Polymorphism and Ligand Binding Modulate Fast Dynamics of Human Telomeric G-Quadruplexes
Abstract
1. Introduction
2. Results
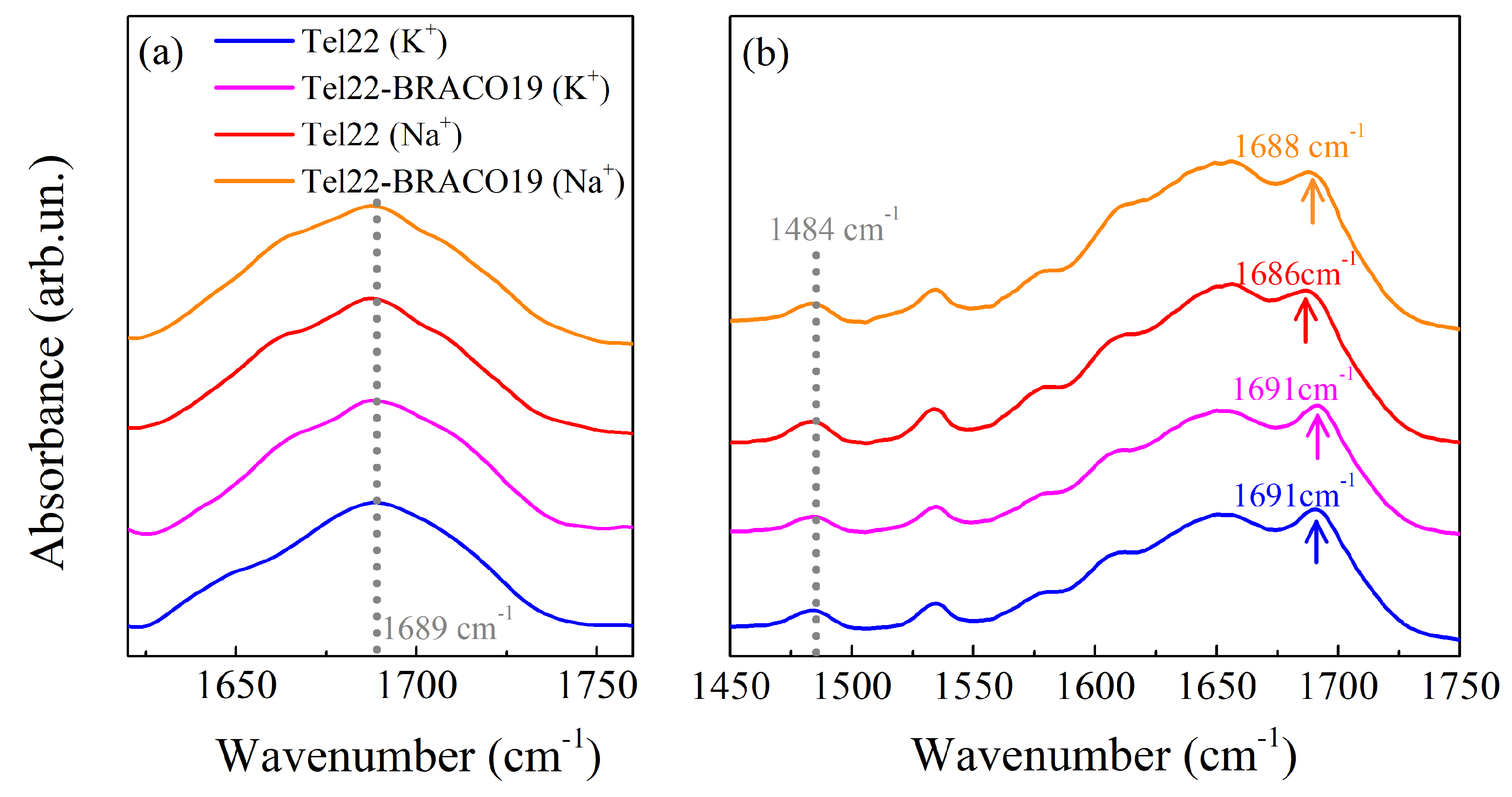
2.1. Infrared Measurements
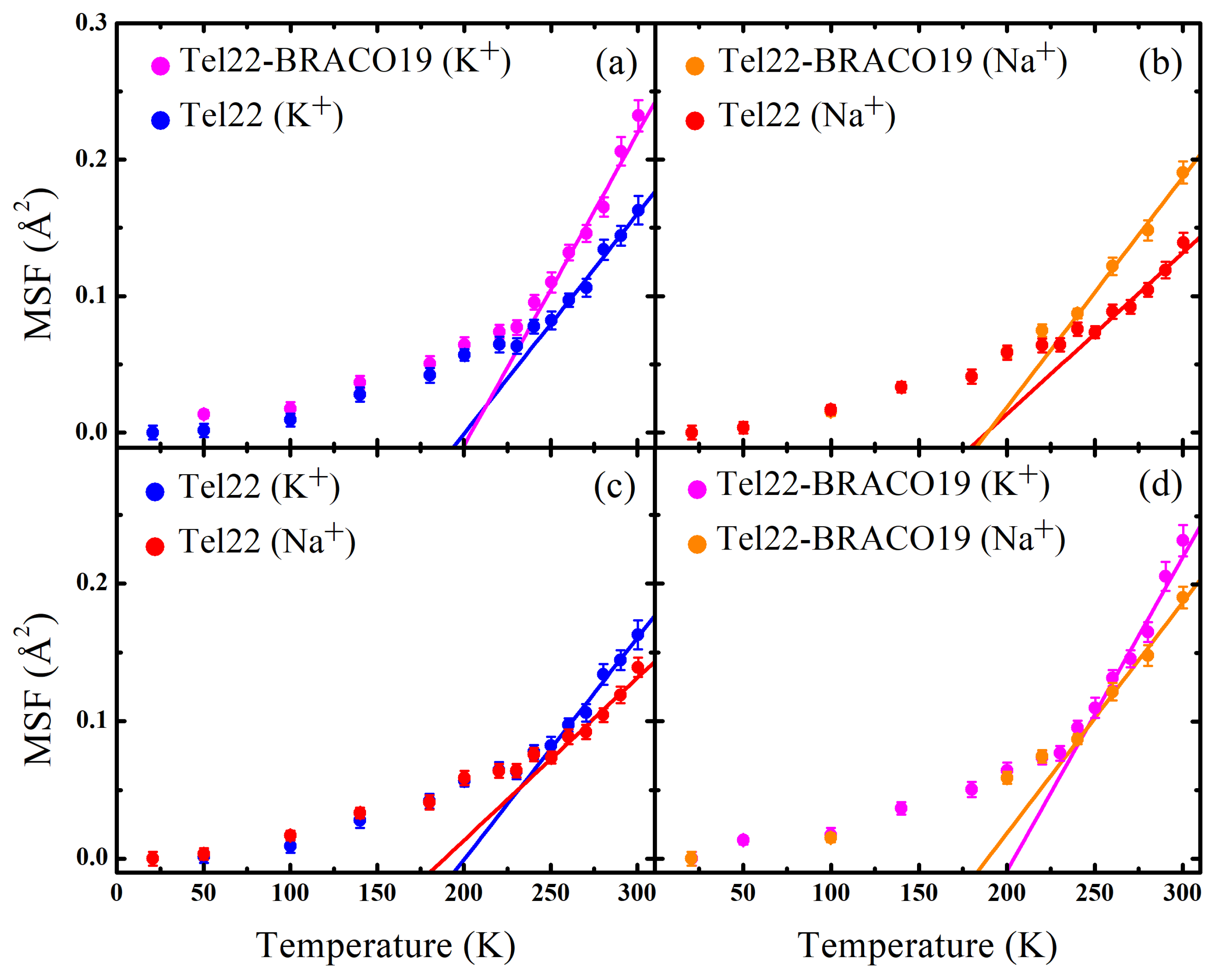
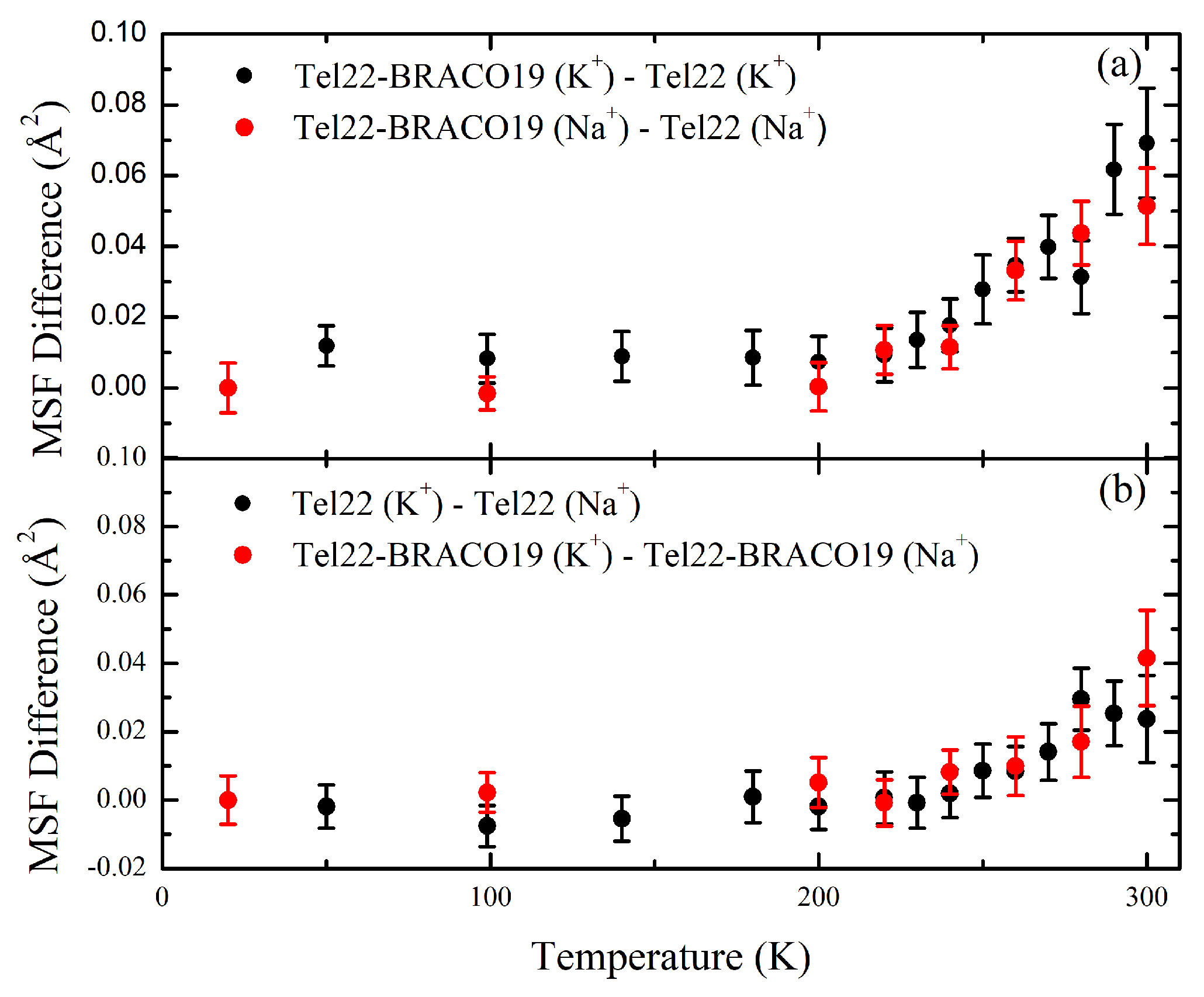
2.2. EINS Measurements
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
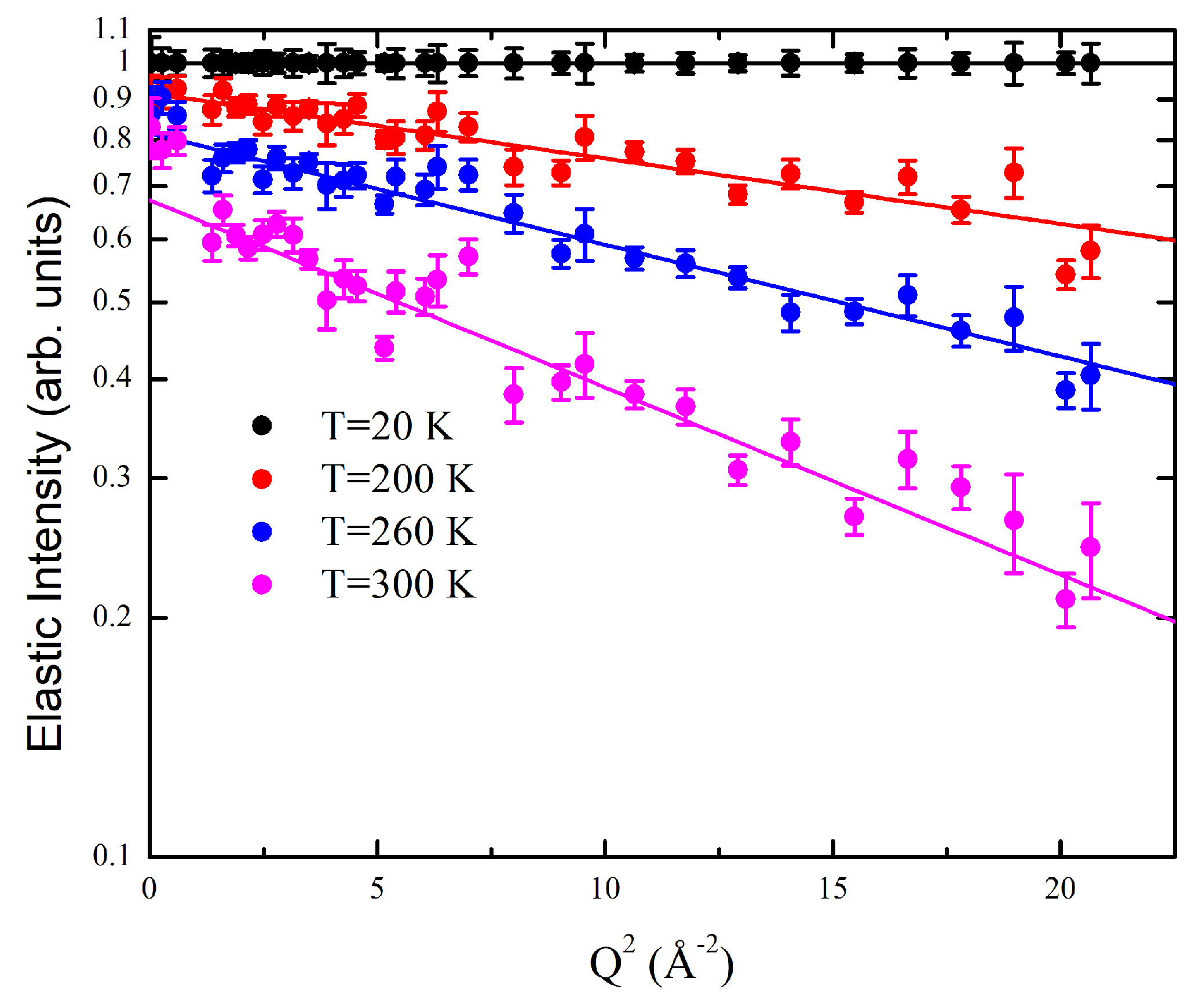
4.2. Neutron Scattering Experiments and Theoretical Model
4.3. Infrared Spectroscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neidle, S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017, 1, 41. [Google Scholar] [CrossRef]
- Collie, G.W.; Parkinson, G.N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011, 40, 5867–5892. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2010, 277, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovská, I.; Renčiuk, D.; Vorlíčková, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef]
- Gray, R.D.; Li, J.; Chaires, J.B. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B 2009, 113, 2676–2683. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The structure and function of DNA G-quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human telomere, oncogenic promoter and 5’-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef]
- Lipps, H.J.; Rhodes, D. G-quadruplex structures: In Vivo evidence and function. Trends Cell Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.q.; Chen, Z.; Zheng, K.w.; Chen, C.y.; Hao, Y.h.; Tan, Z. G-quadruplex formation at the 3’ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011, 39, 6229–6237. [Google Scholar] [CrossRef] [PubMed]
