Circulatory miR-411-5p as a Novel Prognostic Biomarker for Major Adverse Cardiovascular Events in Patients with Atrial Fibrillation
Abstract
1. Introduction
2. Results
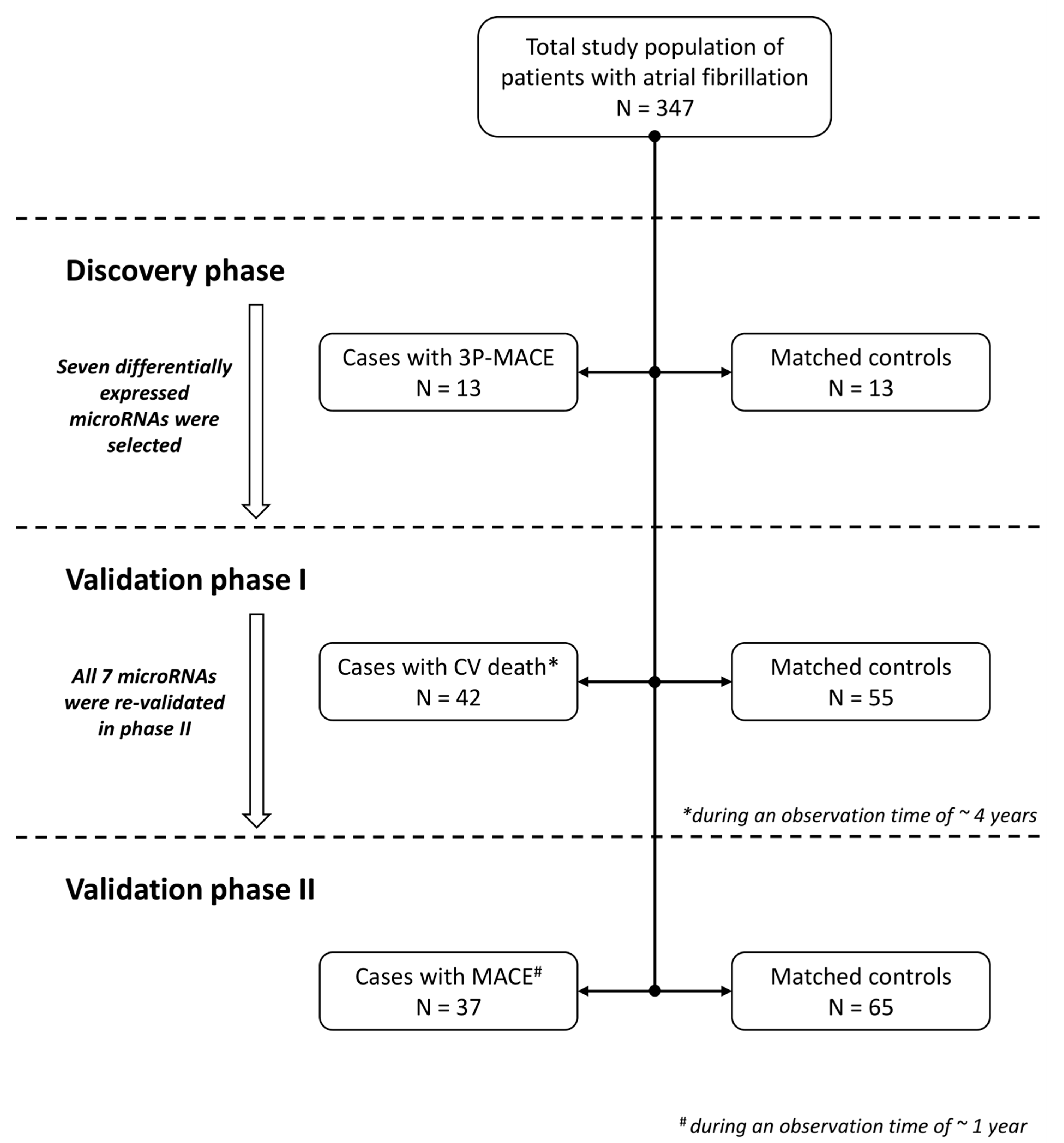
2.1. Study Population
2.2. Selection of Candidate microRNAs in the Discovery Cohort
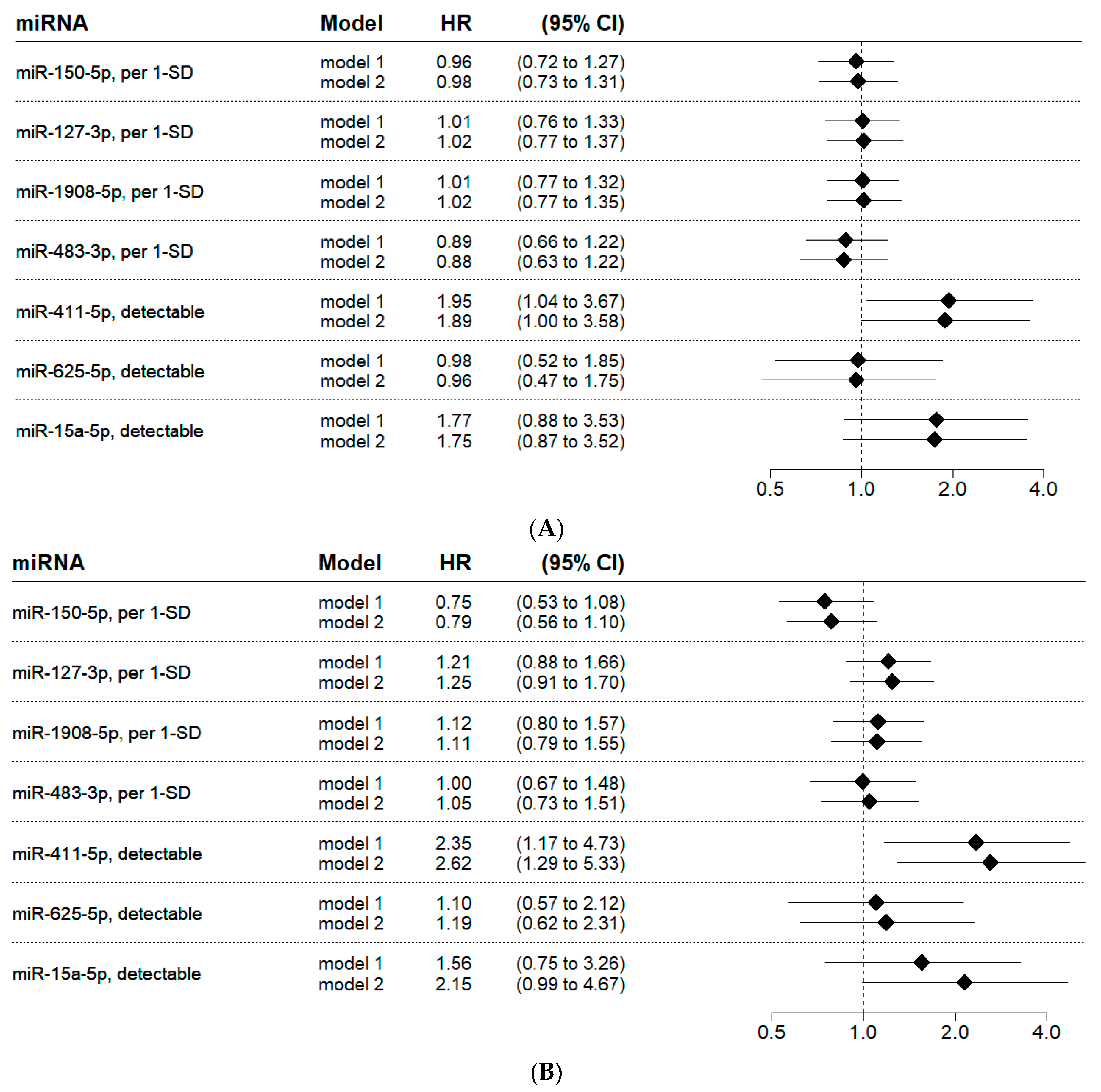
2.3. Validation of Candidate microRNAs for Cardiovascular Death
2.4. Validation of Candidate microRNAs for Major Adverse Cardiovascular Events
3. Discussion
4. Materials and Methods
4.1. Study Population and Data Source
4.2. Study Design
4.3. Discovery Phase
4.4. Validation Phase I: Patients with Cardiovascular Death
4.5. Validation Phase II: Patients with MACE at One Year
4.6. Laboratory and Statistical Methods
4.6.1. Sample Collection
4.6.2. Discovery Phase: Small RNA Sequencing
4.6.3. Discovery Phase: Statistical Analysis
4.6.4. Validation Phases: Quantification Using RT-qPCR
4.6.5. Validation Phases: Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gomez-Outes, A.; Suarez-Gea, M.L.; Garcia-Pinilla, J.M. Causes of death in atrial fibrillation: Challenges and opportunities. Trends Cardiovasc. Med. 2017, 27, 494–503. [Google Scholar] [CrossRef]
- Chen, J.Y.; Zhang, A.D.; Lu, H.Y.; Guo, J.; Wang, F.F.; Li, Z.C. CHADS2 versus CHA2DS2-VASc score in assessing the stroke and thromboembolism risk stratification in patients with atrial fibrillation: A systematic review and meta-analysis. J. Geriatr. Cardiol. 2013, 10, 258–266. [Google Scholar] [CrossRef]
- Stojkovic, S.; Nossent, A.Y.; Haller, P.; Jager, B.; Vargas, K.G.; Wojta, J.; Huber, K. MicroRNAs as Regulators and Biomarkers of Platelet Function and Activity in Coronary Artery Disease. Thromb. Haemost. 2019, 119, 1563–1572. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef]
- Mens, M.M.J.; Heshmatollah, A.; Fani, L.; Ikram, M.A.; Ikram, M.K.; Ghanbari, M. Circulatory MicroRNAs as Potential Biomarkers for Stroke Risk: The Rotterdam Study. Stroke 2021, 52, 945–953. [Google Scholar] [CrossRef]
- Li, P.; Teng, F.; Gao, F.; Zhang, M.; Wu, J.; Zhang, C. Identification of circulating microRNAs as potential biomarkers for detecting acute ischemic stroke. Cell. Mol. Neurobiol. 2015, 35, 433–447. [Google Scholar] [CrossRef]
