Comparative Analysis of miRNA-mRNA Regulation in the Testes of Gobiocypris rarus following 17α-Methyltestosterone Exposure
Abstract
:1. Introduction
2. Results
2.1. Morphological Changes
2.2. Sex-Steroid Hormone Activity
2.3. RNA- and miRNA-Seq Analyses
2.4. STEM Analysis
2.5. qRT-PCR Validation of DEGs and DEMs
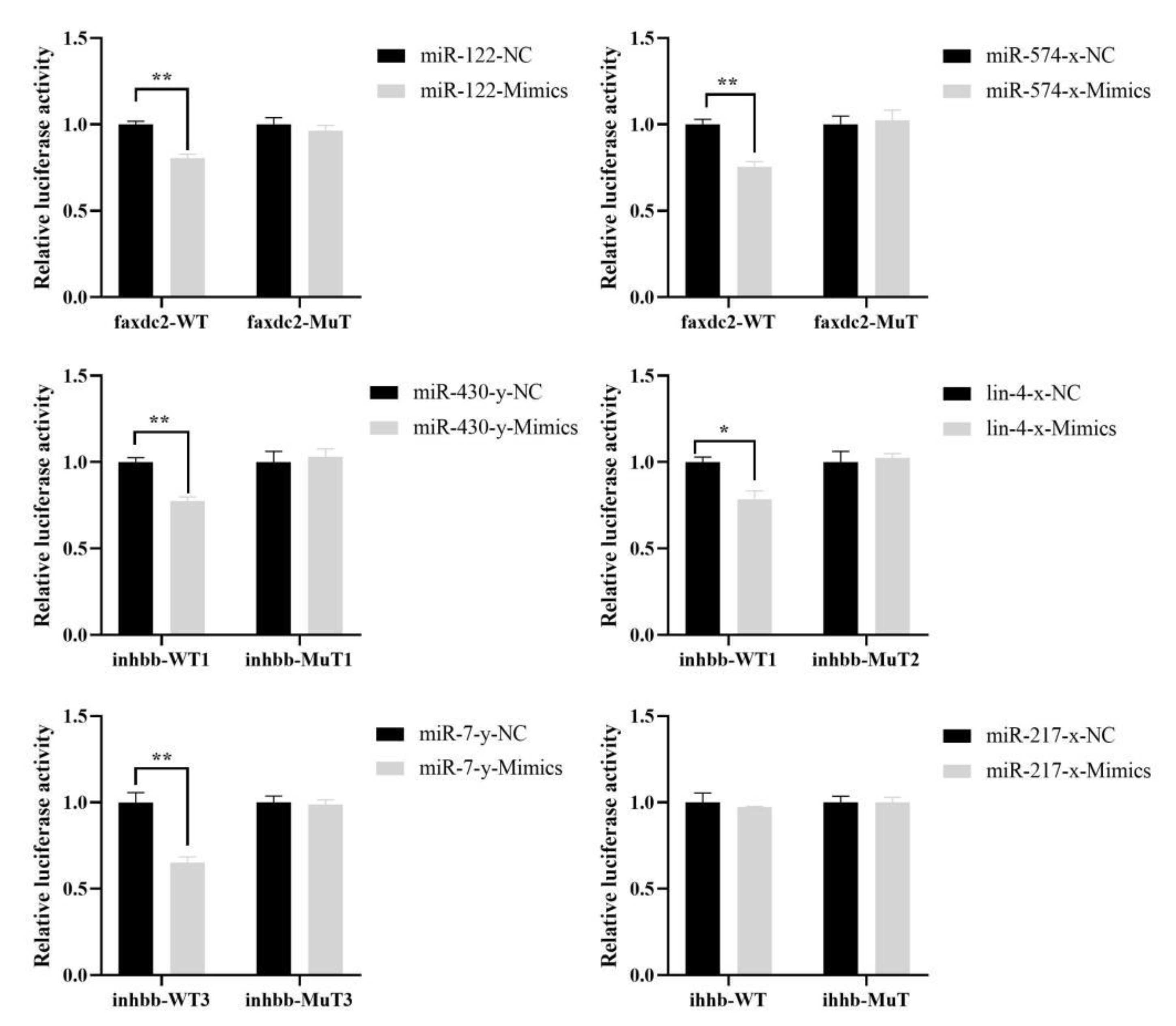
2.6. Targeting Relationship Validations
3. Discussion
4. Material and Methods
4.1. Ethics Statement
4.2. Experimental Animals
4.3. Measurement and Sampling of Biological Indicators
4.3.1. Morphometry and Gonadal Histological Examination
4.3.2. Detecting Steroid Hormone Levels
4.3.3. RNA Sample Collection and Qualification Test
4.4. RNA- and miRNA-Seq
4.5. Differentially Expressed Genes (DEGs) and DE miRNAs (DEMs)
4.6. qRT-PCR Verification of Selected miRNAs and mRNAs
4.7. Dual Luciferase Reporter Assay
4.8. Data Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Kortner, T.M.; Arukwe, A. Effects of 17alpha-methyltestosterone exposure on steroidogenesis and cyclin-B mRNA expression in previtellogenic oocytes of Atlantic cod (Gadus morhua). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007, 146, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.L.J.; Horsfield, J.A.; Black, M.A.; Rutherford, K.; Fisher, A.; Gemmell, N.J. Histological and transcriptomic effects of 17α-methyltestosterone on zebrafish gonad development. BMC Genom. 2017, 18, 557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyu, Q.; Hu, J.; Yang, X.; Liu, X.; Chen, Y.; Xiao, L.; Liu, Y.; Wang, Q.; Chen, J.; Huang, M.; et al. Expression profiles of dmrts and foxls during gonadal development and sex reversal induced by 17α-methyltestosterone in the orange-spotted grouper. Gen. Comp. Endocrinol. 2019, 274, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, S.; Zhang, Y.; Yang, Y.; Yuan, C.; Chen, S.; Wang, Z. Effects of 17 α-methyltestosterone on transcriptome, gonadal histology and sex steroid hormones in rare minnow Gobiocypris rarus. Comp. Biochem. Physiol. Part D Genom. Proteom. 2015, 15, 20–27. [Google Scholar] [CrossRef]
- Liu, S.; Wang, L.; Qin, F.; Zheng, Y.; Li, M.; Zhang, Y.; Yuan, C.; Wang, Z. Gonadal development and transcript profiling of steroidogenic enzymes in response to 17α-methyltestosterone in the rare minnow Gobiocypris rarus. J. Steroid Biochem. Mol. Biol. 2014, 143, 223–232. [Google Scholar] [CrossRef]
- Rutherford, R.; Lister, A.; MacLatchy, D. Comparison of steroidogenic gene expression in mummichog (Fundulus heteroclitus) testis tissue following exposure to aromatizable or non-aromatizable androgens. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2019, 227, 39–49. [Google Scholar] [CrossRef]