- Testorelli, C. Telomerase and cancer. J. Exp. Clin. Cancer Res. CR 2003, 22, 165–169. [Google Scholar] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Sagne, C.; Marcel, V.; Bota, M.; Martel-Planche, G.; Nobrega, A.; Palmero, E.I.; Perriaud, L.; Boniol, M.; Vagner, S.; Cox, D.G.; et al. Age at cancer onset in germline TP53 mutation carriers: Association with polymorphisms in predicted G-quadruplex structures. Carcinogenesis 2014, 35, 807–815. [Google Scholar] [CrossRef]
- Teng, F.Y.; Jiang, Z.Z.; Guo, M.; Tan, X.Z.; Chen, F.; Xi, X.G.; Xu, Y. G-quadruplex DNA: A novel target for drug design. Cell. Mol. Life Sci. 2021, 78, 6557–6583. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.K.; Jackson, P.J.; Rahman, K.M.; Thurston, D.E. Recent advances in targeting the telomeric G-quadruplex DNA sequence with small molecules as a strategy for anticancer therapies. Future Med. Chem. 2016, 8, 1259–1290. [Google Scholar] [CrossRef] [PubMed]
- Awadasseid, A.; Ma, X.; Wu, Y.; Zhang, W. G-quadruplex stabilization via small-molecules as a potential anti-cancer strategy. Biomed. Pharmacother. 2021, 139, 111550. [Google Scholar] [CrossRef]
- Hsu, S.T.D.; Varnai, P.; Bugaut, A.; Reszka, A.P.; Neidle, S.; Balasubramanian, S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 2009, 131, 13399–13409. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.W.; Alberti, P.; Guedin, A.; Lacroix, L.; Riou, J.F.; Royle, N.J.; Mergny, J.L.; Phan, A.T. Sequence variant (CTAGGG) n in the human telomere favors a G-quadruplex structure containing a G· C· G· C tetrad. Nucleic Acids Res. 2009, 37, 6239–6248. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.F.; Li, M.H.; Hsu, S.T.D.; Chang, T.C. Structural basis of sodium–potassium exchange of a human telomeric DNA quadruplex without topological conversion. Nucleic Acids Res. 2014, 42, 4723–4733. [Google Scholar] [CrossRef] [PubMed]