- Tiedt, S.; Prestel, M.; Malik, R.; Schieferdecker, N.; Duering, M.; Kautzky, V.; Stoycheva, I.; Bock, J.; Northoff, B.H.; Klein, M.; et al. RNA-Seq Identifies Circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as Potential Biomarkers for Acute Ischemic Stroke. Circ. Res. 2017, 121, 970–980. [Google Scholar] [CrossRef]
- Escate, R.; Padro, T.; Suades, R.; Camino, S.; Muniz, O.; Diaz-Diaz, J.L.; Sionis, A.; Mata, P.; Badimon, L. High miR-133a levels in the circulation anticipates presentation of clinical events in familial hypercholesterolaemia patients. Cardiovasc. Res. 2021, 117, 109–122. [Google Scholar] [CrossRef]
- Karakas, M.; Schulte, C.; Appelbaum, S.; Ojeda, F.; Lackner, K.J.; Munzel, T.; Schnabel, R.B.; Blankenberg, S.; Zeller, T. Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease-results from the large AtheroGene study. Eur. Heart J. 2017, 38, 516–523. [Google Scholar] [CrossRef]
- Shen, M.; Xu, X.; Liu, X.; Wang, Q.; Li, W.; You, X.; Peng, R.; Yuan, Y.; Long, P.; Niu, R.; et al. Prospective Study on Plasma MicroRNA-4286 and Incident Acute Coronary Syndrome. J. Am. Heart Assoc. 2021, 10, e018999. [Google Scholar] [CrossRef]
- Su, Y.; Sun, Y.; Tang, Y.; Li, H.; Wang, X.; Pan, X.; Liu, W.; Zhang, X.; Zhang, F.; Xu, Y.; et al. Circulating miR-19b-3p as a Novel Prognostic Biomarker for Acute Heart Failure. J. Am. Heart Assoc. 2021, 10, e022304. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, Y.; Wang, N.; Pan, Z.; Gao, X.; Zhang, F.; Zhang, Y.; Shan, H.; Luo, X.; Bai, Y.; et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation 2010, 122, 2378–2387. [Google Scholar] [CrossRef]
- Ling, T.Y.; Wang, X.L.; Chai, Q.; Lau, T.W.; Koestler, C.M.; Park, S.J.; Daly, R.C.; Greason, K.L.; Jen, J.; Wu, L.Q.; et al. Regulation of the SK3 channel by microRNA-499--potential role in atrial fibrillation. Heart Rhythm 2013, 10, 1001–1009. [Google Scholar] [CrossRef]
- Wang, J.; Bai, Y.; Li, N.; Ye, W.; Zhang, M.; Greene, S.B.; Tao, Y.; Chen, Y.; Wehrens, X.H.; Martin, J.F. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc. Natl. Acad. Sci. USA 2014, 111, 9181–9186. [Google Scholar] [CrossRef] [PubMed]
- Chiang, D.Y.; Kongchan, N.; Beavers, D.L.; Alsina, K.M.; Voigt, N.; Neilson, J.R.; Jakob, H.; Martin, J.F.; Dobrev, D.; Wehrens, X.H.; et al. Loss of microRNA-106b-25 cluster promotes atrial fibrillation by enhancing ryanodine receptor type-2 expression and calcium release. Circ. Arrhythmia Electrophysiol. 2014, 7, 1214–1222. [Google Scholar] [CrossRef] [PubMed]