- Kelly, A.D.; Hill, K.E.; Correll, M.; Hu, L.; Wang, Y.E.; Rubio, R.; Duan, S.; Quackenbush, J.; Spentzos, D. Next-generation sequencing and microarray-based interrogation of microRNAs from formalin-fixed, paraffin-embedded tissue: Preliminary assessment of cross-platform concordance. Genomics 2013, 102, 8–14. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zhang, G.; Yin, S.; Li, Z.; Wang, Q.; Chen, S.; Zhou, G. Integrated analysis of mRNA-seq and miRNA-seq reveals the potential roles of sex-biased miRNA-mRNA pairs in gonad tissue of dark sleeper (Odontobutis potamophila). BMC Genom. 2017, 18, 613. [Google Scholar] [CrossRef]
- Zhang, G.; Yin, S.; Mao, J.; Liang, F.; Zhao, C.; Li., P.; Zhou, G.; Chen, S.; Tang, Z. Integrated analysis of mRNA-seq and miRNA-seq in the liver of Pelteobagrus vachelli in response to hypoxia. Sci. Rep. 2016, 6, 22907. [Google Scholar] [CrossRef] [Green Version]
- Renaud, L.; Da Silveira, W.A.; Glen, W.B., Jr.; Hazard, E.S.; Hardiman, G. Interplay between micrornas and targeted genes in cellular homeostasis of adult zebrafish (Danio rerio). Curr. Genom. 2018, 19, 615–629. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, W.; Xu, Z.; Pan, J.; Zhao, Z.; Ren, Q. Eriocheir sinensis microRNA-7 targets crab Myd88 to enhance white spot syndrome virus replication. Fish Shellfish Immunol. 2018, 79, 274–283. [Google Scholar] [CrossRef]
- Bizuayehu, T.T.; Mommens, M.; Sundaram, A.Y.M.; Dhanasiri, A.K.S.; Babiak, I. Postovulatory maternal transcriptome in Atlantic salmon and its relation to developmental potential of embryos. BMC Genom. 2019, 20, 315. [Google Scholar] [CrossRef]
- Fischer, P.; Chen, H.; Pacho, F.; Rieder, D.; Kimmel, R.A.; Meyer, D. FoxH1 represses miR-430 during early embryonic development of zebrafish via non-canonical regulation. BMC Biol. 2019, 17, 61. [Google Scholar] [CrossRef] [Green Version]
- He, P.; Wei, P.; Chen, X.; Lin, Y.; Peng, J. Identification and characterization of microRNAs in the gonad of Trachinotus ovatus using Solexa sequencing. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 30, 312–320. [Google Scholar] [CrossRef]
- Wang, F.; Yang, Q.-W.; Zhao, W.-J.; Du, Q.-Y.; Chang, Z.-J. Effects of short-time exposure to atrazine on miRNA expression profiles in the gonad of common carp (Cyprinus carpio). BMC Genom. 2019, 20, 587. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Peng, C.; Ye, Z.; Tang, Z.; Li, S.; Xiao, L.; Liu, S.; Yang, Y.; Zhao, M.; Zhang, Y.; et al. An estradiol-17β/miRNA-26a/cyp19a1a regulatory feedback loop in the protogynous hermaphroditic fish, Epinephelus coioides. Mol. Cell. Endocrinol. 2019, 504, 110689. [Google Scholar] [CrossRef]
- Lan, T.; Chen, Y.-L.; Gul, Y.; Zhao, B.-W.; Gao, Z.-X. Comparative expression analysis of let-7 microRNAs during ovary development in Megalobrama amblycephala. Fish Physiol. Biochem. 2019, 45, 1101–1115. [Google Scholar] [CrossRef]
