Integrating Transcriptomics and Hormones Dynamics Reveal Seed Germination and Emergence Process in Polygonatum cyrtonema Hua
Abstract
1. Introduction
2. Results
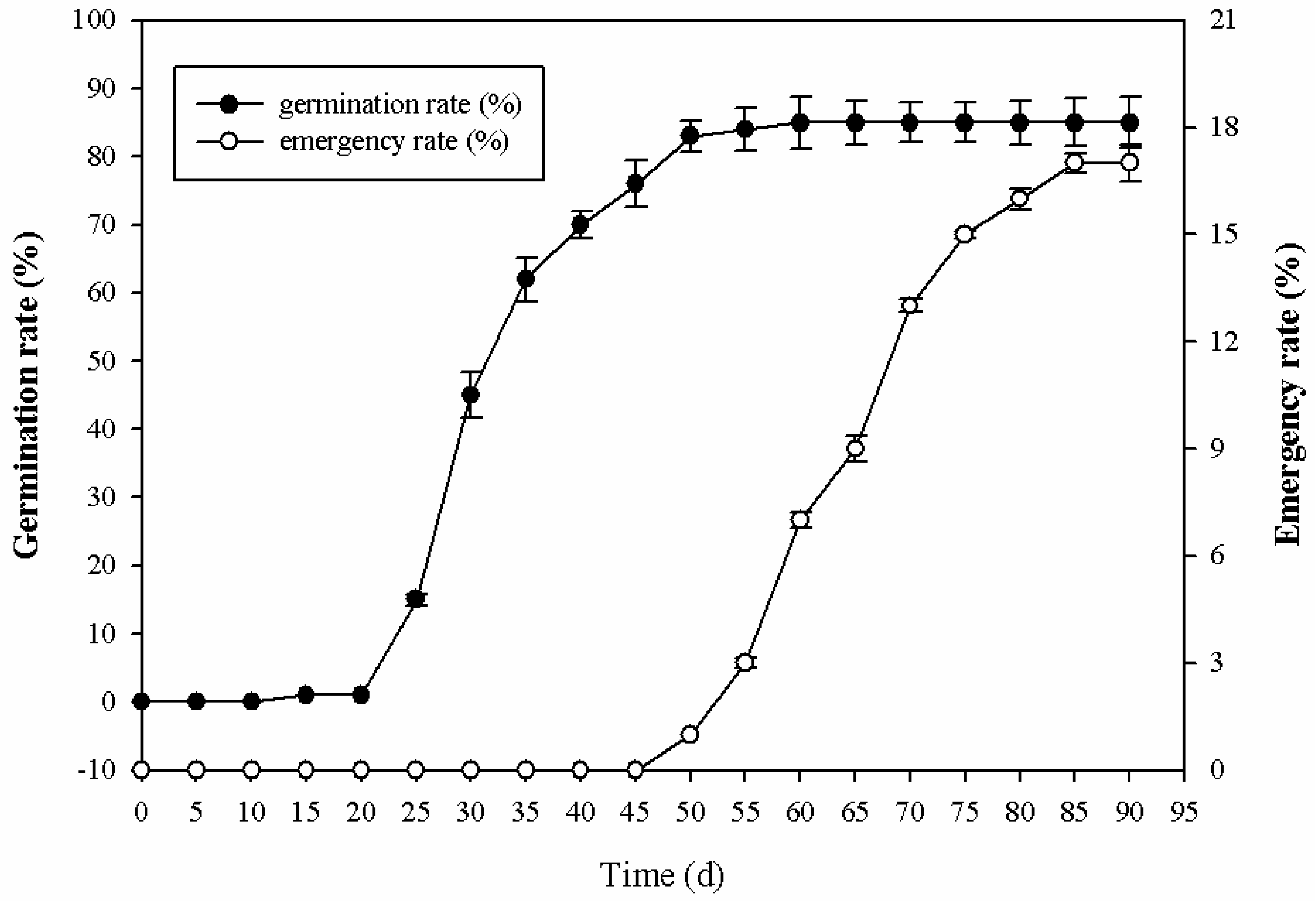
2.1. Morphological Changes during Seed Germination Process in P. cyrtonema Hua
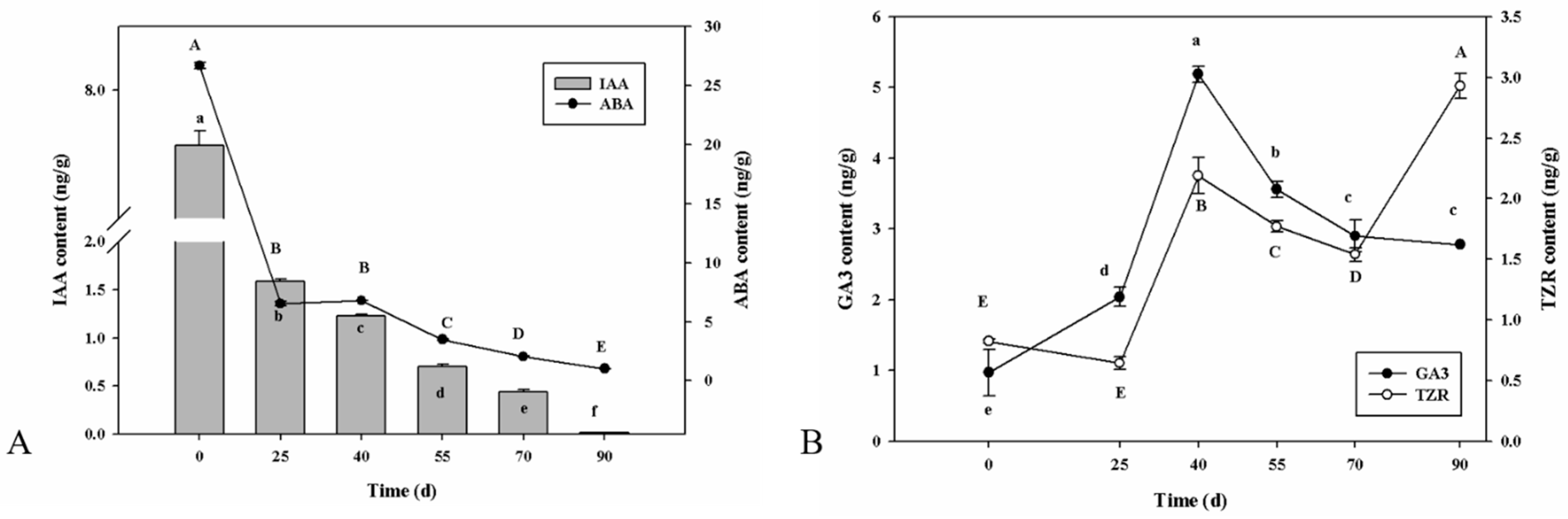
2.2. Phytohormonal Dynamics of P. cyrtonema Hua during Seed Germination Process
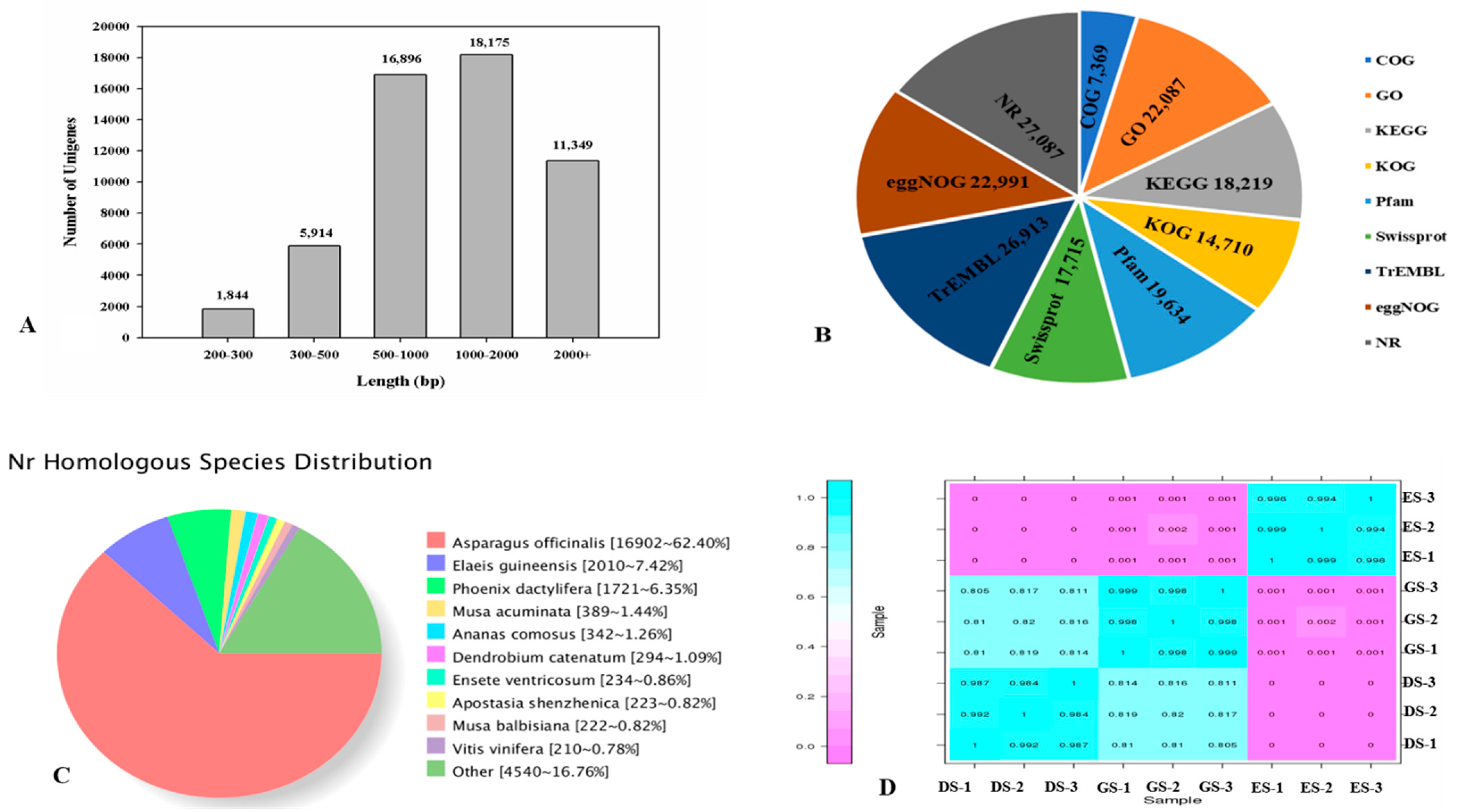
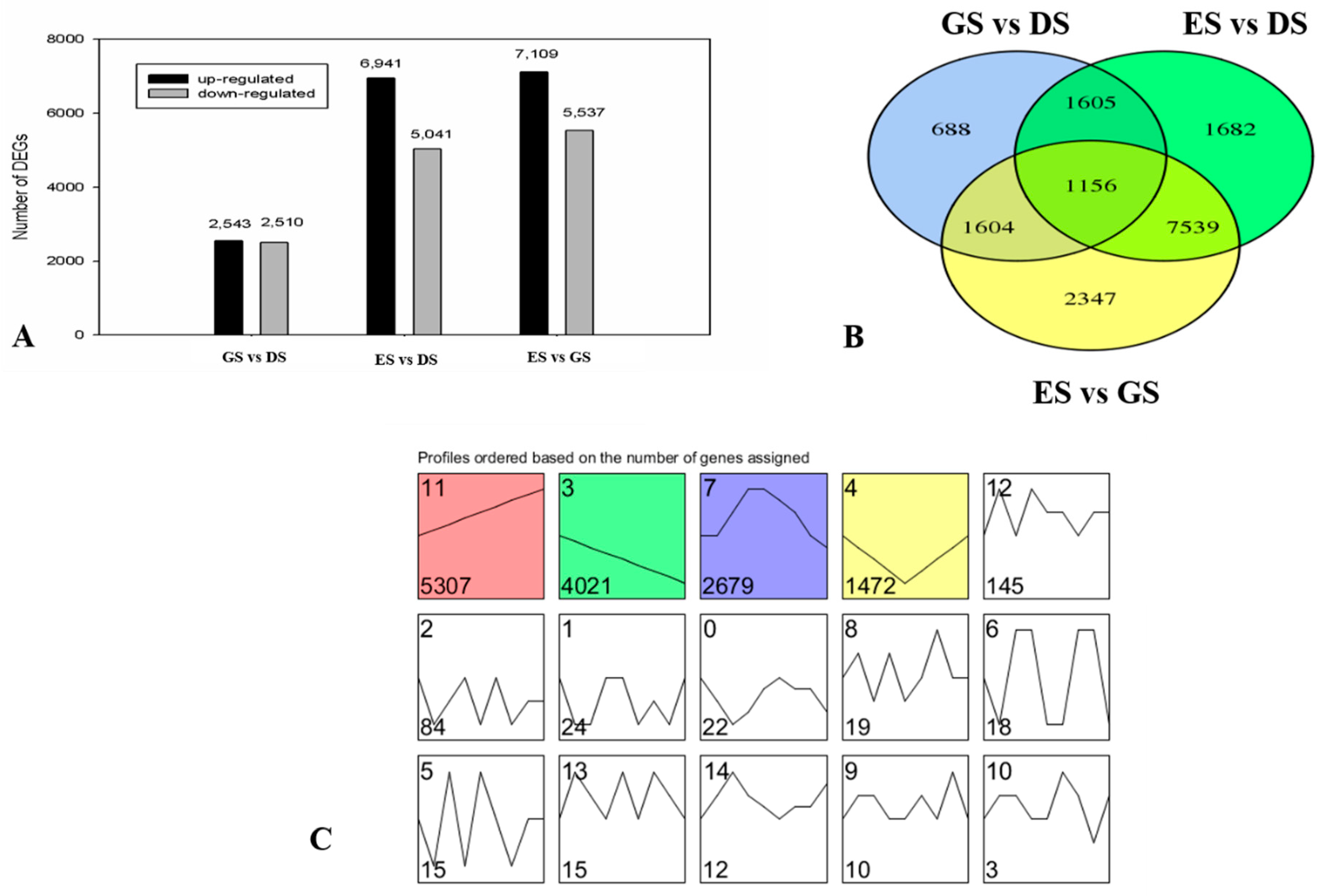
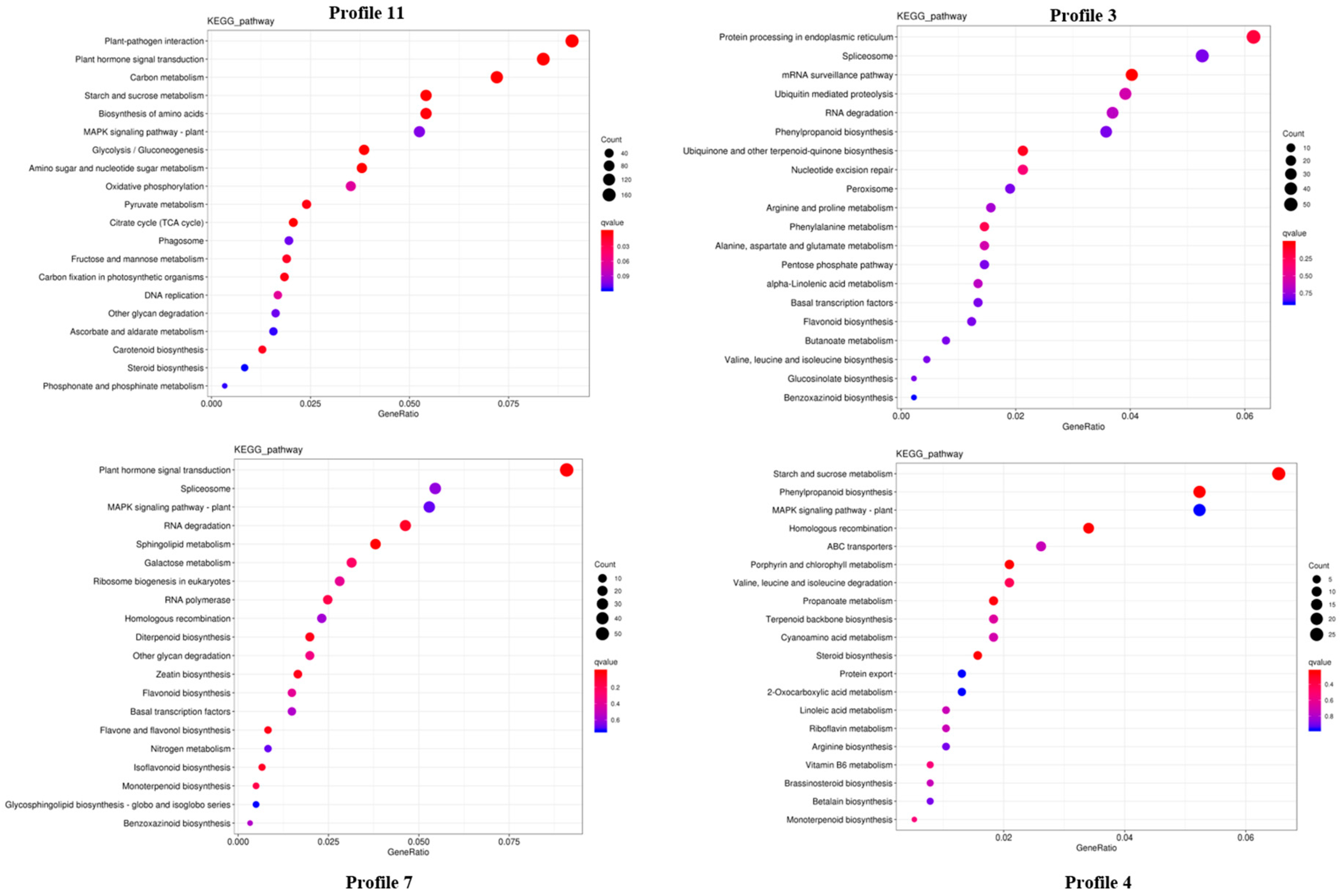
2.3. Transcriptome Analysis and Identification of Differentially Expressed Genes (DEGs) in P. cyrtonema Hua during Seed Germination and Emergence Process
| Sample ID | Base Number | GC Content | % > Q30 | Clean Reads | Mapping Rate (%) |
|---|---|---|---|---|---|
| DS−1 | 5,866,481,748 | 48.87% | 94.26% | 19,597,195 | 62.69% |
| DS−2 | 6,411,437,502 | 49.45% | 94.34% | 21,413,251 | 65.74% |
| DS−3 | 6,471,178,576 | 49.09% | 93.72% | 21,619,346 | 64.35% |
| GS−1 | 6,905,162,498 | 48.20% | 94.47% | 23,066,585 | 62.39% |
| GS−2 | 7,091,894,902 | 48.13% | 94.11% | 23,688,736 | 62.47% |
| GS−3 | 6,177,043,546 | 48.02% | 94.02% | 20,633,298 | 61.89% |
| ES−1 | 6,139,693,670 | 48.68% | 94.42% | 20,505,088 | 62.84% |
| ES−2 | 6,293,346,204 | 48.47% | 93.95% | 21,019,827 | 62.12% |
| ES−3 | 6,307,247,984 | 48.63% | 94.11% | 21,065,521 | 62.98% |
| Total Number | 200−500 bp | 501−1000 bp | 1001−2000 bp | >2000 bp | Total Number | Total Length | N50 Length | Mean Length bp |
|---|---|---|---|---|---|---|---|---|
| 54,178 | 7758 | 16,896 | 18,175 | 11,349 | 54,178 | 75,328,238 | 1847 | 1390.38 |
2.4. Expression Profiles of Plant Hormones and Signal Transduction Genes in P. cyrtonema Hua during Seed Germination
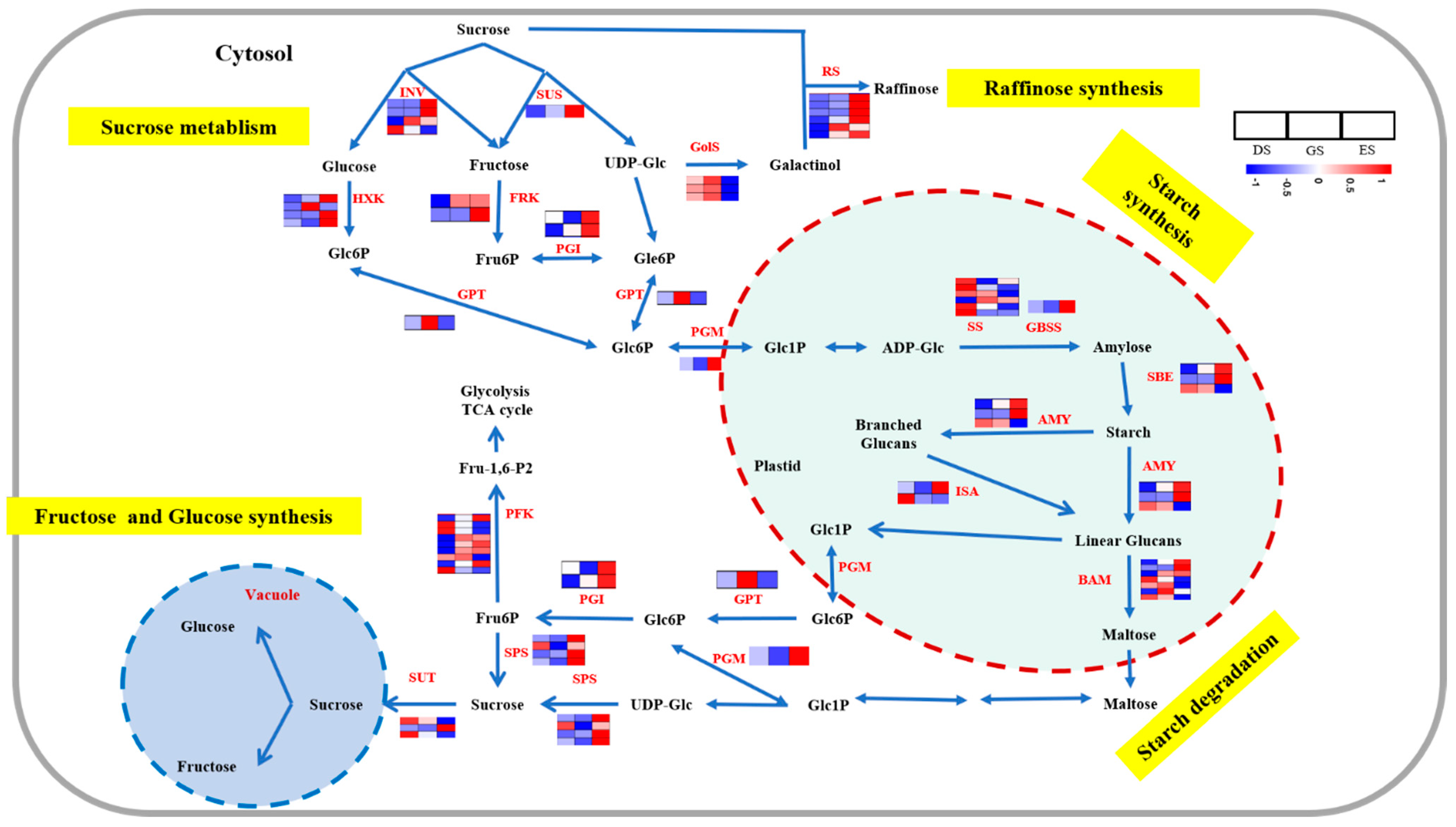
2.5. Expression Profiles of Carbohydrate Metabolism in P. cyrtonema Hua during Seed Germination and Emergence Processes
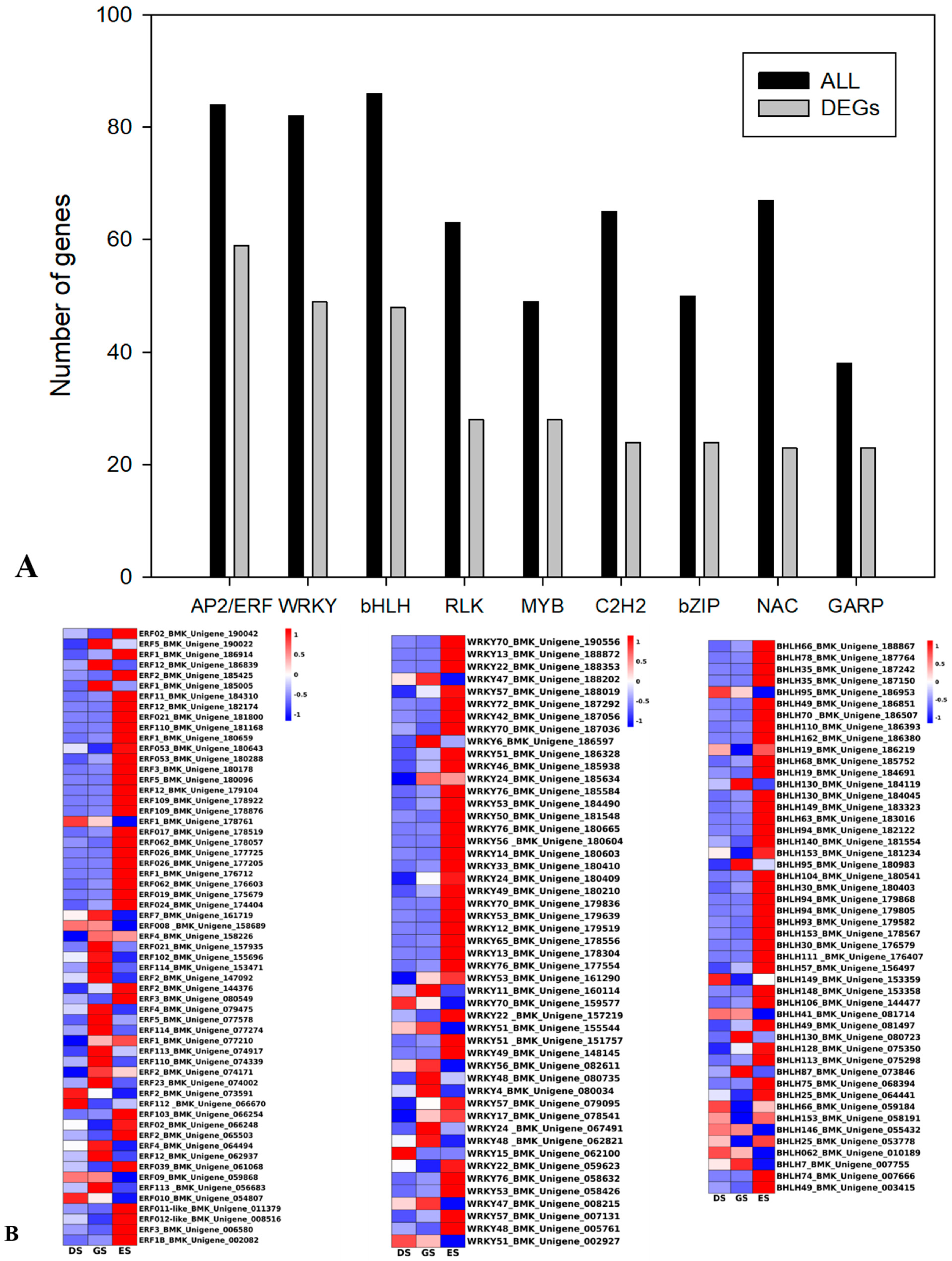
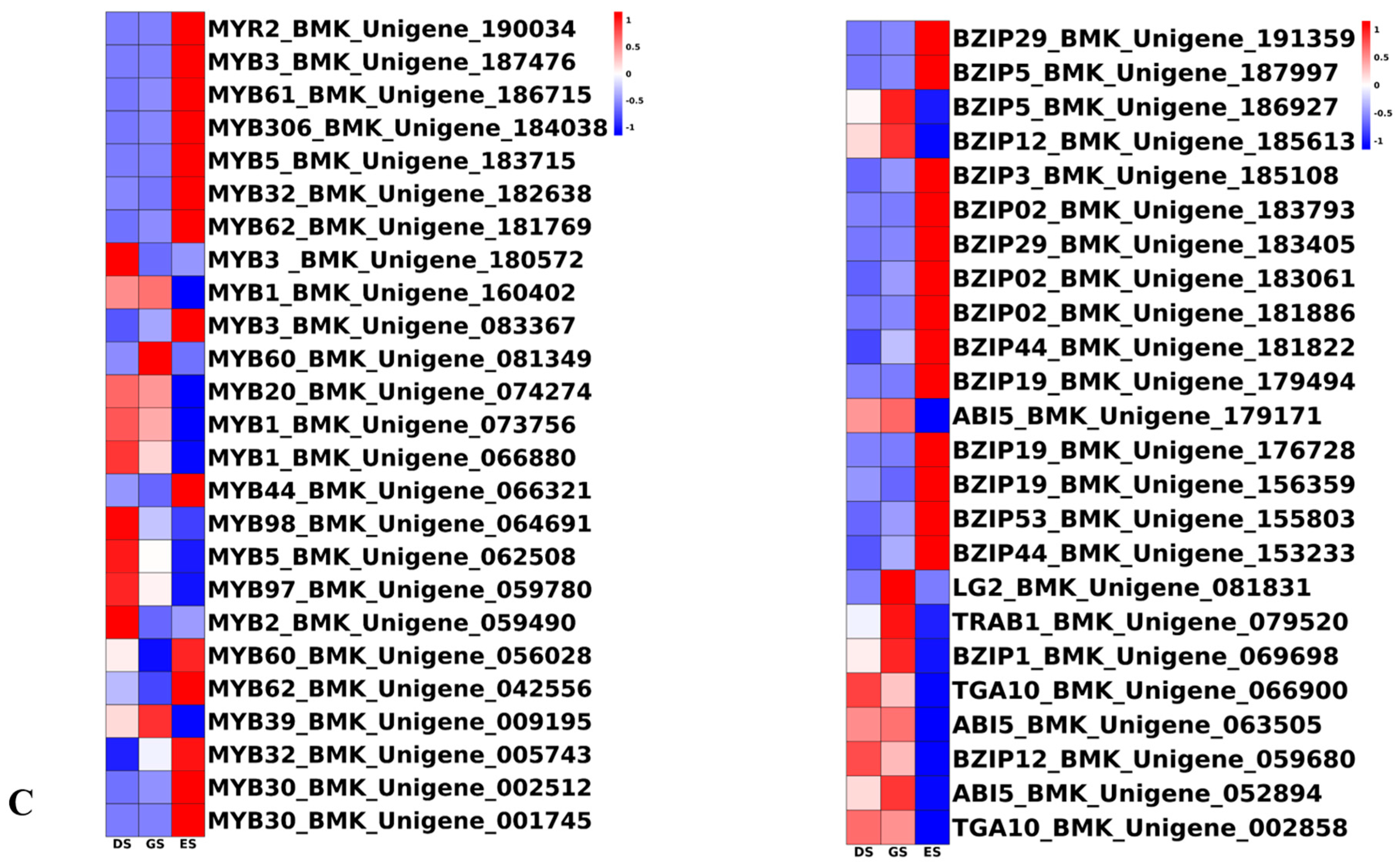