- Welten, S.M.; Goossens, E.A.; Quax, P.H.; Nossent, A.Y. The multifactorial nature of microRNAs in vascular remodelling. Cardiovasc. Res. 2016, 110, 6–22. [Google Scholar] [CrossRef] [PubMed]
- Pordzik, J.; Pisarz, K.; De Rosa, S.; Jones, A.D.; Eyileten, C.; Indolfi, C.; Malek, L.; Postula, M. The Potential Role of Platelet-Related microRNAs in the Development of Cardiovascular Events in High-Risk Populations, Including Diabetic Patients: A Review. Front. Endocrinol. 2018, 9, 74. [Google Scholar] [CrossRef] [PubMed]
- Nikpay, M.; Beehler, K.; Valsesia, A.; Hager, J.; Harper, M.E.; Dent, R.; McPherson, R. Genome-wide identification of circulating-miRNA expression quantitative trait loci reveals the role of several miRNAs in the regulation of cardiometabolic phenotypes. Cardiovasc. Res. 2019, 115, 1629–1645. [Google Scholar] [CrossRef]
- Kemp, J.R.; Unal, H.; Desnoyer, R.; Yue, H.; Bhatnagar, A.; Karnik, S.S. Angiotensin II-regulated microRNA 483-3p directly targets multiple components of the renin-angiotensin system. J. Mol. Cell. Cardiol. 2014, 75, 25–39. [Google Scholar] [CrossRef]
- Cai, K.; Chen, H. MiR-625-5p Inhibits Cardiac Hypertrophy Through Targeting STAT3 and CaMKII. Hum. Gene Ther. Clin. Dev. 2019, 30, 182–191. [Google Scholar] [CrossRef]
- Spinetti, G.; Fortunato, O.; Caporali, A.; Shantikumar, S.; Marchetti, M.; Meloni, M.; Descamps, B.; Floris, I.; Sangalli, E.; Vono, R.; et al. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ. Res. 2013, 112, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, C.J.; Wharton, J.; Boon, R.A.; Roexe, T.; Tsang, H.; Wojciak-Stothard, B.; Chakrabarti, A.; Howard, L.S.; Gibbs, J.S.; Lawrie, A.; et al. Reduced microRNA-150 is associated with poor survival in pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 2013, 187, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.J.; Olsen, K.; Hamblin, M.; Zhang, J.; Schwendeman, S.P.; Chen, Y.E. Vascular endothelial cell-specific microRNA-15a inhibits angiogenesis in hindlimb ischemia. J. Biol. Chem. 2012, 287, 27055–27064. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Willeit, P.; Tilling, L.; Drozdov, I.; Prokopi, M.; Renard, J.M.; Mayr, A.; Weger, S.; Schett, G.; Shah, A.; et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J. Am. Coll. Cardiol. 2012, 60, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Bye, A.; Rosjo, H.; Nauman, J.; Silva, G.J.; Follestad, T.; Omland, T.; Wisloff, U. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals—The HUNT study. J. Mol. Cell. Cardiol. 2016, 97, 162–168. [Google Scholar] [CrossRef]
- Mick, E.; Shah, R.; Tanriverdi, K.; Murthy, V.; Gerstein, M.; Rozowsky, J.; Kitchen, R.; Larson, M.G.; Levy, D.; Freedman, J.E. Stroke and Circulating Extracellular RNAs. Stroke 2017, 48, 828–834. [Google Scholar] [CrossRef]
- Yan, B.; Yao, J.; Liu, J.Y.; Li, X.M.; Wang, X.Q.; Li, Y.J.; Tao, Z.F.; Song, Y.C.; Chen, Q.; Jiang, Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015, 116, 1143–1156. [Google Scholar] [CrossRef]
- van der Kwast, R.; Woudenberg, T.; Quax, P.H.A.; Nossent, A.Y. MicroRNA-411 and Its 5′-IsomiR Have Distinct Targets and Functions and Are Differentially Regulated in the Vasculature under Ischemia. Mol. Ther. 2020, 28, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Welten, S.M.; Bastiaansen, A.J.; de Jong, R.C.; de Vries, M.R.; Peters, E.A.; Boonstra, M.C.; Sheikh, S.P.; La Monica, N.; Kandimalla, E.R.; Quax, P.H.; et al. Inhibition of 14q32 MicroRNAs miR-329, miR-487b, miR-494, and miR-495 increases neovascularization and blood flow recovery after ischemia. Circ. Res. 2014, 115, 696–708. [Google Scholar] [CrossRef]
- Goossens, E.A.C.; de Vries, M.R.; Simons, K.H.; Putter, H.; Quax, P.H.A.; Nossent, A.Y. miRMap: Profiling 14q32 microRNA Expression and DNA Methylation Throughout the Human Vasculature. Front. Cardiovasc. Med. 2019, 6, 113. [Google Scholar] [CrossRef]
- Downie Ruiz Velasco, A.; Welten, S.M.J.; Goossens, E.A.C.; Quax, P.H.A.; Rappsilber, J.; Michlewski, G.; Nossent, A.Y. Posttranscriptional Regulation of 14q32 MicroRNAs by the CIRBP and HADHB during Vascular Regeneration after Ischemia. Mol. Ther. Nucleic Acids 2019, 14, 329–338. [Google Scholar] [CrossRef]