- Libera, V.; Bianchi, F.; Rossi, B.; D’Amico, F.; Masciovecchio, C.; Petrillo, C.; Sacchetti, F.; Paciaroni, A.; Comez, L. Solvent vibrations as a proxy of the telomere G-quadruplex rearrangements across thermal unfolding. Int. J. Mol. Sci. 2022, 23, 5123. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d [AG3 (T2AG3) 3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef]
- Williamson, J.R.; Raghuraman, M.; Cech, T.R. Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell 1989, 59, 871–880. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Dvorkin, S.A.; Karsisiotis, A.I.; Webba da Silva, M. Encoding canonical DNA quadruplex structure. Sci. Adv. 2018, 4, eaat3007. [Google Scholar] [CrossRef]
- Monsen, R.C.; Maguire, J.M.; DeLeeuw, L.W.; Chaires, J.B.; Trent, J.O. Drug discovery of small molecules targeting the higher-order hTERT promoter G-quadruplex. PLoS ONE 2022, 17, e0270165. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.; Comez, L.; Biehl, R.; D’Amico, F.; Gessini, A.; Longo, M.; Masciovecchio, C.; Petrillo, C.; Radulescu, A.; Rossi, B.; et al. Structure of human telomere G-quadruplex in the presence of a model drug along the thermal unfolding pathway. Nucleic Acids Res. 2018, 46, 11927–11938. [Google Scholar] [CrossRef] [PubMed]
- Di Fonzo, S.; Amato, J.; D’Aria, F.; Caterino, M.; D’Amico, F.; Gessini, A.; Brady, J.W.; Cesàro, A.; Pagano, B.; Giancola, C. Ligand binding to G-quadruplex DNA: New insights from ultraviolet resonance Raman spectroscopy. Phys. Chem. Chem. Phys. 2020, 22, 8128–8140. [Google Scholar] [CrossRef] [PubMed]
- Comez, L.; Bianchi, F.; Libera, V.; Longo, M.; Petrillo, C.; Sacchetti, F.; Sebastiani, F.; D’Amico, F.; Rossi, B.; Gessini, A.; et al. Polymorphism of human telomeric quadruplexes with drugs: A multi-technique biophysical study. Phys. Chem. Chem. Phys. 2020, 22, 11583–11592. [Google Scholar] [CrossRef]