2.6. TFs Were Differentially Expressed in P. cyrtonema Hua during Seed Germination and Emergence Process
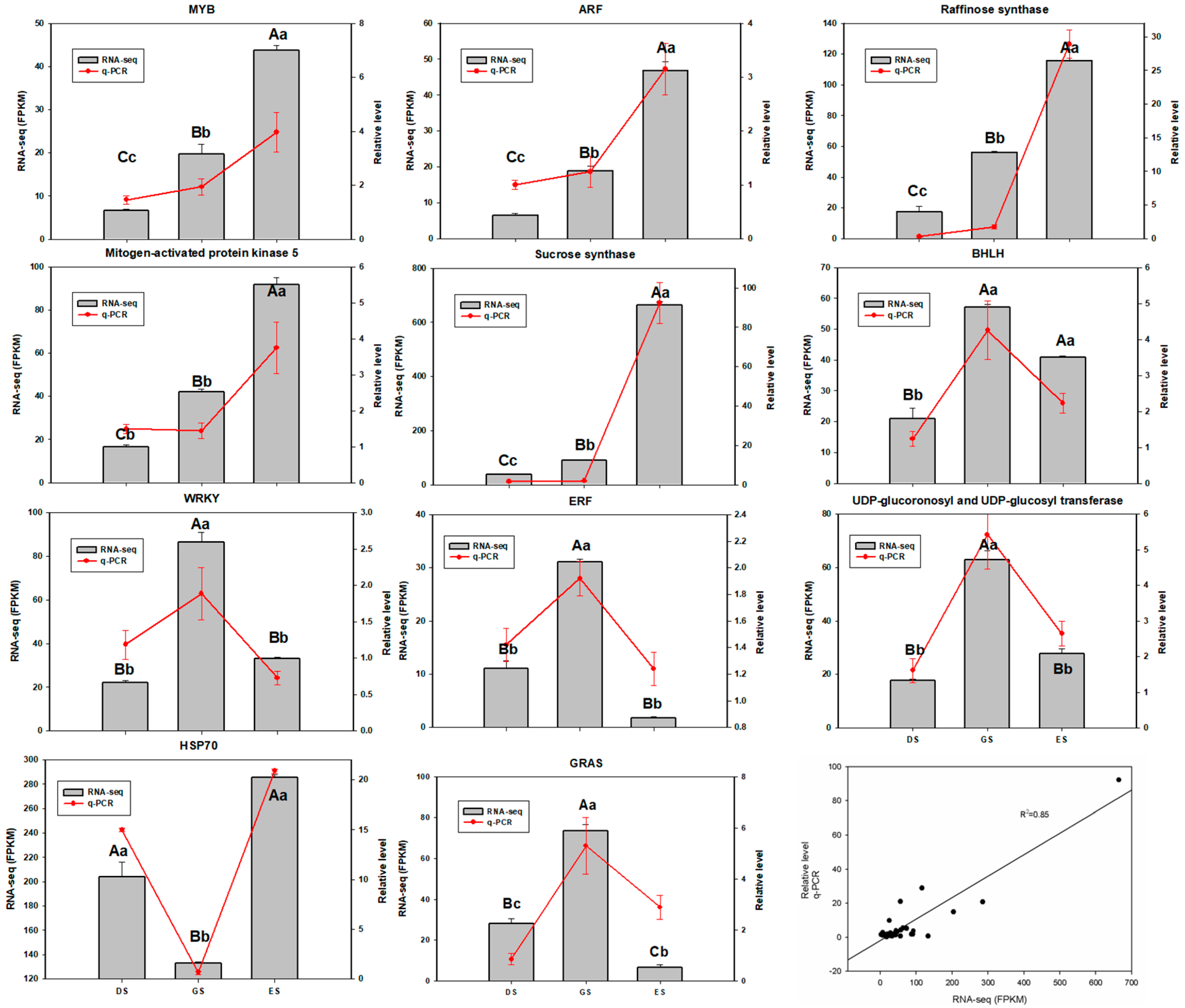
2.7. Validation of RNA-Seq by qRT-PCR Analysis
3. Discussion
3.1. Regulation of Plant Hormones in P. cyrtonema Hua during Seed Germination and Emergence Progression
3.2. Regulation of Carbohydrate in P. cyrtonema Hua during Seed Germination and Emergence Progression
3.3. Transcription Factors Play a Pivotal Role in P. cyrtonema Hua Seed Germination and Emergence Progression
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Measurements of Hormone Contents
4.3. RNA Extraction and Transcriptome Sequencing
4.4. Differential Expression Analysis
4.5. Quantitative Real-Time PCR Validation Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, X.Y.; Li, J. Chemical constituents of the genus polygonatum and their role in medicinal treatment. Nat. Prod. Commun. 2015, 10, 683–688. [Google Scholar] [CrossRef]
- Che, Y.Y.; Qian, Y.; Wu, Y.; Qu, W.; Liang, J.Y. Two new homoisoflavanones from the rhizome of Polygonatum odoratum. Chem. Nat. Compd. 2015, 51, 54–56. [Google Scholar] [CrossRef]
- Jza, B.; Hc, A.; Lan, L.B.; Zz, B.; Yw, A.; Tg, A.; Lian, Y.B.; Teng, P.A.; Mw, B. Structures of fructan and galactan from Polygonatum cyrtonema and their utilization by probiotic bacteria. Carbohydr. Polym. 2021, 267, 118219. [Google Scholar]
- Fu, L.F.; Ding, Z.M.; Ma, C.D.; Wei, J.H.; Li, X.E.; Qi, J.H. Studies on Germination and Emergence Characteristic of Polygonatum sibiricum Red Seed. Mod. Chin. Med. 2017, 19, 1151–1156. (In Chinese) [Google Scholar]
- Vishal, B.; Kumar, P.P. Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Front. Plant Sci. 2018, 9, 838. [Google Scholar]
