CrossRef]
- Parish, A.; Schwaederle, M.; Daniels, G.; Piccioni, D.; Fanta, P.; Schwab, R.; Shimabukuro, K.; Parker, B.A.; Helsten, T.; Kurzrock, R. Fibroblast growth factor family aberrations in cancers: Clinical and molecular characteristics. Cell Cycle 2015, 14, 2121–2128. [Google Scholar] [CrossRef] [PubMed]
- Touat, M.; Ileana, E.; Postel-Vinay, S.; André, F.; Soria, J.C. Targeting FGFR Signaling in Cancer. Clin. Cancer Res. 2015, 21, 2684–2694. [Google Scholar] [CrossRef]
- Astolfi, A.; Pantaleo, M.A.; Indio, V.; Urbini, M.; Nannini, M. The Emerging Role of the FGF/FGFR Pathway in Gastrointestinal Stromal Tumor. Int. J. Mol. Sci. 2020, 21, 3313. [Google Scholar] [CrossRef]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Rodon, J.; Prat, A.; Perez-Garcia, J.; Adamo, B.; Felip, E.; Cortes, J.; Iafrate, A.J.; Nuciforo, P.; Tabernero, J. Genomic aberrations in the FGFR pathway: Opportunities for targeted therapies in solid tumors. Ann. Oncol. 2014, 25, 552–563. [Google Scholar] [CrossRef]
- Dickson, C.; Spencer-Dene, B.; Dillon, C.; Fantl, V. Tyrosine kinase signalling in breast cancer: Fibroblast growth factors and their receptors. Breast Cancer Res. 2000, 2, 191–196. [Google Scholar] [CrossRef]
- Grose, R.; Dickson, C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005, 16, 179–186. [Google Scholar] [CrossRef]
- Champème, M.H.; Bièche, I.; Hacène, K.; Lidereau, R. Int-2/FGF3 amplification is a better independent predictor of relapse than c-myc and c-erbB-2/neu amplifications in primary human breast cancer. Mod. Pathol. 1994, 7, 900–905. [Google Scholar] [PubMed]
- Kitagawa, Y.; Ueda, M.; Ando, N.; Shinozawa, Y.; Shimizu, N.; Abe, O. Significance of int-2/hst-1 coamplification as a prognostic factor in patients with esophageal squamous carcinoma. Cancer Res. 1991, 51, 1504–1508. [Google Scholar] [PubMed]
- Liscia, D.S.; Merlo, G.R.; Garrett, C.; French, D.; Mariani-Costantini, R.; Callahan, R. Expression of int-2 mRNA in human tumors amplified at the int-2 locus. Oncogene 1989, 4, 1219–1224. [Google Scholar]
- Arao, T.; Ueshima, K.; Matsumoto, K.; Nagai, T.; Kimura, H.; Hagiwara, S.; Sakurai, T.; Haji, S.; Kanazawa, A.; Hidaka, H.; et al. FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology 2013, 57, 1407–1415. [Google Scholar] [CrossRef]
- Hu, L.; Sham, J.S.; Xie, D.; Wen, J.M.; Wang, W.S.; Wang, Y.; Guan, X.Y. Up-regulation of fibroblast growth factor 3 is associated with tumor metastasis and recurrence in human hepatocellular carcinoma. Cancer Lett. 2007, 252, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Q.; Li, J.J.; Moscatelli, D.; Basilico, C.; Nicolaides, A.; Zhang, W.G.; Poiesz, B.J.; Friedman-Kien, A.E. Expression of int-2 oncogene in Kaposi’s sarcoma lesions. J. Clin. Investig. 1993, 91, 1191–1197. [Google Scholar] [CrossRef]