- Libera, V.; Andreeva, E.A.; Martel, A.; Thureau, A.; Longo, M.; Petrillo, C.; Paciaroni, A.; Schirò, G.; Comez, L. Porphyrin Binding and Irradiation Promote G-Quadruplex DNA Dimeric Structure. J. Phys. Chem. Lett. 2021, 12, 8096–8102. [Google Scholar] [CrossRef] [PubMed]
- Frauenfelder, H. The Physics of Proteins: An Introduction to Biological Physics and Molecular Biophysics; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Fenimore, P.W.; Frauenfelder, H.; McMahon, B.; Young, R. Bulk-solvent and hydration-shell fluctuations, similar to α-and β-fluctuations in glasses, control protein motions and functions. Proc. Natl. Acad. Sci. USA 2004, 101, 14408–14413. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, J.R.; Halse, M.E.; Blackledge, M.; Emsley, L. Direct observation of hierarchical protein dynamics. Science 2015, 348, 578–581. [Google Scholar] [CrossRef]
- Henzler-Wildman, K.; Kern, D. Dynamic Personalities of Proteins. Nature 2008, 450, 964–972. [Google Scholar] [CrossRef]
- Sun, D.; Thompson, B.; Cathers, B.E.; Salazar, M.; Kerwin, S.M.; Trent, J.O.; Jenkins, T.C.; Neidle, S.; Hurley, L.H. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997, 40, 2113–2116. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, M.R.; Liquier, J.; Brahmachari, S.; Taillandier, E. Characterization of parallel and antiparallel G-tetraplex structures by vibrational spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2006, 64, 495–503. [Google Scholar] [CrossRef]
- Doster, W.; Cusack, S.; Petry, W. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature 1989, 337, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Ngai, K.; Capaccioli, S.; Paciaroni, A. Change of caged dynamics at T g in hydrated proteins: Trend of mean squared displacements after correcting for the methyl-group rotation contribution. J. Chem. Phys. 2013, 138, 06B611_1. [Google Scholar] [CrossRef] [PubMed]
