Atf7ip Inhibits Osteoblast Differentiation via Negative Regulation of the Sp7 Transcription Factor
Abstract
:1. Introduction
2. Results
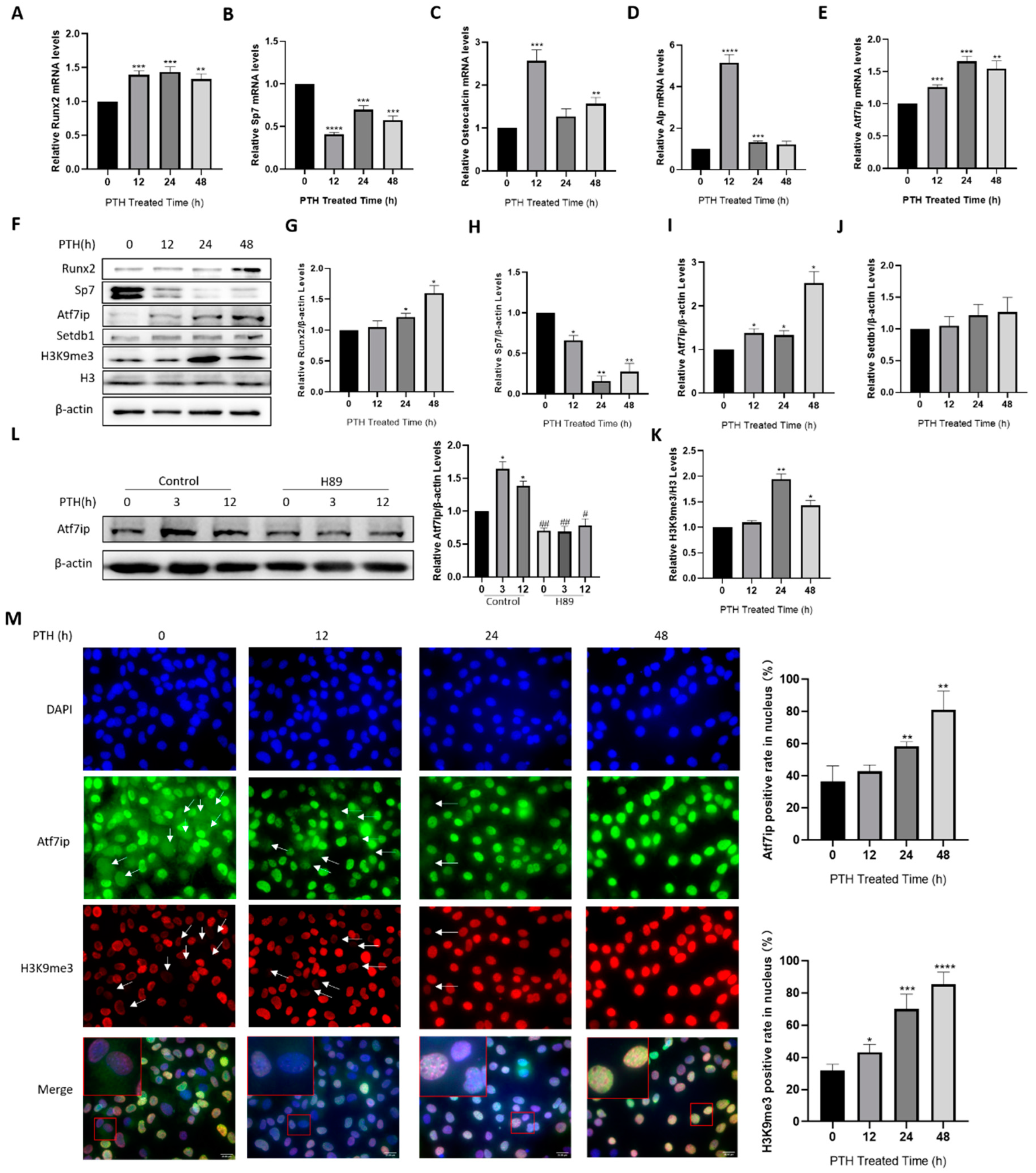
2.1. Atf7ip Expression Increases during Osteoblast Differentiation
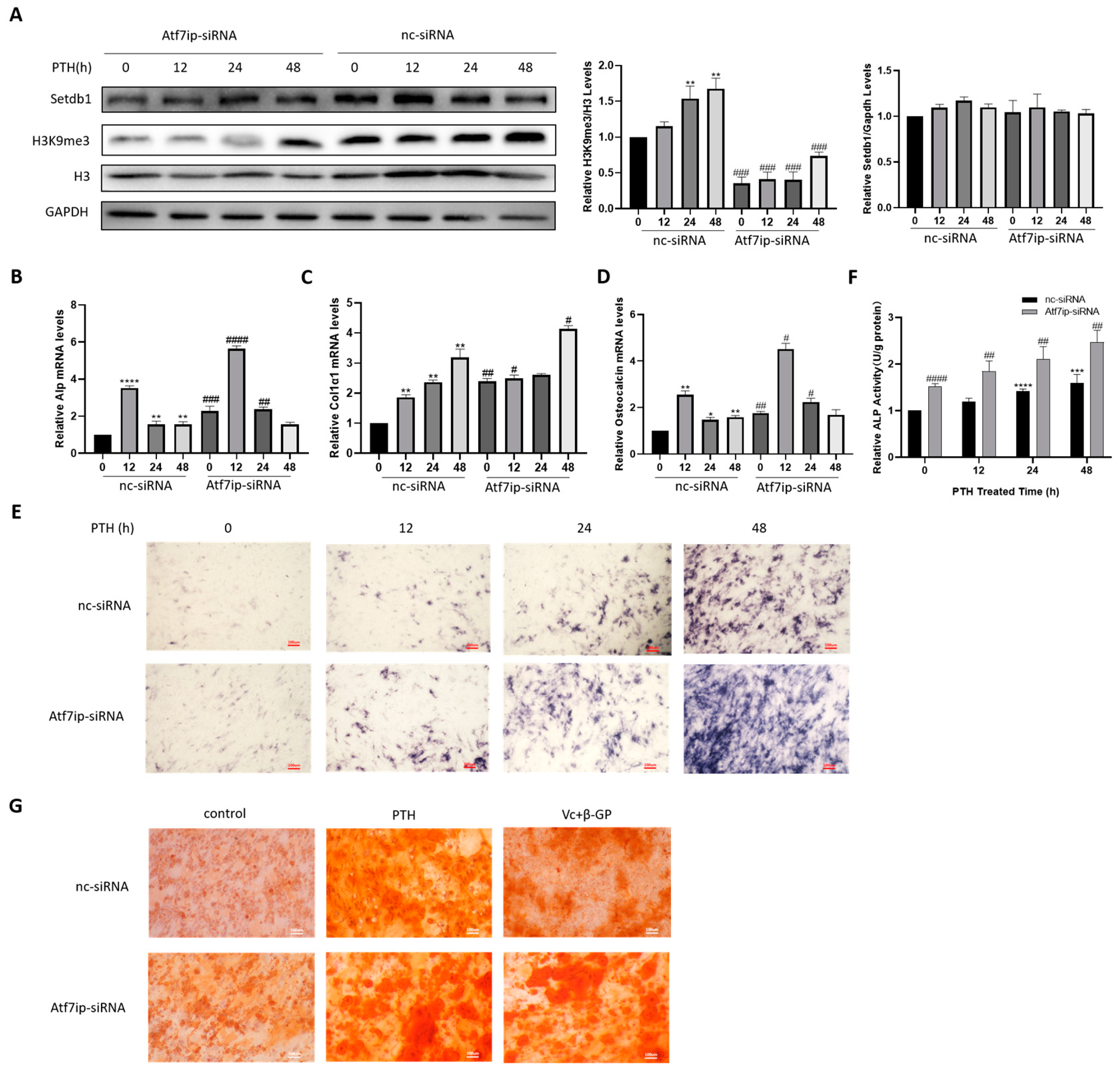
2.2. Silencing of Atf7ip Expression Promotes MC3T3-E1 Osteogenesis
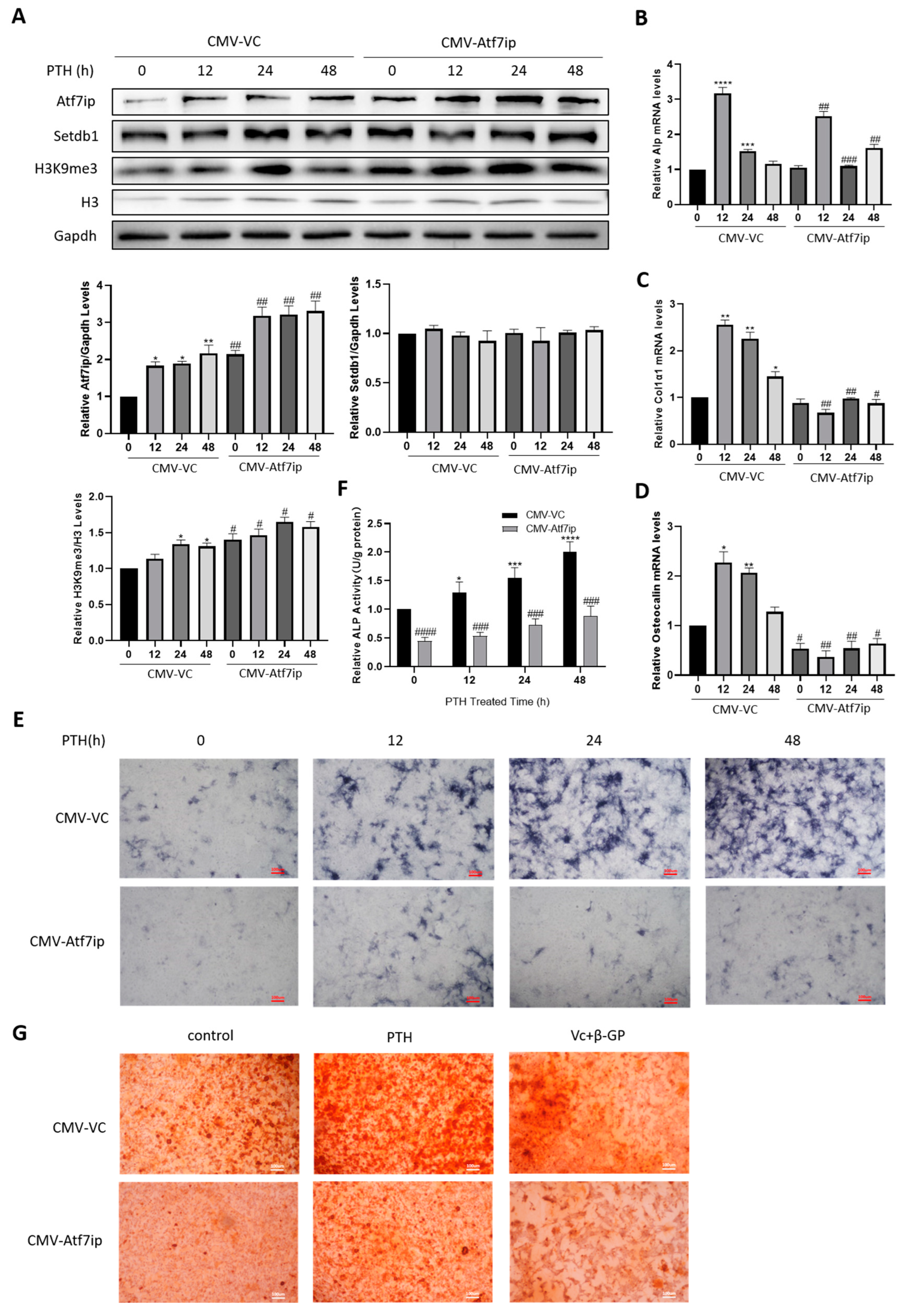
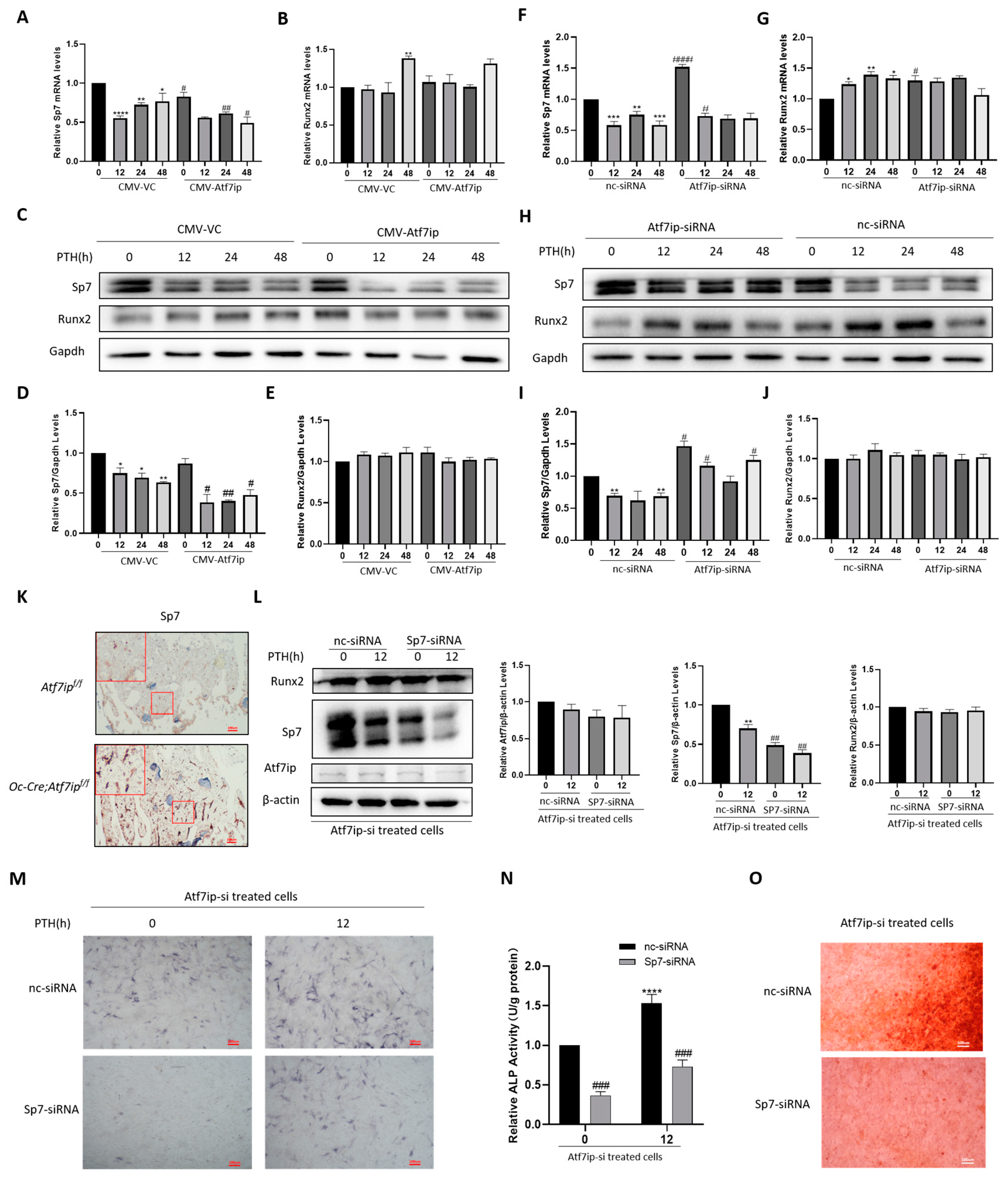
2.3. Atf7ip Overexpression Inhibits the Osteogenic Differentiation of MC3T3-E1 Cells
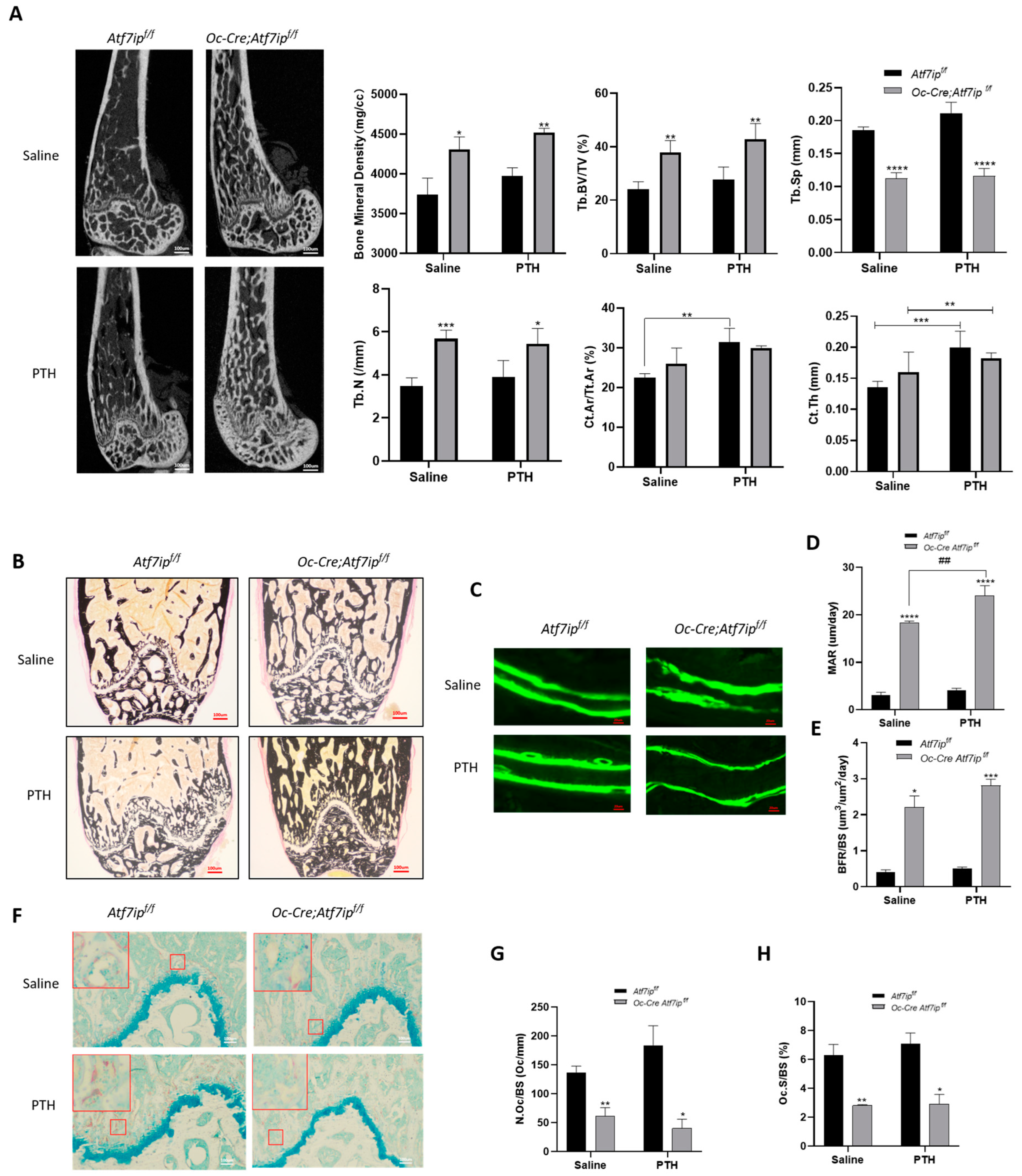
2.4. Osteoblastic Atf7ip Deficiency Mice Show Higher Bone Formation
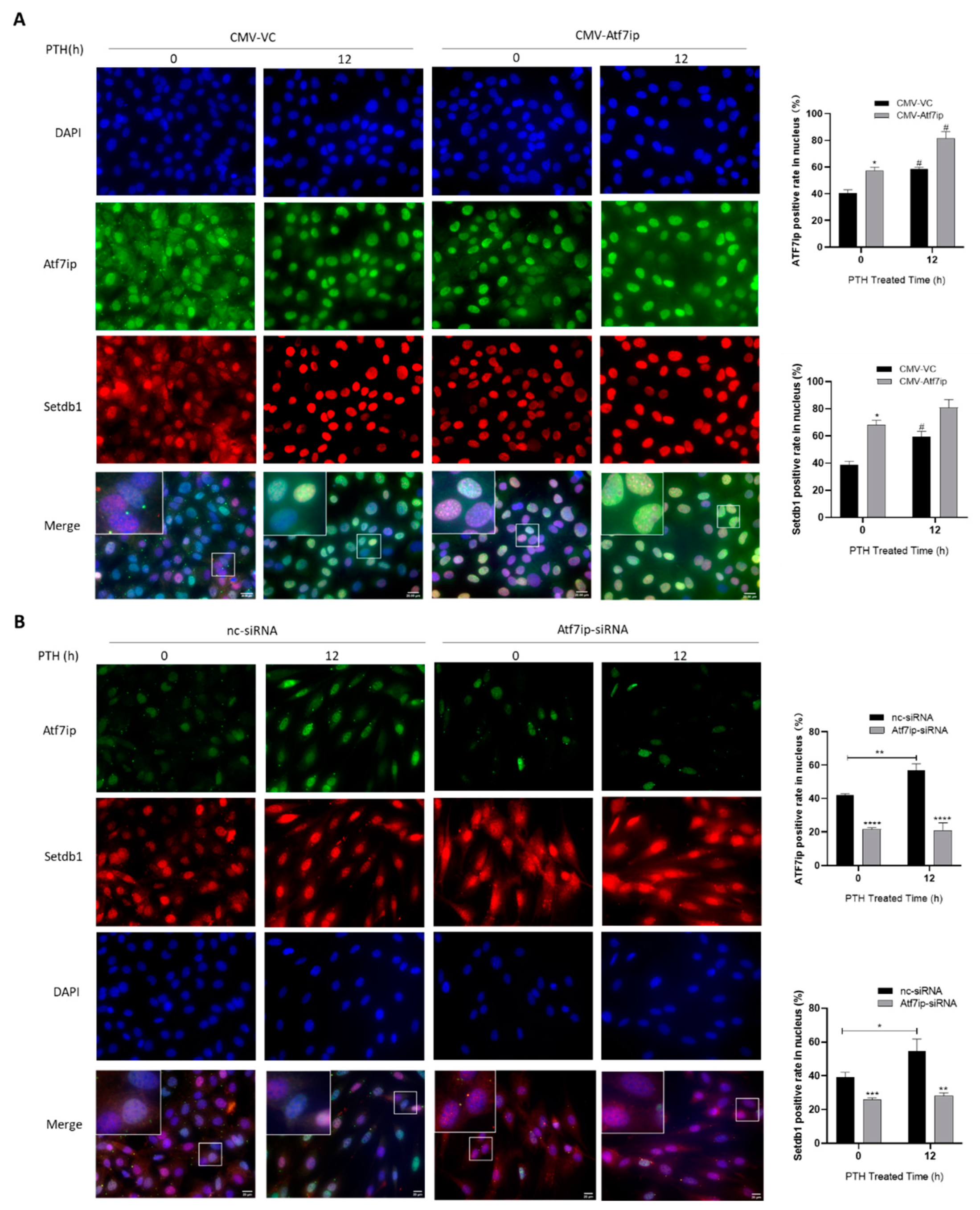
2.5. Expression of Atf7ip Influences the Localization of Setdb1 in the Nucleus
2.6. Inhibition of Sp7 Inhibits Atf7ip–siRNA-Promoting Bone Differentiation
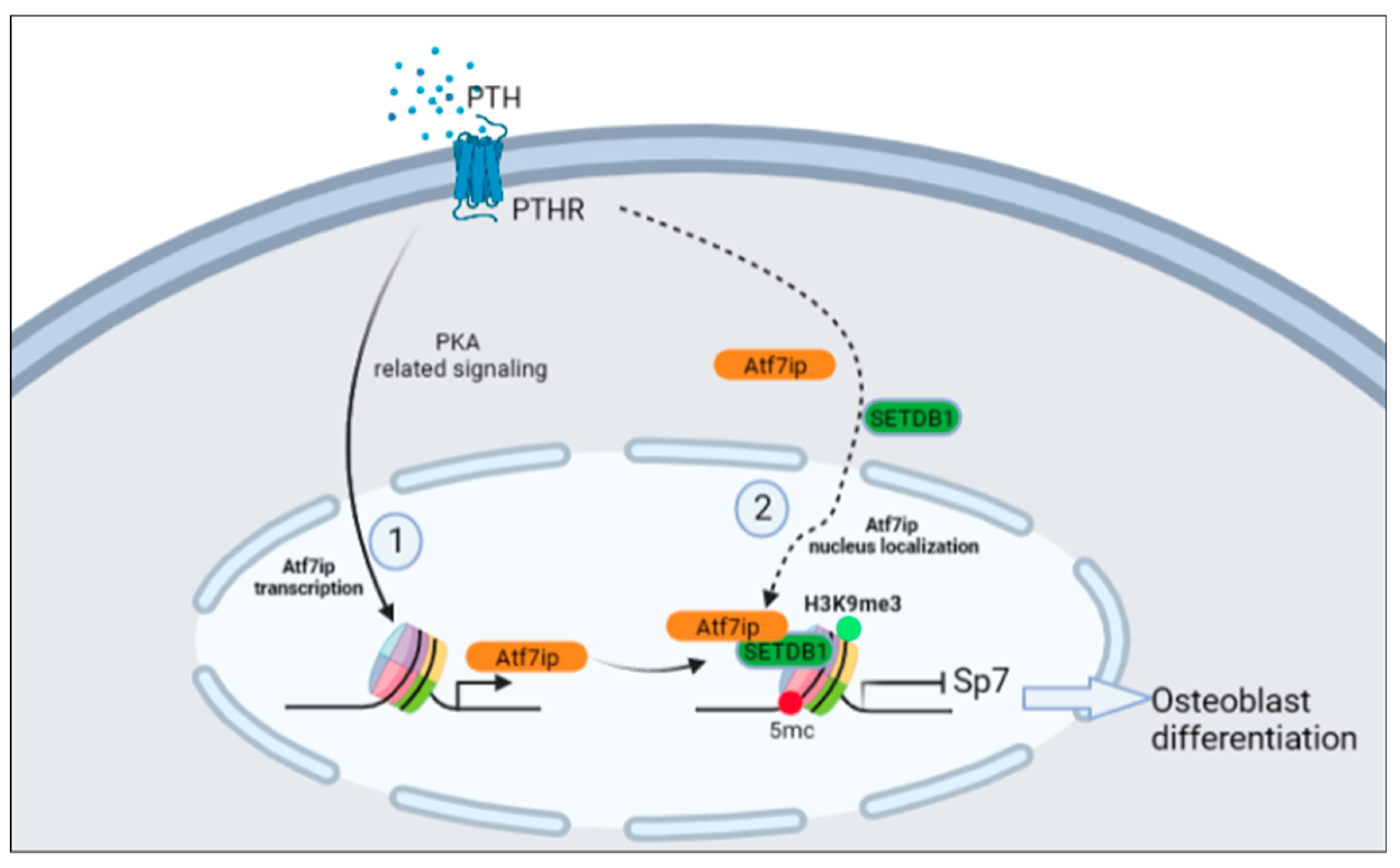
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cell Culture
4.3. Generation of Atf7ip Conditional Mouse Strains and In Vivo Parathyroid Hormone Treatment
4.4. siRNA-Mediated Knockdown
4.5. Construction of Atf7ip Plasmid
4.6. Osteogenic Differentiation
4.7. Adipogenic Differentiation
4.8. RNA Extraction and q-PCR
4.9. Western Blot
4.10. Immunofluorescence Staining
4.11. Alkaline Phosphatase (Alp) Staining and ALP Activity Assay
4.12. Alizarin Red Staining
4.13. Immunohistochemical Staining
4.14. Calcein Double Labeling and Von Kossa Staining
4.15. Micro-CT
4.16. Cell Proliferation
4.17. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hojo, H.; Ohba, S. Gene regulatory landscape in osteoblast differentiation. Bone 2020, 137, 115458. [Google Scholar] [CrossRef]