- Skaftnesmo, K.O.; Edvardsen, R.B.; Furmanek, T.; Crespo, D.; Andersson, E.; Kleppe, L.; Taranger, G.L.; Bogerd, J.; Schulz, R.W.; Wargelius, A. Integrative testis transcriptome analysis reveals differentially expressed miRNAs and their mRNA targets during early puberty in Atlantic salmon. BMC Genom. 2017, 18, 801. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, G.; Sun, Z.; Hou, J.; Wang, Y. microRNA-mRNA analysis in pituitary and hypothalamus of sterile Japanese flounder. Mol. Reprod. Dev. 2019, 86, 727–737. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Colborn, T.; Hayes, T.B.; Heindel, J.J.; Jacobs, D.R., Jr.; Lee, D.-H.; Shioda, T.; Soto, A.M.; vom Saal, F.S.; Welshons, W.V.; et al. Hormones and endocrine-disrupting chemicals: Low-dose effects and nonmonotonic dose responses. Endocr. Rev. 2012, 33, 378–455. [Google Scholar] [CrossRef]
- Hromadová, M.; Hácik, T.; Malatinský, E.; Sklovský, A.; Cervenakov, I. Some measures of lipid metabolism in young sterile males before and after testosterone treatment. Endocrinol. Exp. 1989, 23, 205–211. [Google Scholar] [PubMed]
- Leatherland, J.F. Effects of 17 beta-estradiol and methyltestosterone on the activity of the thyroid gland in rainbow trout, Salmo gairdneri Richardson. Gen. Comp. Endocrinol. 1985, 60, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Janer, G.; Navarro, J.C.; Porte, C. Exposure to TBT increases accumulation of lipids and alters fatty acid homeostasis in the ramshorn snail Marisa cornuarietis. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007, 146, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Xu, Y.; Zang, H.; Yang, L.; Lin, Z.; Li, Y.; Geng, Z. Expression of genes related to lipid transport in meat-type ducks divergent for low or high residual feed intake. Asian-Australas J. Anim. Sci. 2020, 33, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Ren, Y.; Wang, M.; Suraneni, P.K.; Li, D.; Crispino, J.D.; Fan, J.; Huang, Z. Novel function of FAXDC2 in megakaryopoiesis. Blood Cancer J. 2016, 6, e478. [Google Scholar] [CrossRef] [Green Version]
- Bao, J.-W.; Qiang, J.; Tao, Y.-F.; Li, H.-X.; He, J.; Xu, P.; Chen, D.-J. Responses of blood biochemistry, fatty acid composition and expression of microRNAs to heat stress in genetically improved farmed tilapia (Oreochromis niloticus). J. Therm. Biol. 2018, 73, 91–97. [Google Scholar] [CrossRef]
- Qiang, J.; Tao, Y.-F.; He, J.; Xu, P.; Bao, J.-W.; Sun, Y.-L. miR-122 promotes hepatic antioxidant defense of genetically improved farmed tilapia (GIFT, Oreochromis niloticus) exposed to cadmium by directly targeting a metallothionein gene. Aquat. Toxicol. 2017, 182, 39–48. [Google Scholar] [CrossRef]
- Qiang, J.; Tao, Y.F.; Bao, J.W.; Chen, D.J.; Li, H.X.; He, J.; Xu, P. High fat diet-induced miR-122 regulates lipid metabolism and fat deposition in genetically improved farmed tilapia (GIFT, Oreochromis niloticus) liver. Front. Physiol. 2018, 9, 1422. [Google Scholar] [CrossRef] [Green Version]