- Renčiuk, D.; Kejnovská, I.; Školáková, P.; Bednářová, K.; Motlová, J.; Vorlíčková, M. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009, 37, 6625–6634. [Google Scholar] [CrossRef]
- Abu-Ghazalah, R.M.; Rutledge, S.; Lau, L.W.; Dubins, D.N.; Macgregor Jr, R.B.; Helmy, A.S. Concentration-dependent structural transitions of human telomeric DNA sequences. Biochemistry 2012, 51, 7357–7366. [Google Scholar] [CrossRef]
- Machireddy, B.; Kalra, G.; Jonnalagadda, S.; Ramanujachary, K.; Wu, C. Probing the binding pathway of BRACO19 to a parallel-stranded human telomeric G-quadruplex using molecular dynamics binding simulation with AMBER DNA OL15 and ligand GAFF2 force fields. J. Chem. Inf. Model. 2017, 57, 2846–2864. [Google Scholar] [CrossRef] [PubMed]
- Machireddy, B.; Sullivan, H.J.; Wu, C. Binding of BRACO19 to a telomeric G-quadruplex DNA probed by all-atom molecular dynamics simulations with explicit solvent. Molecules 2019, 24, 1010. [Google Scholar] [CrossRef]
- Ghosh, A.; Trajkovski, M.; Teulade-Fichou, M.P.; Gabelica, V.; Plavec, J. Phen-DC3 Induces Refolding of Human Telomeric DNA into a Chair-Type Antiparallel G-Quadruplex through Ligand Intercalation. Angew. Chem. 2022, 134, e202207384. [Google Scholar] [CrossRef]
- Fenimore, P.W.; Frauenfelder, H.; McMahon, B.H.; Parak, F.G. Slaving: Solvent fluctuations dominate protein dynamics and functions. Proc. Natl. Acad. Sci. USA 2002, 99, 16047–16051. [Google Scholar] [CrossRef]
- Li, K.; Yatsunyk, L.; Neidle, S. Water spines and networks in G-quadruplex structures. Nucleic Acids Res. 2021, 49, 519–528. [Google Scholar] [CrossRef]