- Sepulveda, H.; Villagra, A.; Montecino, M. Tet-Mediated DNA Demethylation Is Required for SWI/SNF-Dependent Chromatin Remodeling and Histone-Modifying Activities That Trigger Expression of the Sp7 Osteoblast Master Gene during Mesenchymal Lineage Commitment. Mol. Cell Biol. 2017, 37, e00177-17. [Google Scholar] [CrossRef] [Green Version]
- Komori, T. Regulation of Proliferation, Differentiation and Functions of Osteoblasts by Runx2. Int. J. Mol. Sci. 2019, 20, 1694. [Google Scholar] [CrossRef] [Green Version]
- Long, F. Building strong bones: Molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 2011, 13, 27–38. [Google Scholar] [CrossRef]
- Montecino, M.; Carrasco, M.E.; Nardocci, G. Epigenetic Control of Osteogenic Lineage Commitment. Front. Cell Dev. Biol. 2020, 8, 611197. [Google Scholar] [CrossRef]
- Padeken, J.; Methot, S.P.; Gasser, S.M. Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance. Nat. Rev. Mol. Cell Biol. 2022, 23, 623–640. [Google Scholar] [CrossRef]
- Blackburn, M.L.; Chansky, H.A.; Zielinska-Kwiatkowska, A.; Matsui, Y.; Yang, L. Genomic structure and expression of the mouse ESET gene encoding an ERG-associated histone methyltransferase with a SET domain. Biochim. Biophys. Acta 2003, 1629, 8–14. [Google Scholar] [CrossRef]
- Markouli, M.; Strepkos, D.; Piperi, C. Structure, Activity and Function of the SETDB1 Protein Methyltransferase. Life 2021, 11, 817. [Google Scholar] [CrossRef]