- Ai, P.; Shen, B.; Pan, H.; Chen, K.; Zheng, J.; Liu, F. MiR-411 suppressed vein wall fibrosis by downregulating MMP-2 via targeting HIF-1alpha. J. Thromb. Thrombolysis 2018, 45, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Stather, P.W.; Sylvius, N.; Sidloff, D.A.; Dattani, N.; Verissimo, A.; Wild, J.B.; Butt, H.Z.; Choke, E.; Sayers, R.D.; Bown, M.J. Identification of microRNAs associated with abdominal aortic aneurysms and peripheral arterial disease. Br. J. Surg. 2015, 102, 755–766. [Google Scholar] [CrossRef]
- Benetatos, L.; Hatzimichael, E.; Londin, E.; Vartholomatos, G.; Loher, P.; Rigoutsos, I.; Briasoulis, E. The microRNAs within the DLK1-DIO3 genomic region: Involvement in disease pathogenesis. Cell. Mol. Life Sci. 2013, 70, 795–814. [Google Scholar] [CrossRef] [PubMed]
- Dimmeler, S.; Yla-Herttuala, S. 14q32 miRNA cluster takes center stage in neovascularization. Circ. Res. 2014, 115, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Wezel, A.; Welten, S.M.; Razawy, W.; Lagraauw, H.M.; de Vries, M.R.; Goossens, E.A.; Boonstra, M.C.; Hamming, J.F.; Kandimalla, E.R.; Kuiper, J.; et al. Inhibition of MicroRNA-494 Reduces Carotid Artery Atherosclerotic Lesion Development and Increases Plaque Stability. Ann. Surg. 2015, 262, 841–847; discussion 847–848. [Google Scholar] [CrossRef] [PubMed]
- Welten, S.M.J.; de Jong, R.C.M.; Wezel, A.; de Vries, M.R.; Boonstra, M.C.; Parma, L.; Jukema, J.W.; van der Sluis, T.C.; Arens, R.; Bot, I.; et al. Inhibition of 14q32 microRNA miR-495 reduces lesion formation, intimal hyperplasia and plasma cholesterol levels in experimental restenosis. Atherosclerosis 2017, 261, 26–36. [Google Scholar] [CrossRef]
- Muiwo, P.; Pandey, P.; Ahmad, H.M.; Ramachandran, S.S.; Bhattacharya, A. IsomiR processing during differentiation of myelogenous leukemic cell line K562 by phorbol ester PMA. Gene 2018, 641, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Li, J.L.; Yuan, X.W. MicroRNA-411 plays a protective role in diabetic retinopathy through targeted regulating Robo4. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9171–9179. [Google Scholar] [CrossRef]
- Shan, D.; Shang, Y.; Hu, T. MicroRNA-411 Inhibits Cervical Cancer Progression by Directly Targeting STAT3. Oncol. Res. 2019, 27, 349–358. [Google Scholar] [CrossRef]