- Gallina, M.E.; Sassi, P.; Paolantoni, M.; Morresi, A.; Cataliotti, R.S. Vibrational analysis of molecular interactions in aqueous glucose solutions. Temperature and concentration effects. J. Phys. Chem. B 2006, 110, 8856–8864. [Google Scholar] [CrossRef]
- Freda, M.; Piluso, A.; Santucci, A.; Sassi, P. Transmittance Fourier transform infrared spectra of liquid water in the whole mid-infrared region: Temperature dependence and structural analysis. Appl. Spectrosc. 2005, 59, 1155–1159. [Google Scholar] [CrossRef]
- Bertini, L.; Libera, V.; Ripanti, F.; Seydel, T.; Paolantoni, M.; Orecchini, A.; Petrillo, C.; Comez, L.; Paciaroni, A. Role of fast dynamics in the complexation of G-quadruplexes with small molecules. Phys. Chem. Chem. Phys. 2022, 24, 29232–29240. [Google Scholar] [CrossRef] [PubMed]
- Roh, J.; Novikov, V.; Gregory, R.; Curtis, J.; Chowdhuri, Z.; Sokolov, A. Onsets of anharmonicity in protein dynamics. Phys. Rev. Lett. 2005, 95, 038101. [Google Scholar] [CrossRef]
- Fitter, J. The temperature dependence of internal molecular motions in hydrated and dry α-amylase: The role of hydration water in the dynamical transition of proteins. Biophys. J. 1999, 76, 1034–1042. [Google Scholar] [CrossRef]
- Paciaroni, A.; Cinelli, S.; Onori, G. Effect of the environment on the protein dynamical transition: A neutron scattering study. Biophys. J. 2002, 83, 1157–1164. [Google Scholar] [CrossRef]
- Neidle, S. Structured waters mediate small molecule binding to G-quadruplex nucleic acids. Pharmaceuticals 2022, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Lovesey, S.W. Theory of Neutron Scattering from Condensed Matter; Clarendon Oxford: Oxford, UK, 1984. [Google Scholar]
- Bée, M. Quasielastic Neutron Scattering; Adam & Hilger: London, UK, 1988. [Google Scholar]