- Lawson, K.A.; Teteak, C.J.; Gao, J.; Li, N.; Hacquebord, J.; Ghatan, A.; Zielinska-Kwiatkowska, A.; Song, G.; Chansky, H.A.; Yang, L. ESET histone methyltransferase regulates osteoblastic differentiation of mesenchymal stem cells during postnatal bone development. FEBS Lett. 2013, 587, 3961–3967. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; An, W.; Cao, R.; Xia, L.; Erdjument-Bromage, H.; Chatton, B.; Tempst, P.; Roeder, R.G.; Zhang, Y. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 2003, 12, 475–487. [Google Scholar] [CrossRef]
- Ichimura, T.; Watanabe, S.; Sakamoto, Y.; Aoto, T.; Fujita, N.; Nakao, M. Transcriptional repression and heterochromatin formation by MBD1 and MCAF/AM family proteins. J. Biol. Chem. 2005, 280, 13928–13935. [Google Scholar] [CrossRef] [Green Version]
- Minkovsky, A.; Sahakyan, A.; Rankin-Gee, E.; Bonora, G.; Patel, S.; Plath, K. The Mbd1-Atf7ip-Setdb1 pathway contributes to the maintenance of X chromosome inactivation. Epigenetics Chromatin 2014, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Basavapathruni, A.; Gureasko, J.; Porter Scott, M.; Hermans, W.; Godbole, A.; Leland, P.A.; Boriack-Sjodin, P.A.; Wigle, T.J.; Copeland, R.A.; Riera, T.V. Characterization of the Enzymatic Activity of SETDB1 and Its 1:1 Complex with ATF7IP. Biochemistry 2016, 55, 1645–1651. [Google Scholar] [CrossRef] [PubMed]
- Timms, R.T.; Tchasovnikarova, I.A.; Antrobus, R.; Dougan, G.; Lehner, P.J. ATF7IP-Mediated Stabilization of the Histone Methyltransferase SETDB1 Is Essential for Heterochromatin Formation by the HUSH Complex. Cell Rep. 2016, 17, 653–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsusaka, T.; Shimura, C.; Shinkai, Y. ATF7IP regulates SETDB1 nuclear localization and increases its ubiquitination. EMBO Rep. 2019, 20, e48297. [Google Scholar] [CrossRef] [PubMed]