- Palomar, V.M.; Garciarrubio, A.; Garay-Arroyo, A.; Martínez-Martínez, C.; RosasBringas, O.; Reyes, J.L.; Covarrubias, A.A. The canonical RdDM pathway mediates the control of seed germination timing under salinity. Plant J. 2021, 105, 691–707. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Deng, X.; Xiao, Q.; Han, Y.; Zhu, S.; Chen, J. IAA priming improves the germination and seedling growth in cotton (Gossypium hirsutum L.) via regulating the endogenous phytohormones and enhancing the sucrose metabolism. Ind. Crops Prod. 2020, 155, 112788. [Google Scholar] [CrossRef]
- Ruggiero, A.; Landi, S.; Punzo, P.; Possenti, M.; Van Oosten, M.J.; Costa, A.; Morelli, G.; Maggio, A.; Grillo, S.; Batelli, G. Salinity and ABA seed responses in pepper: Expression and interaction of ABA core signaling components. Front. Plant Sci. 2019, 10, 304. [Google Scholar] [CrossRef] [PubMed]
- Hyoung, S.; Cho, S.H.; Chung, J.H.; So, W.M.; Cui, M.H.; Shin, J.S. Cytokinin oxidase PpCKX1 plays regulatory roles in development and enhances dehydration and salt tolerance in Physcomitrella patens. Plant Cell Rep. 2020, 39, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Di, F.; Jian, H.; Wang, T.; Chen, X.; Ding, Y.; Du, H.; Lu, K.; Li, J.; Liu, L. Genome-wide analysis of the PYL gene family and identification of PYL genes that respond to abiotic stress in Brassica napus. Genes 2018, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, F.Q.; Chen, S.S.; Li, H.; Zhao, Z. Study on the changes of endogenous hormones content in Polygonatum cyrtonema seed germination. J. Chin. Med. Mater. 2020, 43, 523–527. (In Chinese) [Google Scholar]
- Liu, R.; Lu, J.; Xing, J.; Du, M.; Wang, M.; Zhang, L.; Li, Y.; Zhang, C.; Wu, Y. Transcriptome and metabolome analyses revealing the potential mechanism of seed germination in Polygonatum cyrtonema. Sci. Rep. 2021, 11, 12161. [Google Scholar] [CrossRef] [PubMed]
- Cantoro, R.; Crocco, C.D.; Benech-Arnold, R.L.; Rodriguez, M.V. In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: Possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 2013, 64, 5721–5735. [Google Scholar] [CrossRef] [PubMed]