- Florczyk, M.; Brzuzan, P.; Krom, J.; Woźny, M.; Łakomiak, A. miR-122-5p as a plasma biomarker of liver injury in fish exposed to microcystin-LR. J. Fish Dis. 2016, 39, 741–751. [Google Scholar] [CrossRef]
- Feng, Y.; Ma, J.; Xiang, R.; Li, X. Alterations in microRNA expression in the tissues of silver carp (Hypophthalmichthys molitrix) following microcystin-LR exposure. Toxicon 2017, 128, 15–22. [Google Scholar] [CrossRef]
- Laudadio, I.; Manfroid, I.; Achouri, Y.; Schmidt, D.; Wilson, M.D.; Cordi, S.; Thorrez, L.; Knoops, L.; Jacquemin, P.; Schuit, F.; et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology 2012, 142, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Shao, F.; Yu, J.; Jiang, H.; Gong, D.; Gu, Z. MicroRNA-122 targets genes related to liver metabolism in chickens. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2015, 184, 29–35. [Google Scholar] [CrossRef]
- Ma, J.; Li, Y.; Wu, M.; Zhang, C.; Che, Y.; Li, W.; Li, X. Serum immune responses in common carp (Cyprinus carpio L.) to paraquat exposure: The traditional parameters and circulating microRNAs. Fish Shellfish Immunol. 2018, 76, 133–142. [Google Scholar] [CrossRef]
- Sun, J.; Liu, Q.; Zhao, L.; Cui, C.; Wu, H.; Liao, L.; Tang, G.; Yang, S.; Yang, S. Potential regulation by miRNAs on glucose metabolism in liver of common carp (Cyprinus carpio) at different temperatures. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 32, 100628. [Google Scholar] [CrossRef]
- Rasal, K.; Iquebal, M.A.; Pandey, A.; Behera, P.; Jaiswal, S.; Vasam, M.; Dixit, S.; Raza, M.; Sahoo, L.; Nandi, S.; et al. Revealing liver specific microRNAs linked with carbohydrate metabolism of farmed carp, Labeo rohita (Hamilton, 1822). Genomics 2020, 112, 32–44. [Google Scholar] [CrossRef]
- Zhu, T.; Corraze, G.; Plagnes-Juan, E.; Skiba-Cassy, S. Circulating miRNA measurements are reflective of cholesterol-based changes in rainbow trout (Oncorhynchus mykiss). PLoS ONE 2018, 13, e0206727. [Google Scholar] [CrossRef] [Green Version]
- Jia, Z.-L.; Cen, J.; Wang, J.-B.; Zhang, F.; Xia, Q.; Wang, X.; Chen, X.-Q.; Wang, R.-C.; Hsiao, C.-D.; Liu, K.-C.; et al. Mechanism of isoniazid-induced hepatotoxicity in zebrafish larvae: Activation of ROS-mediated ERS, apoptosis and the Nrf2 pathway. Chemosphere 2019, 227, 541–550. [Google Scholar] [CrossRef]
- Tang, T.; Hu, Y.; Peng, M.; Chu, W.; Hu, Y.; Zhong, L. Effects of high-fat diet on growth performance, lipid accumulation and lipid metabolism-related MicroRNA/gene expression in the liver of grass carp (Ctenopharyngodon idella). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2019, 234, 34–40. [Google Scholar] [CrossRef]