- Sin, J.H.; Zuckerman, C.; Cortez, J.T.; Eckalbar, W.L.; Erle, D.J.; Anderson, M.S.; Waterfield, M.R. The epigenetic regulator ATF7ip inhibits Il2 expression, regulating Th17 responses. J. Exp. Med. 2019, 216, 2024–2037. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xu, L.; Zhang, X.; Wang, K.; Tan, Y.; Li, G.; Wang, Y.; Xue, T.; Sun, Q.; Cao, X.; et al. Methyltransferase Setdb1 Promotes Osteoblast Proliferation by Epigenetically Silencing Macrod2 with the Assistance of Atf7ip. Cells 2022, 11, 2580. [Google Scholar] [CrossRef]
- Silva, B.C.; Bilezikian, J.P. Parathyroid hormone: Anabolic and catabolic actions on the skeleton. Curr. Opin. Pharmacol. 2015, 22, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.H.; Lu, X.; Nanes, M.S.; Mitchell, J. Regulation of osterix (Osx, Sp7) and the Osx promoter by parathyroid hormone in osteoblasts. J. Mol. Endocrinol. 2009, 43, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Oton-Gonzalez, L.; Mazziotta, C.; Iaquinta, M.R.; Mazzoni, E.; Nocini, R.; Trevisiol, L.; D’Agostino, A.; Tognon, M.; Rotondo, J.C.; Martini, F. Genetics and Epigenetics of Bone Remodeling and Metabolic Bone Diseases. Int. J. Mol. Sci. 2022, 23, 1500. [Google Scholar] [CrossRef]
- Guo, Y.; Mao, X.; Xiong, L.; Xia, A.; You, J.; Lin, G.; Wu, C.; Huang, L.; Wang, Y.; Yang, S. Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain. Angew. Chem. Int. Ed. Engl. 2021, 60, 8760–8765. [Google Scholar] [CrossRef] [PubMed]
- Grossel, M.J.; Hinds, P.W. Beyond the cell cycle: A new role for Cdk6 in differentiation. J. Cell Biochem. 2006, 97, 485–493. [Google Scholar] [CrossRef]