- Matilla, A.J. Seed Dormancy: Molecular Control of Its Induction and Alleviation. Plants 2020, 9, 1402. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Kuo, S.R.; Chien, C.T. Roles of gibberellins and abscisic acid in dormancy and germination of red bayberry (Myrica rubra) seeds. Tree Physiol. 2008, 28, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.; Perez, M.; Steinbrecher, T.; Gawthrop, F.; Pavlovic, I.; Novak, O.; Tarkowska, D.; Strnad, M.; Marone, F.; Nakabayashi, K.; et al. Molecular mechanisms and hormonal regulation underpinning morphological dormancy: A case study using Apium graveolens (Apiaceae). Plant J. 2021, 108, 1020–1036. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.C.; Zhao, B.B.; Liu, S.; Lu, Z.G.; Chang, B.; Jiang, H.R.; Cui, H.; He, Q.S.; Li, W.X.; Jin, B.; et al. Embryo transcriptome and miRNA analyses reveal the regulatory network of seed dormancy in Ginkgo biloba. Tree Physiol. 2021, 41, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qian, J.Y.; Bian, Y.H.; Liu, X.; Wang, C.L. Transcriptome and Metabolite Conjoint Analysis Reveals the Seed Dormancy Release Process in Callery Pear. Int. J. Mol Sci. 2022, 23, 2186. [Google Scholar] [CrossRef] [PubMed]
- Seiler, C.; Harshavardhan, T.V.; Rajesh, K.; Reddy, S.; Strickert, M.; Rolletschek, H.; Scholz, U.; Wobus, U.; Sreenivasulu, N. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 2011, 62, 2615–2632. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 80-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.; Xie, Q.; He, Z. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Cheng, J.P.; Wang, J.K.; Cheng, Y.H.; He, Y.Q.; Zhang, H.S.; Wang, Z.F. Physiological characteristics of cold stratification on seed dormancy release in rice. Plant Growth Regul. 2019, 89, 131–141. [Google Scholar] [CrossRef]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef]
- Dai, C.; Xue, H.W. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010, 29, 1916–1927. [Google Scholar] [CrossRef] [PubMed]