- Wang, X.; Yin, D.; Li, P.; Yin, S.; Wang, L.; Jia, Y.; Shu, X. MicroRNA-sequence profiling reveals novel osmoregulatory microRNA expression patterns in catadromous eel Anguilla marmorata. PLoS ONE 2015, 10, e0136383. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Chu, Q.; Xu, T. miR-122 involved in the regulation of toll-like receptor signaling pathway after Vibrio anguillarum infection by targeting TLR14 in miiuy croaker. Fish Shellfish Immunol. 2016, 58, 67–72. [Google Scholar] [CrossRef]
- Han, J.; Chu, Q.; Huo, R.; Xu, T. Inducible microRNA-122 modulates RIG-I signaling pathway via targeting DAK in miiuy croaker after poly(I:C) stimulation. Dev. Comp. Immunol. 2018, 78, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Ahi, E.P.; Lecaudey, L.A.; Ziegelbecker, A.; Steiner, O.; Glabonjat, R.; Goessler, W.; Hois, V.; Wagner, C.; Lass, A.; Sefc, K.M. Comparative transcriptomics reveals candidate carotenoid color genes in an East African cichlid fish. BMC Genom. 2020, 21, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Zhao, H.; Zhang, L.; Liu, C.; Feng, S.; Ma, J.; Li, Q.; Ke, H.; Wang, X.; Liu, L.; et al. Integrated analysis of RNA-seq and microRNA-seq depicts miRNA-mRNA networks involved in stripe patterns of Botia superciliaris skin. Funct. Integr. Genom. 2019, 19, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Luckenbach, J.A.; Dickey, J.T.; Swanson, P. Follicle-stimulating hormone regulation of ovarian transcripts for steroidogenesis-related proteins and cell survival, growth and differentiation factors in vitro during early secondary oocyte growth in coho salmon. Gen. Comp. Endocrinol. 2011, 171, 52–63. [Google Scholar] [CrossRef]
- Chen, W.; Liu, L.; Ge, W. Expression analysis of growth differentiation factor 9 (Gdf9/gdf9), anti-müllerian hormone (Amh/amh) and aromatase (Cyp19a1a/cyp19a1a) during gonadal differentiation of the zebrafish, Danio rerio. Biol. Reprod. 2017, 96, 401–413. [Google Scholar] [CrossRef] [Green Version]
- Nudelman, G.; Frasca, A.; Kent, B.; Sadler, K.C.; Sealfon, S.C.; Walsh, M.J.; Zaslavsky, E. High resolution annotation of zebrafish transcriptome using long-read sequencing. Genome Res. 2018, 28, 1415–1425. [Google Scholar] [CrossRef] [Green Version]
- Laue, K.; Rajshekar, S.; Courtney, A.J.; Lewis, Z.A.; Goll, M.G. The maternal to zygotic transition regulates genome-wide heterochromatin establishment in the zebrafish embryo. Nat. Commun. 2019, 10, 1551. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.H.; Tang, Y.; Miao, L.; Darwich-Codore, H.; Vejnar, C.E.; Beaudoin, J.-D.; Musaev, D.; Fernandez, J.P.; Benitez, M.D.; Bazzini, A.A.; et al. Brd4 and P300 confer transcriptional competency during zygotic genome activation. Dev. Cell. 2019, 49, 867–881.e8. [Google Scholar] [CrossRef]
- Woldemariam, N.T.; Agafonov, O.; Høyheim, B.; Houston, R.D.; Taggart, J.B.; Andreassen, R. Expanding the miRNA repertoire in Atlantic Salmon; discovery of isomirs and miRNAs highly expressed in different tissues and developmental stages. Cells 2019, 8, E42. [Google Scholar] [CrossRef] [Green Version]