- Ishikawa, M.; Iwamoto, T.; Fukumoto, S.; Yamada, Y. Pannexin 3 inhibits proliferation of osteoprogenitor cells by regulating Wnt and p21 signaling. J. Biol. Chem. 2014, 289, 2839–2851. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.C.; Huang, T.W.; Chuang, P.Y.; Yang, T.Y.; Chang, S.F. Zoledronate induces cell cycle arrest and differentiation by upregulating p21 in mouse MC3T3-E1 preosteoblasts. Int. J. Med. Sci. 2019, 16, 751–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ideno, H.; Nakashima, K.; Komatsu, K.; Araki, R.; Abe, M.; Arai, Y.; Kimura, H.; Shinkai, Y.; Tachibana, M.; Nifuji, A. G9a is involved in the regulation of cranial bone formation through activation of Runx2 function during development. Bone 2020, 137, 115332. [Google Scholar] [CrossRef] [PubMed]
- Rummukainen, P.; Tarkkonen, K.; Dudakovic, A.; Al-Majidi, R.; Nieminen-Pihala, V.; Valensisi, C.; Hawkins, R.D.; van Wijnen, A.J.; Kiviranta, R. Lysine-Specific Demethylase 1 (LSD1) epigenetically controls osteoblast differentiation. PLoS ONE 2022, 17, e0265027. [Google Scholar] [CrossRef]
- Qin, G.; Li, Y.; Wang, H.; Yang, J.; Chen, Q.; Tang, H.; Wang, Y.; Zhang, M.; Jiang, T.; Lin, S.; et al. Lysine-Specific Demethylase 4A Regulates Osteogenic Differentiation via Regulating the Binding Ability of H3K9me3 with the Promoters of Runx2, Osterix and Osteocalcin. J. Biomed. Nanotechnol. 2020, 16, 899–909. [Google Scholar] [CrossRef]
- Qi, Q.; Wang, Y.; Wang, X.; Yang, J.; Xie, Y.; Zhou, J.; Li, X.; Wang, B. Histone demethylase KDM4A regulates adipogenic and osteogenic differentiation via epigenetic regulation of C/EBPalpha and canonical Wnt signaling. Cell Mol. Life Sci. 2020, 77, 2407–2421. [Google Scholar] [CrossRef]