- Pellizzaro, A.; Neveu, M.; Lalanne, D.; Vu, B.L.; Kanno, Y.; Seo, M.; Leprince, O.; Buitink, J. A role for auxin signaling in the acquisition of longevity during seed maturation. New Phytol. 2020, 225, 284–296. [Google Scholar] [CrossRef]
- Lavy, M.; Estelle, M. Mechanisms of auxin signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef]
- Kendrick, M.D.; Chang, C. Ethylene signaling: New levels of complexity and regulation. Curr. Opin. Plant Biol. 2008, 11, 479–485. [Google Scholar] [CrossRef]
- Bakshi, A.; Piya, S.; Fernandez, J.C.; Chervin, C.; Hewezi, T.; Binder, B. Ethylene receptors signal via a noncanonical pathway to regulate abscisic acid response. Plant Physiol. 2018, 176, 910–929. [Google Scholar] [CrossRef]
- Li, Q.F.; Xiong, M.; Xu, P.; Huang, L.C.; Zhang, C.Q.; Liu, Q.Q. Dissection of brassinosteroid-regulated proteins in rice embryos during germination by quantitative proteomics. Sci. Rep. 2016, 6, 34583. [Google Scholar] [CrossRef]
- Steber, G.M.; McCourt, P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001, 125, 763–769. [Google Scholar] [CrossRef]
- Yang, X.; Bai, Y.; Shang, J.X.; Xin, R.J.; Tang, W.Q. The antagonistic regulation of abscisic acid-inhibited root growth by brassinosteroids is partially mediated via direct suppression of ABSCISIC ACID INSENSITIVE 5 expression by BRASSINAZOLE RESISTANT1. Plant Cell Environ. 2016, 39, 1994–2003. [Google Scholar] [CrossRef]
- Avramova, Z. The Jasmonic acid-signaling and abscisic acid-signaling pathways cross talk during one, but not repeated, dehydration stress: A non-specific panicky or a meaningful response? Plant Cell Environ. 2017, 40, 1704–1710. [Google Scholar] [CrossRef]
- Liu, A.; Gao, F.; Kanno, Y.; Jordan, M.C.; Kamiya, Y.; Seo, M.; Ayele, B.T. Regulation of wheat seed dormancy by after-ripening is mediated by specific transcriptional switches that induce changes in seed hormone metabolism and signaling. PLoS ONE 2013, 8, e56570. [Google Scholar] [CrossRef]