- Romney, A.L.T.; Podrabsky, J.E. Small noncoding RNA profiles along alternative developmental trajectories in an annual killifish. Sci. Rep. 2018, 8, 13364. [Google Scholar] [CrossRef] [Green Version]
- Veil, M.; Schaechtle, M.A.; Gao, M.; Kirner, V.; Buryanova, L.; Grethen, R.; Onichtchouk, D. Maternal Nanog is required for zebrafish embryo architecture and for cell viability during gastrulation. Development 2018, 145, dev155366. [Google Scholar] [CrossRef] [Green Version]
- Ouchi, Y.; Yamamoto, J.; Iwamoto, T. The heterochronic genes lin-28a and lin-28b play an essential and evolutionarily conserved role in early zebrafish development. PLoS ONE 2014, 9, e88086. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Chi, X.; Luo, W.; Yu, S.; Zhang, J.; Guo, Y.; Ren, Q.; Zhang, W. Lung myofibroblast transition and fibrosis is regulated by circ0044226. Int. J. Biochem. Cell Biol. 2020, 118, 105660. [Google Scholar] [CrossRef]
- Sarangdhar, M.A.; Chaubey, D.; Srikakulam, N.; Pillai, B. Parentally inherited long non-coding RNA Cyrano is involved in zebrafish neurodevelopment. Nucleic Acids Res. 2018, 46, 9726–9735. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Lin, C.-Y.; Hsu, C.-C.; Yeh, K.-Y.; Her, G.M. Liver-directed microRNA-7a depletion induces nonalcoholic fatty liver disease by stabilizing YY1-mediated lipogenic pathways in zebrafish. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018, 1863, 844–856. [Google Scholar] [CrossRef]
- Zhou, X.; Chang, Y.; Zhan, Y.; Wang, X.; Lin, K. Integrative mRNA-miRNA interaction analysis associate with immune response of sea cucumber Apostichopus japonicus based on transcriptome database. Fish Shellfish Immunol. 2018, 72, 69–76. [Google Scholar] [CrossRef]
- Zhao, M.-R.; Meng, C.; Xie, X.-L.; Li, C.-H.; Liu, H.-P. Characterization of microRNAs by deep sequencing in red claw crayfish Cherax quadricarinatus haematopoietic tissue cells after white spot syndrome virus infection. Fish Shellfish Immunol. 2016, 59, 469–483. [Google Scholar] [CrossRef]
- Wu, Y.; Li, W.; Yuan, M.; Liu, X. The synthetic pyrethroid deltamethrin impairs zebrafish (Danio rerio) swim bladder development. Sci. Total Environ. 2020, 701, 134870. [Google Scholar] [CrossRef]
- Kaneko, T.; Freeha, K.; Wu, X.; Mogi, M.; Uji, S.; Yokoi, H.; Suzuki, T. Role of notochord cells and sclerotome-derived cells in vertebral column development in fugu, Takifugu rubripes: Histological and gene expression analyses. Cell Tissue Res. 2016, 366, 37–49. [Google Scholar] [CrossRef]
- Wang, S.; Furmanek, T.; Kryvi, H.; Krossøy, C.; Totland, G.K.; Grotmol, S.; Wargelius, A. Transcriptome sequencing of Atlantic salmon (Salmo salar L.) notochord prior to development of the vertebrae provides clues to regulation of positional fate, chordoblast lineage and mineralisation. BMC Genom. 2014, 15, 141. [Google Scholar] [CrossRef] [Green Version]