- Natali, F.; Peters, J.; Russo, D.; Barbieri, S.; Chiapponi, C.; Cupane, A.; Deriu, A.; Di Bari, M.; Farhi, E.; Gerelli, Y.; et al. IN13 backscattering spectrometer at ILL: Looking for motions in biological macromolecules and organisms. Neutron. News 2008, 19, 14–18. [Google Scholar]
- Richard, D.; Ferrand, M.; Kearley, G. Lamp, the large array manipulation program. J. Neutron. Res. 1996, 4, 33–39. [Google Scholar] [CrossRef]
- Yi, Z.; Miao, Y.; Baudry, J.; Jain, N.; Smith, J.C. Derivation of Mean-Square Displacements for Protein Dynamics from Elastic Incoherent Neutron Scattering. J. Phys. Chem. B 2012, 116, 5028–5036. [Google Scholar] [CrossRef]
- Gabel, F.; Bicout, D.; Lehnert, U.; Tehei, M.; Weik, M.; Zaccai, G. Protein dynamics studied by neutron scattering. Q. Rev. Biophys. 2002, 35, 327–367. [Google Scholar] [CrossRef]
- Bicout, D.; Zaccai, G. Protein flexibility from the dynamical transition: A force constant analysis. Biophys. J. 2001, 80, 1115–1123. [Google Scholar] [CrossRef]
- Katava, M.; Stirnemann, G.; Zanatta, M.; Capaccioli, S.; Pachetti, M.; Ngai, K.; Sterpone, F.; Paciaroni, A. Critical structural fluctuations of proteins upon thermal unfolding challenge the Lindemann criterion. Proc. Natl. Acad. Sci. USA 2017, 114, 9361–9366. [Google Scholar] [CrossRef] [PubMed]
- Rosi, B.P.; D’Angelo, A.; Buratti, E.; Zanatta, M.; Tavagnacco, L.; Natali, F.; Zamponi, M.; Noferini, D.; Corezzi, S.; Zaccarelli, E.; et al. Impact of the Environment on the PNIPAM Dynamical Transition Probed by Elastic Neutron Scattering. Macromolecules 2022, 55, 4752–4765. [Google Scholar] [CrossRef]
- Zeller, D.; Telling, M.; Zamponi, M.; Garcia Sakai, V.; Peters, J. Analysis of elastic incoherent neutron scattering data beyond the Gaussian approximation. J. Chem. Phys. 2018, 149, 234908. [Google Scholar] [CrossRef] [PubMed]