- Ge, W.; Liu, Y.; Chen, T.; Zhang, X.; Lv, L.; Jin, C.; Jiang, Y.; Shi, L.; Zhou, Y. The epigenetic promotion of osteogenic differentiation of human adipose-derived stem cells by the genetic and chemical blockade of histone demethylase LSD1. Biomaterials 2014, 35, 6015–6025. [Google Scholar] [CrossRef]
- Lv, L.; Ge, W.; Liu, Y.; Lai, G.; Liu, H.; Li, W.; Zhou, Y. Lysine-specific demethylase 1 inhibitor rescues the osteogenic ability of mesenchymal stem cells under osteoporotic conditions by modulating H3K4 methylation. Bone Res. 2016, 4, 16037. [Google Scholar] [CrossRef] [Green Version]
- Young, N.L.; Dere, R. Mechanistic insights into KDM4A driven genomic instability. Biochem. Soc. Trans. 2021, 49, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Hillringhaus, L.; Yue, W.W.; Rose, N.R.; Ng, S.S.; Gileadi, C.; Loenarz, C.; Bello, S.H.; Bray, J.E.; Schofield, C.J.; Oppermann, U. Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J. Biol. Chem. 2011, 286, 41616–41625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudakovic, A.; Camilleri, E.T.; Paradise, C.R.; Samsonraj, R.M.; Gluscevic, M.; Paggi, C.A.; Begun, D.L.; Khani, F.; Pichurin, O.; Ahmed, F.S.; et al. Enhancer of zeste homolog 2 (Ezh2) controls bone formation and cell cycle progression during osteogenesis in mice. J. Biol. Chem. 2018, 293, 12894–12907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Link, D.C. Targeting of Mesenchymal Stromal Cells by Cre-Recombinase Transgenes Commonly Used to Target Osteoblast Lineage Cells. J. Bone Miner. Res. 2016, 31, 2001–2007. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.E.; Nakashima, K.; de Crombrugghe, B. Transgenic mice expressing a ligand-inducible cre recombinase in osteoblasts and odontoblasts: A new tool to examine physiology and disease of postnatal bone and tooth. Am. J. Pathol. 2004, 165, 1875–1882. [Google Scholar] [CrossRef]
- Choi, H.; Magyar, C.E.; Nervina, J.M.; Tetradis, S. Different duration of parathyroid hormone exposure distinctively regulates primary response genes Nurr1 and RANKL in osteoblasts. PLoS ONE 2018, 13, e0208514. [Google Scholar] [CrossRef] [Green Version]
- Maridas, D.E.; Rendina-Ruedy, E.; Le, P.T.; Rosen, C.J. Isolation, Culture, and Differentiation of Bone Marrow Stromal Cells and Osteoclast Progenitors from Mice. J. Vis. Exp. 2018, 131, e56750. [Google Scholar] [CrossRef]
- Brown, S.D.; Moore, M.W. Towards an encyclopaedia of mammalian gene function: The International Mouse Phenotyping Consortium. Dis. Model Mech. 2012, 5, 289–292. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Spandidos, A.; Wang, H.; Seed, B. PrimerBank: A PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012, 40, D1144–D1149. [Google Scholar] [CrossRef] [Green Version]
| Gene | Primer Sequences (5′→3′) |
|---|---|
| Mouse Atf7ip | FP: CGGGAGCCATGAGAATGGAG |
| RP: GCATACAAGGGGTCTCTTTCC | |
| Mouse Alp | FP: CCAACTCTTTTGTGCCAGAGA |
| RP: GGCTACATTGGTGTTGAGCTTTT | |
| Mouse Runx2 | FP: ATGCTTCATTCGCCTCACAAA |
| RP: GCACTCACTGACTCGGTTGG | |
| Mouse Sp7 | FP: CCTCTGCGGGACTCAACAAC |
| RP: AGCCCATTAGTGCTTGTAAAGG | |
| Mouse Osteocalcin | FP: CTGACCTCACAGATCCCAAGC |
| RP: TGGTCTGATAGCTCGTCACAAG | |