- Wen, W.; Pillai-Kastoori, L.; Wilson, S.G.; Morris, A.C. Sox4 regulates choroid fissure closure by limiting Hedgehog signaling during ocular morphogenesis. Dev. Biol. 2015, 399, 139–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, Q.; Sun, Y.; Bi, D.; Cui, J.; Xu, T. Up-regulated of miR-8159-5p and miR-217-5p by LPS stimulation negatively co-regulate TLR1 in miiuy croaker. Dev. Comp. Immunol. 2017, 67, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, S.; Niu, H.; Chang, J.; Hu, Z.; Han, Y. Influence of dietary linseed oil as substitution of fish oil on whole fish fatty acid composition, lipid metabolism and oxidative status of juvenile Manchurian trout, Brachymystax lenok. Sci. Rep. 2019, 9, 13846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, N.-Y.; Chan, Y.-J.; Ding, S.-T.; Lee, Y.-H.; HuangFu, W.-C.; Liu, I.-H. Sterol O-Acyltransferase 2 Contributes to the Yolk Cholesterol Trafficking during Zebrafish Embryogenesis. PLoS ONE 2016, 11, e0167644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, T.; Hassan, A.; Thompson, B.M.; McDonald, J.G.; Wang, J.; Li, X. Structural basis for human sterol isomerase in cholesterol biosynthesis and multidrug recognition. Nat. Commun. 2019, 10, 2452. [Google Scholar] [CrossRef] [Green Version]
- Borchel, A.; Verleih, M.; Kühn, C.; Rebl, A.; Goldammer, T. Evolutionary expression differences of creatine synthesis-related genes: Implications for skeletal muscle metabolism in fish. Sci. Rep. 2019, 9, 5429. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, Y.; Shao, M.; Zhang, H. Spatiotemporal expression of the creatine metabolism related genes agat, gamt and ct1 during zebrafish embryogenesis. Int. J. Dev. Biol. 2007, 51, 247–253. [Google Scholar] [CrossRef]
- Mortensen, A.S.; Letcher, R.J.; Cangialosi, M.V.; Chu, S.; Arukwe, A. Tissue bioaccumulation patterns, xenobiotic biotransformation and steroid hormone levels in Atlantic salmon (Salmo salar) fed a diet containing perfluoroactane sulfonic or perfluorooctane carboxylic acids. Chemosphere 2011, 83, 1035–1044. [Google Scholar] [CrossRef]
- Gao, J.; Liu, S.; Zhang, Y.; Yuan, C.; Yang, Y.; Wang, Z. Hepatic expression patterns of aryl hydrocarbon receptor, pregnane X receptor, two cytochrome P450s and five phase II metabolism genes responsive to 17alpha-methyltestosterone in rare minnow Gobiocypris rarus. Environ. Toxicol. Pharmacol. 2014, 37, 1157–1168. [Google Scholar] [CrossRef]