- Nadal, E.; Zhong, J.; Lin, J.; Reddy, R.M.; Ramnath, N.; Orringer, M.B.; Chang, A.C.; Beer, D.G.; Chen, G. A MicroRNA cluster at 14q32 drives aggressive lung adenocarcinoma. Clin. Cancer Res. 2014, 20, 3107–3117. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.L.; Sheng, J.F.; Huang, M.L.; Zou, Y.; Wang, Y.P.; Wang, F.; Zeng, F.; Hua, Q.Q.; Chen, S.M. Integrated analysis of deregulation microRNA expression in head and neck squamous cell carcinoma. Medicine 2021, 100, e24618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, H.; Liu, X.; Hu, Y.; Ding, L.; Zhang, X.; Sun, Q.; Li, Y. Oncogenic microRNA-411 promotes lung carcinogenesis by directly targeting suppressor genes SPRY4 and TXNIP. Oncogene 2019, 38, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, H.; Deng, M.; He, L.; Ping, F.; He, Y.; Fan, Z.; Cheng, B.; Xia, J. Upregulated miR4115p levels promote lymph node metastasis by targeting RYBP in head and neck squamous cell carcinoma. Int. J. Mol. Med. 2021, 47, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, G.; Liu, G.; Ye, Y.; Zhang, C.; Fan, C.; Wang, H.; Cai, H.; Xiao, R.; Huang, Z.; et al. miR-411-5p inhibits proliferation and metastasis of breast cancer cell via targeting GRB2. Biochem. Biophys. Res. Commun. 2016, 476, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Shen, J.; Wu, Y.; Zhong, C.; Fang, L.; Zhu, F.; Duan, S. Dysregulation of miR-411 in cancer: Causative factor for pathogenesis, diagnosis and prognosis. Biomed. Pharmacother. 2022, 149, 112896. [Google Scholar] [CrossRef]
- Konigsbrugge, O.; Simon, A.; Domanovits, H.; Pabinger, I.; Ay, C. Thromboembolic events, bleeding, and drug discontinuation in patients with atrial fibrillation on anticoagulation: A prospective hospital-based registry. BMC Cardiovasc. Disord. 2016, 16, 254. [Google Scholar] [CrossRef]
- Aparicio-Puerta, E.; Lebron, R.; Rueda, A.; Gomez-Martin, C.; Giannoukakos, S.; Jaspez, D.; Medina, J.M.; Zubkovic, A.; Jurak, I.; Fromm, B.; et al. sRNAbench and sRNAtoolbox 2019: Intuitive fast small RNA profiling and differential expression. Nucleic Acids Res. 2019, 47, W530–W535. [Google Scholar] [CrossRef]
- Roberts, T.C.; Coenen-Stass, A.M.; Betts, C.A.; Wood, M.J. Detection and quantification of extracellular microRNAs in murine biofluids. Biol. Proced. Online 2014, 16, 5. [Google Scholar] [CrossRef]
- de Ronde, M.W.J.; Ruijter, J.M.; Lanfear, D.; Bayes-Genis, A.; Kok, M.G.M.; Creemers, E.E.; Pinto, Y.M.; Pinto-Sietsma, S.J. Practical data handling pipeline improves performance of qPCR-based circulating miRNA measurements. RNA 2017, 23, 811–821. [Google Scholar] [CrossRef]