| Mouse Setdb1 | FP: CCTGGGTGCATGAGTTTGG |
| RP: TGTACTGACGAAGTTCCTCCATA | |
| Mouse Col1a1 | FP: GCTCCTCTTAGGGGCCACT |
| RP: CCACGTCTCACCATTGGGG | |
| Mouse β-actin | FP: GGCTGTATTCCCCTCCATCG |
| RP: CCAGTTGGTAACAATGCCATGT | |
| Mouse Pparγ | FP: TCGCTGATGCACTGCCTATG |
| RP: GAGAGGTCCACAGAGCTGATT | |
| Mouse Cebp/α | FP: CAAGAACAGCAACGAGTACCG |
| RP: GTCACTGGTCAACTCCAGCAC | |
| Mouse Twist | FP: GGACAAGCTGAGCAAGATTCA |
| RP: CGGAGAAGGCGTAGCTGAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, G.; Shi, X.; Qu, X.; Han, C.; Hu, A.; Jia, Z.; Yang, J.; Liu, H.; Wu, Y. Atf7ip Inhibits Osteoblast Differentiation via Negative Regulation of the Sp7 Transcription Factor. Int. J. Mol. Sci. 2023, 24, 4305. https://doi.org/10.3390/ijms24054305
Hu G, Shi X, Qu X, Han C, Hu A, Jia Z, Yang J, Liu H, Wu Y. Atf7ip Inhibits Osteoblast Differentiation via Negative Regulation of the Sp7 Transcription Factor. International Journal of Molecular Sciences. 2023; 24(5):4305. https://doi.org/10.3390/ijms24054305
Chicago/Turabian StyleHu, Guoqin, Xian Shi, Xiuxia Qu, Chunqing Han, Anran Hu, Zhongtang Jia, Jiatao Yang, Huanliang Liu, and Yu Wu. 2023. "Atf7ip Inhibits Osteoblast Differentiation via Negative Regulation of the Sp7 Transcription Factor" International Journal of Molecular Sciences 24, no. 5: 4305. https://doi.org/10.3390/ijms24054305
APA StyleHu, G., Shi, X., Qu, X., Han, C., Hu, A., Jia, Z., Yang, J., Liu, H., & Wu, Y. (2023). Atf7ip Inhibits Osteoblast Differentiation via Negative Regulation of the Sp7 Transcription Factor. International Journal of Molecular Sciences, 24(5), 4305. https://doi.org/10.3390/ijms24054305