- Lange, A.; Katsu, Y.; Ichikawa, R.; Paull, G.C.; Chidgey, L.L.; Coe, T.S.; Iguchi, T.; Tyler, C.R. Altered sexual development in roach (Rutilus rutilus) exposed to environmental concentrations of the pharmaceutical 17a-ethinylestradiol and associated expression dynamics of aromatases and estrogen receptors. Toxicol. Sci. 2008, 106, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Filteau, M.; Pavey, S.A.; St-Cyr, J.; Bernatchez, L. Gene coexpression networks reveal key drivers of phenotypic divergence in lake whitefish. Mol. Biol. Evol. 2013, 30, 1384–1396. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Jian, T.; Wu, Y.; Zuo, Y.; Li, J.; Lv, H.; Ma, L.; Ren, B.; Zhao, L.; Li, W.; et al. Ellagic acid ameliorates oxidative stress and insulin resistance in high glucose-treated HepG2 cells via miR-223/keap1-Nrf2 pathway. Biomed. Pharmacother. 2019, 110, 85–94. [Google Scholar] [CrossRef]
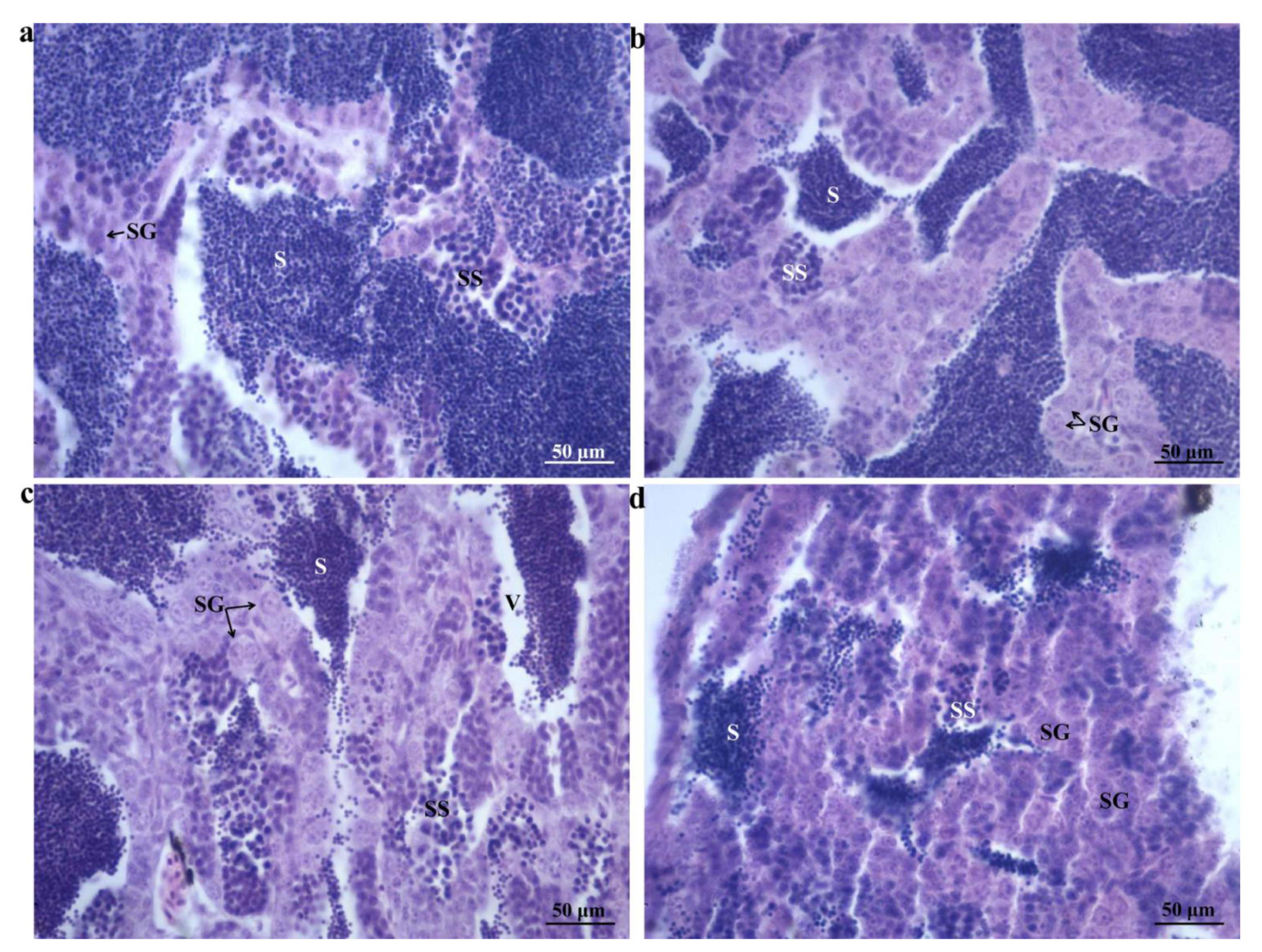
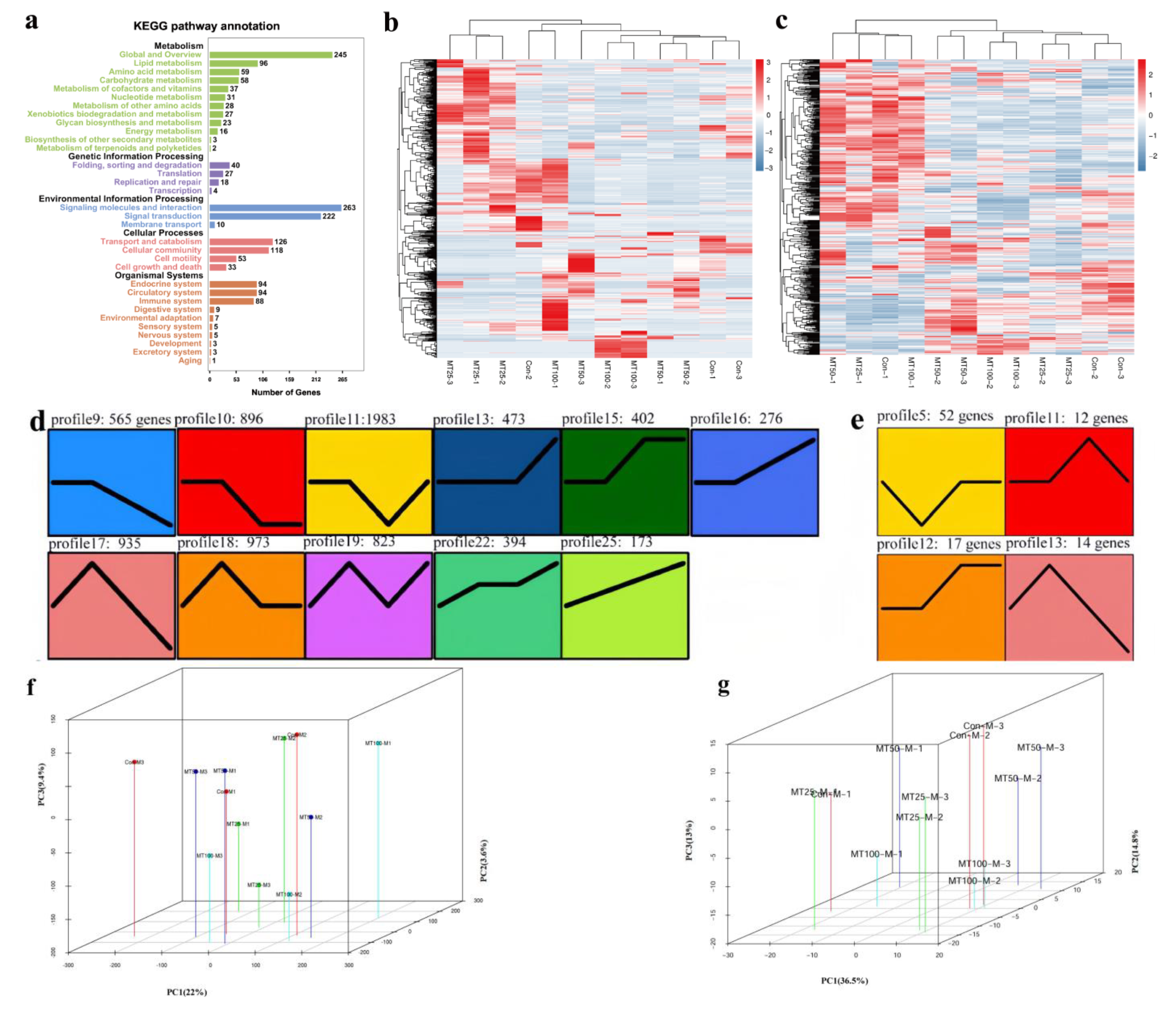
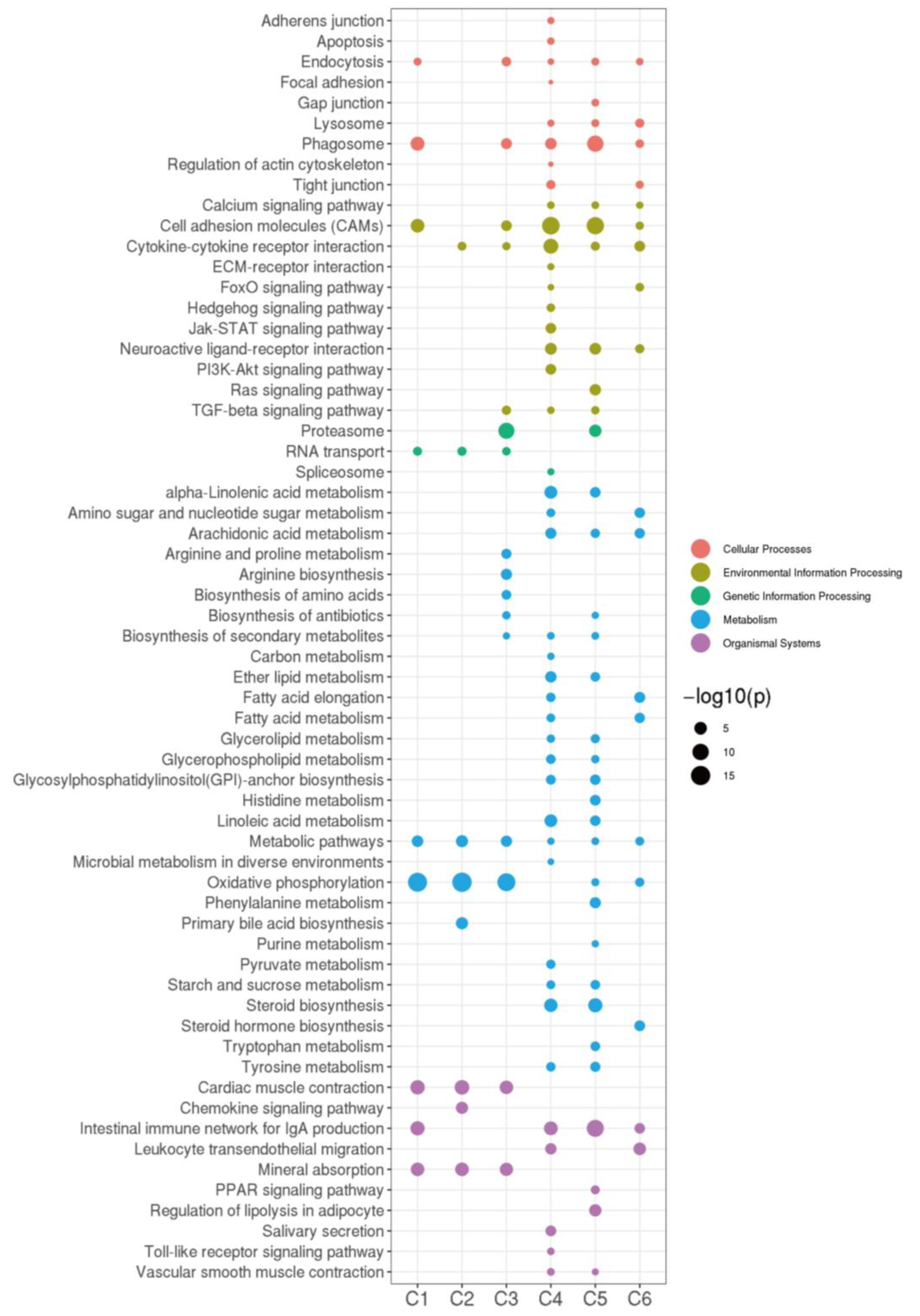

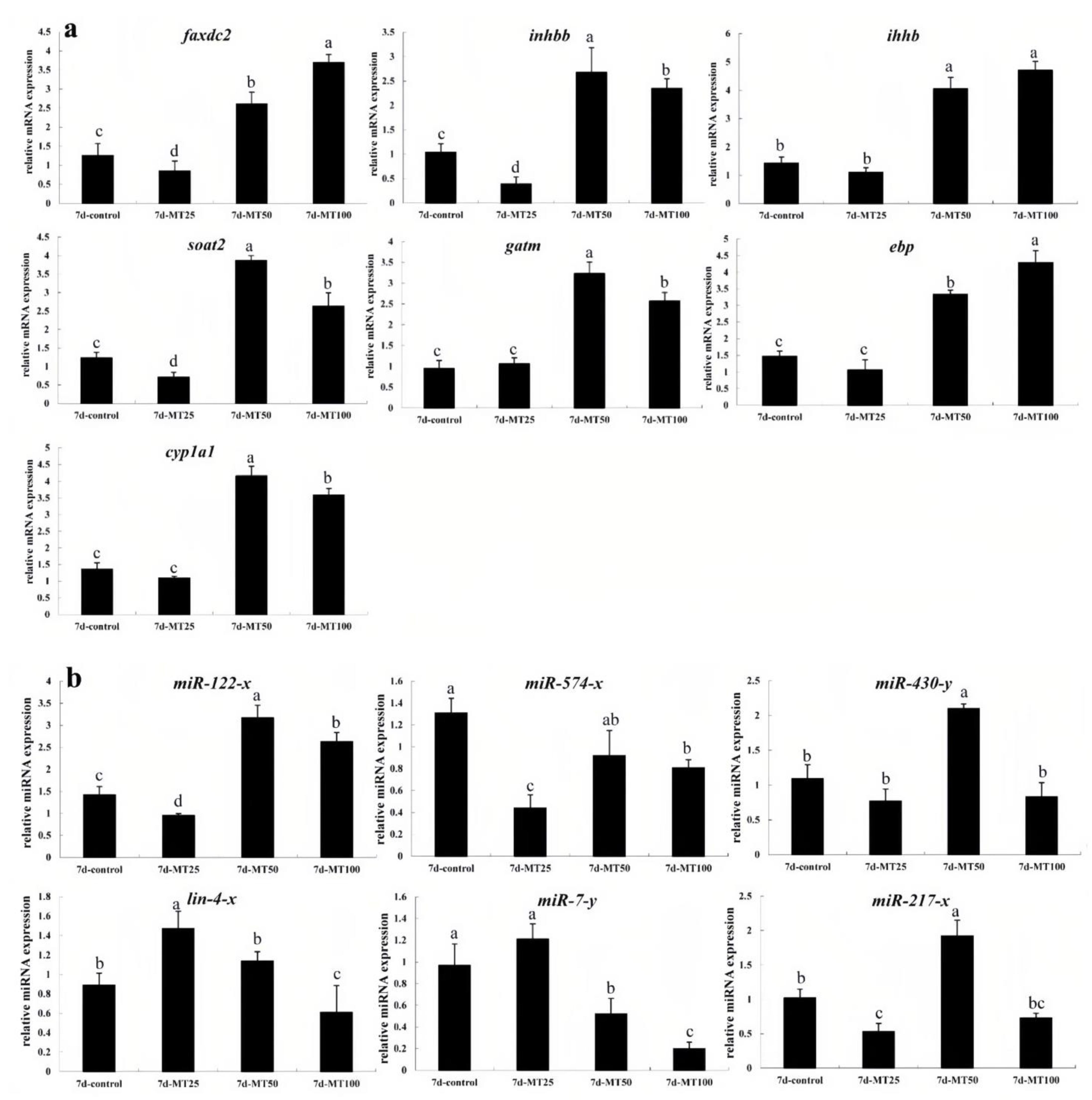
| Group | Body Length (cm) | Total Length (cm) | Total Weight | VTG (ng/mg) | FSH (mIU/mg) | 11-KT (pg/mg) | E2 (pg/mg) | LH (mIU/mg) | T (pg/mg) |
|---|---|---|---|---|---|---|---|---|---|
| Control | 3.64 ± 0.43 | 5.22 ± 0.31 | 1.36 ± 0.32 | 282.12 ± 15.05 | 61.49 ± 0.96 | 43.56 ± 2.04 | 5.37 ± 0.23 | 19.98 ± 0.72 | 78.31 ± 3.09 |
| 25 ng/L MT | 3.52 ± 0.10 | 4.86 ± 0.27 | 1.37 ± 0.26 | 303.70 ± 18.13 | 75.91 ± 0.68 *↑ | 51.99 ± 0.65 **↑ | 7.10 ± 0.06 **↑ | 22.61 ± 0.68 | 75.83 ± 1.72 |
| 50 ng/L MT | 3.44 ± 0.47 | 4.80 ± 0.63 | 1.44 ± 0.65 | 209.75 ± 10.14 *↓ | 41.65 ± 3.70 **↓ | 31.80 ± 0.32 **↓ | 4.26 ± 0.18 **↓ | 11.15 ± 0.91 **↓ | 70.20 ± 7.21 |
| 100 ng/L MT | 3.38 ± 0.33 | 1.24 ± 0.35 | 3.30 ± 0.33 | 185.37 ± 4.21 **↓ | 46.53 ± 2.60 *↓ | 36.11 ± 1.129 **↓ | 4.31 ± 0.20 *↓ | 9.22 ± 1.23 **↓ | 55.20 ± 0.95 *↓ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Zhou, J.; Yang, Q.; Chen, Y.; Liu, Q.; Wang, W.; Song, J.; Wang, X.; Liu, Y. Comparative Analysis of miRNA-mRNA Regulation in the Testes of Gobiocypris rarus following 17α-Methyltestosterone Exposure. Int. J. Mol. Sci. 2023, 24, 4239. https://doi.org/10.3390/ijms24044239
Liu S, Zhou J, Yang Q, Chen Y, Liu Q, Wang W, Song J, Wang X, Liu Y. Comparative Analysis of miRNA-mRNA Regulation in the Testes of Gobiocypris rarus following 17α-Methyltestosterone Exposure. International Journal of Molecular Sciences. 2023; 24(4):4239. https://doi.org/10.3390/ijms24044239
Chicago/Turabian StyleLiu, Shaozhen, Junliang Zhou, Qiong Yang, Yue Chen, Qing Liu, Weiwei Wang, Jing Song, Xianzong Wang, and Yu Liu. 2023. "Comparative Analysis of miRNA-mRNA Regulation in the Testes of Gobiocypris rarus following 17α-Methyltestosterone Exposure" International Journal of Molecular Sciences 24, no. 4: 4239. https://doi.org/10.3390/ijms24044239
APA StyleLiu, S., Zhou, J., Yang, Q., Chen, Y., Liu, Q., Wang, W., Song, J., Wang, X., & Liu, Y. (2023). Comparative Analysis of miRNA-mRNA Regulation in the Testes of Gobiocypris rarus following 17α-Methyltestosterone Exposure. International Journal of Molecular Sciences, 24(4), 4239. https://doi.org/10.3390/ijms24044239