- Sun, M.; Tuan, P.A.; Izydorczyk, M.S.; Ayele, B.T. Ethylene regulates post-germination seedling growth in wheat through spatial and temporal modulation of ABA/GA balance. J. Exp. Bot. 2020, 71, 1985–2004. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.L.; Tang, G.Y.; Cui, W.P.; Chen, G.X.; Ma, C.L.; Zhu, J.Q.; Li, P.X.; Shan, L.; Liu, Z.J.; Wan, S.B. Transcriptional Differences in Peanut (Arachis hypogaea L.) Seeds at the Freshly Harvested, After-ripening and Newly Germinated Seed Stages: Insights into the Regulatory Networks of Seed Dormancy Release and Germination. PLoS ONE 2020, 15, e0219413. [Google Scholar] [CrossRef] [PubMed]
- Bakhshy, E.; Fatemeh, Z.; Mehrdad, M.N. Structural and quantitative changes of starch in seed of Trigonella persica during germination. International journal of biological macromolecules. Int. J. Biol. Macromol. 2020, 7, 262. [Google Scholar]
- Zhu, J.; Yu, W.; Zhang, C.; Zhu, Y.; Liu, Q. New insights into amylose and amylopectin biosynthesis in rice endosperm. Carbohydr. Polym. 2019, 230, 115656. [Google Scholar] [CrossRef]
- Zhao, Y.; Tang, J.; Brummell, D.A.; Song, C.; Qi, S.; Lin, Q.; Bi, J.; Duan, Y. Abscisic acid alleviates chilling injury in cold-stored peach fruit by regulating the metabolism of sucrose. Sci. Hortic. 2022, 298, 111000. [Google Scholar] [CrossRef]
- Velasco, D.; Fita, I.; Guinovart, J.; Garcia-Amoros, J.; Ferrer, C. Selective photoregulation of the activity of glycogen synthase and glycogen phosphorylase, two key enzymes in glycogen metabolism. Org. Biomol. Chem. 2015, 13, 7282. [Google Scholar]
- Xie, Z.; Zhang, Z.L.; Hanzlik, S.; Cook, E.; Chen, Q.J. Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol. Biol. 2007, 64, 293–303. [Google Scholar] [CrossRef]
- Chen, P.W.; Chiang, C.M.; Tseng, T.H.; Yu, S.M. Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of a-amylase genes. Plant Cell 2006, 18, 2326–2340. [Google Scholar] [CrossRef] [PubMed]