| Cases with Cardiovascular Death (n = 42) | Controls (n = 55) | |
|---|---|---|
| Demographics | ||
| Age, years | 77.0 (71.3–80.8) | 76.0 (71.0–80.0) |
| Male sex | 28 (66.7%) | 37 (67.3%) |
| BMI, kg/m2 [2] | 26.6 (24.6–29.0) | 26.9 (25.3–30.7) |
| Information on atrial fibrillation, n (%) | ||
| Type of atrial fibrillation [2] | ||
| First onset | 4 (9.5%) | 6 (10.9%) |
| Paroxysmal | 18 (42.9%) | 26 (47.3%) |
| Persistent | 3 (7.1%) | 2 (3.6%) |
| Permanent | 16 (38.1%) | 20 (36.4%) |
| History of electrical cardioversion | 10 (23.8%) | 20 (36.4%) |
| History of ablation | 6 (14.3%) | 4 (7.3%) |
| Comorbidities | ||
| Arterial hypertension | 38 (90.5%) | 50 (90.9%) |
| Diabetes mellitus type 2 | 14 (33.3%) | 18 (32.7%) |
| Coronary artery disease | 15 (35.7%) | 17 (30.9%) |
| Peripheral artery disease | 6 (14.3%) | 3 (5.5%) |
| Heart failure | 20 (47.6%) | 13 (23.6%) |
| History of myocardial infarction | 9 (21.4%) | 7 (12.7%) |
| History of stroke, TIA, and systemic embolism | 8 (19.0%) | 15 (27.3%) |
| History of cancer | 8 (19.0%) | 14 (25.5%) |
| History of bleeding | 9 (21.4%) | 10 (18.2%) |
| CHA2DS2-VASc score, median (IQR) | 4 (3–6) | 4 (3–5) |
| HAS-BLED score, median (IQR) | 2 (1–3) | 2 (1–2) |
| Medication | ||
| Anticoagulant therapy | ||
| DOAC | 18 (42.9%) | 26 (47.3%) |
| Vitamin K-Antagonist | 23 (54.8%) | 26 (47.3%) |
| Other | 0 (0%) | 3 (5.5%) |
| None | 1 (2.4%) | 0 (0%) |
| Platelet inhibitor therapy | ||
| Acetylsalicylic acid | 10 (23.8%) | 6 (10.9%) |
| P2Y12-Inhibitors | 4 (9.5%) | 3 (5.5%) |
| Cases with MACE at 1 Year (n = 37) | Controls (n = 65) | |
|---|---|---|
| Demographics | ||
| Age, years | 75.0 (69.0–79.0) | 74.0 (67.0–79.0) |
| Male sex | 24 (64.9%) | 41 (64.6%) |
| BMI, kg/m2 [3] | 27.1 (23.8–29.2) | 26.5 (24.7–31.3) |
| Information on atrial fibrillation, n (%) | ||
| Type of atrial fibrillation [5] | ||
| First onset | 5 (13.5%) | 8 (12.3%) |
| Paroxysmal | 15 (40.5%) | 26 (40.0%) |
| Persistent | 1 (2.7%) | 3 (4.6%) |
| Permanent | 15 (40.5%) | 24 (36.9%) |
| History of electrical cardioversion | 14 (37.8%) | 18 (27.7%) |
| History of ablation | 2 (5.4%) | 8 (12.3%) |
| Comorbidities | ||
| Arterial hypertension | 30 (81.1%) | 59 (90.8%) |