- Li, J.J.; Friedman-Kien, A.E.; Cockerell, C.; Nicolaides, A.; Liang, S.L.; Huang, Y.Q. Evaluation of the tumorigenic and angiogenic potential of human fibroblast growth factor FGF3 in nude mice. J. Cancer Res. Clin. Oncol. 1998, 124, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Dengler, V.L.; Galbraith, M.; Espinosa, J.M. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions; deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms; SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
| ID | Sex | Age (Months) | Side | Stage | Metastatic Site | Neoadjuvant Treatment | Surgery | Adjuvant Treatment | RT | Relapse |
|---|---|---|---|---|---|---|---|---|---|---|
| CCSK1 | M | 37 | L | I | - | N | Y | Y | N | N |
| CCSK2 | F | 20 | L | III | - | N | Y | Y | Y | N |
| CCSK3 | M | 27 | R | IV | femur | Y | Y | Y | Y | Iliac wing |
| CCSK4 | M | 14 | R | I | - | N | Y | Y | N | N |
| CCSK5 | M | 21 | L | IV | lung | N | Y | Y | Y | N |
| CCSK6 | M | 27 | L | I | - | N | Y | Y | N | N |
| CCSK7 | F | 17 | L | II | - | Y | Y | Y | Y | N |
| CCSK8 | F | 25 | R | II | - | Y | Y | Y | Y | N |
| CCSK9 | M | 22 | R | IV | diffuse | Y | Y | Y | N | NR |
| ID1 | ITD-BCOR | Samples | WES | WTS | ||
|---|---|---|---|---|---|---|
| Tumor | Normal | Tumor | Normal | |||
| CCSK1 | ITD-3 | FF | PB | Y | Y | Y |
| CCSK2 | ITD-1 | FF | PB | Y | Y | Y |
| CCSK3 | ITD-3 | FF | NA | Y | N | Y |
| CCSK4 | ITD-1 | FF | PB | Y | Y | Y |
| CCSK5 | ITD-3 | FF | PB | Y | Y | Y |
| CCSK6 | ITD-3 | FF | NA | Y | N | Y |
| CCSK7 | ITD-1 | FF | FF | Y | Y | Y |
| CCSK8 | ITD-2 | FF | PB | Y | Y | Y |
| CCSK9 | ITD-3 | FFPE | PB | Y | Y | N |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiore, M.; Taddia, A.; Indio, V.; Bertuccio, S.N.; Messelodi, D.; Serravalle, S.; Bandini, J.; Spreafico, F.; Perotti, D.; Collini, P.; et al. Molecular Signature of Biological Aggressiveness in Clear Cell Sarcoma of the Kidney (CCSK). Int. J. Mol. Sci. 2023, 24, 3743. https://doi.org/10.3390/ijms24043743
Fiore M, Taddia A, Indio V, Bertuccio SN, Messelodi D, Serravalle S, Bandini J, Spreafico F, Perotti D, Collini P, et al. Molecular Signature of Biological Aggressiveness in Clear Cell Sarcoma of the Kidney (CCSK). International Journal of Molecular Sciences. 2023; 24(4):3743. https://doi.org/10.3390/ijms24043743
Chicago/Turabian StyleFiore, Michele, Alberto Taddia, Valentina Indio, Salvatore Nicola Bertuccio, Daria Messelodi, Salvatore Serravalle, Jessica Bandini, Filippo Spreafico, Daniela Perotti, Paola Collini, and et al. 2023. "Molecular Signature of Biological Aggressiveness in Clear Cell Sarcoma of the Kidney (CCSK)" International Journal of Molecular Sciences 24, no. 4: 3743. https://doi.org/10.3390/ijms24043743
APA StyleFiore, M., Taddia, A., Indio, V., Bertuccio, S. N., Messelodi, D., Serravalle, S., Bandini, J., Spreafico, F., Perotti, D., Collini, P., Di Cataldo, A., Pasquinelli, G., Chiarini, F., Fois, M., Melchionda, F., Pession, A., & Astolfi, A. (2023). Molecular Signature of Biological Aggressiveness in Clear Cell Sarcoma of the Kidney (CCSK). International Journal of Molecular Sciences, 24(4), 3743. https://doi.org/10.3390/ijms24043743