- Washio, K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 2003, 133, 850–863. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]

| Time (min) | Velocity of Flow (mL/min) | A% |
|---|---|---|
| 0–1 | 0.3 | 20 |
| 1–3 | 0.3 | Increase from 20 to 50 |
| 3–9 | 0.3 | Increase from 50 to 80 |
| 9–10.5 | 0.3 | 80 |
| 10.5–10.6 | 0.3 | Decrease from 80 to 20 |
| 10.6–13.5 | 0.3 | 20 |
| Hormone | Polarity | Parent Ion (m/z) | Daughter (m/z) | De-Clustering Voltage (V) | Collision Energy (V) |
|---|---|---|---|---|---|
| GA3 | − | 345.2 | 143.0 */239.0 | −80 | −30/−33 |
| ABA | − | 263.1 | 153.1 */204.2 | −60 | −14/−27 |
| TZR | + | 352.2 | 220.1 */136/202.1 | 90 | 25/40/32 |
| IAA | + | 176.1 | 130.1 */102.9 | 65 | 12/42 |
| Gene Id | Primer Name | Sequence (5′ to 3′) | Annotation |
|---|---|---|---|
| Reference gene | 18S-F | CAACCATAAACGATGCCGA | 18S |
| Reference gene | 18S-R | AGCCTTGCGACCATACTCC | |
| BMK_Unigene_187871 | BMK_Unigene_187871-F | CATACTGGTTTTCTTCCCCTTTT | MYB |
| BMK_Unigene_187871 | BMK_Unigene_187871-R | TTTCAGCTAAGGAAACAACAGG | |
| BMK_Unigene_187926 | BMK_Unigene_187926-F | GTGGCACTAACAAAGGTGAAAGA | ARF 9-like |
| BMK_Unigene_187926 | BMK_Unigene_187926-R | TTCCTCTAATAATTCCTTGCGTGC | |
| BMK_Unigene_068480 | BMK_Unigene_068480-F | GAAGTAATCAACCAAGCAGTC | Raffinose synthase |
| BMK_Unigene_068480 | BMK_Unigene_068480-R | CATCCAAACCAGTCCAGAA | |
| BMK_Unigene_068480 | BMK_Unigene_175000-F | AAAGGGAAATGACCAGAC | MAPK |
| BMK_Unigene_068480 | BMK_Unigene_175000-R | TCCTCCAGTACAACATCT | |
| BMK_Unigene_185451 | BMK_Unigene_185451-F | CGCTCAGGCTTTGTATTCTAT | Sucrose synthase |
| BMK_Unigene_185451 | BMK_Unigene_185451-R | GTATCCGTTGCTTAACTTCCT | |
| BMK_Unigene_190684 | BMK_Unigene_190684-F | TGTAGATGTTGAGGCTGATG | bHLH-MYC |
| BMK_Unigene_190684 | BMK_Unigene_190684-R | TTGTATTACCTTCGCACACT | |
| BMK_Unigene_067491 | BMK_Unigene_067491-F | GCATTCGCTGAAGTCTCTAA | WRKY |
| BMK_Unigene_067491 | BMK_Unigene_067491-R | TCTCCTCTTCTCTTGACCAA | |