| Diabetes mellitus type 2 | 14 (37.8%) | 21 (32.3%) |
| Coronary artery disease | 11 (29.7%) | 21 (32.3%) |
| Peripheral artery disease | 10 (27.0%) | 5 (7.7%) |
| Heart failure | 19 (51.4%) | 18 (27.7%) |
| History of myocardial infarction | 8 (21.6%) | 8 (12.3%) |
| History of stroke, TIA, systemic embolism | 10 (27.0%) | 16 (24.6%) |
| History of cancer | 9 (24.3%) | 15 (23.1%) |
| History of bleeding | 12 (32.4%) | 13 (20.0%) |
| CHA2DS2-VASc score, median (IQR) | 4 (3–6) | 4 (3–5) |
| HAS-BLED score, median (IQR) | 2 (1–3) | 2 (1–2) |
| Medication | ||
| Anticoagulant therapy | ||
| DOAC | 19 (51.4%) | 35 (53.8%) |
| Vitamin K-Antagonist | 14 (37.8%) | 27 (41.5%) |
| Other | 2 (5.4%) | 1 (1.5%) |
| None | 1 (2.7%) | 2 (3.1%) |
| Platelet inhibitor therapy | ||
| Acetylsalicylic acid | 7 (18.9%) | 10 (15.4%) |
| P2Y12-Inhibitors | 5 (13.5%) | 4 (6.2%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nopp, S.; van der Bent, M.L.; Kraemmer, D.; Königsbrügge, O.; Wojta, J.; Pabinger, I.; Ay, C.; Nossent, A.Y. Circulatory miR-411-5p as a Novel Prognostic Biomarker for Major Adverse Cardiovascular Events in Patients with Atrial Fibrillation. Int. J. Mol. Sci. 2023, 24, 3861. https://doi.org/10.3390/ijms24043861
Nopp S, van der Bent ML, Kraemmer D, Königsbrügge O, Wojta J, Pabinger I, Ay C, Nossent AY. Circulatory miR-411-5p as a Novel Prognostic Biomarker for Major Adverse Cardiovascular Events in Patients with Atrial Fibrillation. International Journal of Molecular Sciences. 2023; 24(4):3861. https://doi.org/10.3390/ijms24043861
Chicago/Turabian StyleNopp, Stephan, M. Leontien van der Bent, Daniel Kraemmer, Oliver Königsbrügge, Johann Wojta, Ingrid Pabinger, Cihan Ay, and Anne Yaël Nossent. 2023. "Circulatory miR-411-5p as a Novel Prognostic Biomarker for Major Adverse Cardiovascular Events in Patients with Atrial Fibrillation" International Journal of Molecular Sciences 24, no. 4: 3861. https://doi.org/10.3390/ijms24043861
APA StyleNopp, S., van der Bent, M. L., Kraemmer, D., Königsbrügge, O., Wojta, J., Pabinger, I., Ay, C., & Nossent, A. Y. (2023). Circulatory miR-411-5p as a Novel Prognostic Biomarker for Major Adverse Cardiovascular Events in Patients with Atrial Fibrillation. International Journal of Molecular Sciences, 24(4), 3861. https://doi.org/10.3390/ijms24043861