- Price, D.A.; Wedamulla, P.; Hill, T.D.; Loth, T.M.; Moran, S.D. The polarization dependence of 2D IR cross-peaks distinguishes parallel-stranded and antiparallel-stranded DNA G-quadruplexes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 267, 120596. [Google Scholar] [CrossRef] [PubMed]

| Tel22 (K) | Tel22 (Na) | Tel22 (K) | Tel22 (Na) | |
|---|---|---|---|---|
| -BRACO19 | -BRACO19 | |||
| (N/m) | 2.9 ± 0.2 | 3.6 ± 0.3 | 2.1 ± 0.3 | 2.7 ± 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertini, L.; Libera, V.; Ripanti, F.; Natali, F.; Paolantoni, M.; Orecchini, A.; Nucara, A.; Petrillo, C.; Comez, L.; Paciaroni, A. Polymorphism and Ligand Binding Modulate Fast Dynamics of Human Telomeric G-Quadruplexes. Int. J. Mol. Sci. 2023, 24, 4280. https://doi.org/10.3390/ijms24054280
Bertini L, Libera V, Ripanti F, Natali F, Paolantoni M, Orecchini A, Nucara A, Petrillo C, Comez L, Paciaroni A. Polymorphism and Ligand Binding Modulate Fast Dynamics of Human Telomeric G-Quadruplexes. International Journal of Molecular Sciences. 2023; 24(5):4280. https://doi.org/10.3390/ijms24054280
Chicago/Turabian StyleBertini, Luca, Valeria Libera, Francesca Ripanti, Francesca Natali, Marco Paolantoni, Andrea Orecchini, Alessandro Nucara, Caterina Petrillo, Lucia Comez, and Alessandro Paciaroni. 2023. "Polymorphism and Ligand Binding Modulate Fast Dynamics of Human Telomeric G-Quadruplexes" International Journal of Molecular Sciences 24, no. 5: 4280. https://doi.org/10.3390/ijms24054280
APA StyleBertini, L., Libera, V., Ripanti, F., Natali, F., Paolantoni, M., Orecchini, A., Nucara, A., Petrillo, C., Comez, L., & Paciaroni, A. (2023). Polymorphism and Ligand Binding Modulate Fast Dynamics of Human Telomeric G-Quadruplexes. International Journal of Molecular Sciences, 24(5), 4280. https://doi.org/10.3390/ijms24054280