| BMK_Unigene_075429 | BMK_Unigene_075429-F | TCTCCTTGAATCCGACACT | ERF |
| BMK_Unigene_075429 | BMK_Unigene_075429-R | GCTACCCACCTCACTATTTAC | |
| BMK_Unigene_079332 | BMK_Unigene_079332-F | GCCCCTTTAACCTTTTGTTG | Zeatin biosynthesis |
| BMK_Unigene_079332 | BMK_Unigene_079332-R | TTGTAGTATCCGCTTATCACG | |
| BMK_Unigene_081582 | BMK_Unigene_081582-F | ATTGACGGTGGAAACAAC | MYB |
| BMK_Unigene_081582 | BMK_Unigene_081582-R | GAAAACGAGAACAGGAGAG | |
| BMK_Unigene_069145 | BMK_Unigene_069145-F | GATGATGAACACGCTGAAGA | HSP70 |
| BMK_Unigene_069145 | BMK_Unigene_069145-R | TCTCTGAACTTGCTGAACAC | |
| BMK_Unigene_003009 | BMK_Unigene_003009-F | TCATCATTCTCTCACTGCTTAGTT | GRAS |
| BMK_Unigene_003009 | BMK_Unigene_003009-R | ACCAGATACCTGAAGGAAACAAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, X.; Jiang, W.; Wu, K.; Chen, J.; Li, Y.; Tao, Z. Integrating Transcriptomics and Hormones Dynamics Reveal Seed Germination and Emergence Process in Polygonatum cyrtonema Hua. Int. J. Mol. Sci. 2023, 24, 3792. https://doi.org/10.3390/ijms24043792
Duan X, Jiang W, Wu K, Chen J, Li Y, Tao Z. Integrating Transcriptomics and Hormones Dynamics Reveal Seed Germination and Emergence Process in Polygonatum cyrtonema Hua. International Journal of Molecular Sciences. 2023; 24(4):3792. https://doi.org/10.3390/ijms24043792
Chicago/Turabian StyleDuan, Xiaojing, Wu Jiang, Kunjing Wu, Jiadong Chen, Yaping Li, and Zhengming Tao. 2023. "Integrating Transcriptomics and Hormones Dynamics Reveal Seed Germination and Emergence Process in Polygonatum cyrtonema Hua" International Journal of Molecular Sciences 24, no. 4: 3792. https://doi.org/10.3390/ijms24043792
APA StyleDuan, X., Jiang, W., Wu, K., Chen, J., Li, Y., & Tao, Z. (2023). Integrating Transcriptomics and Hormones Dynamics Reveal Seed Germination and Emergence Process in Polygonatum cyrtonema Hua. International Journal of Molecular Sciences, 24(4), 3792. https://doi.org/10.3390/ijms24043792





