Proteomic Fingerprint of Lung Fibrosis Progression and Response to Therapy in Bleomycin-Induced Mouse Model
Abstract
1. Introduction
2. Results and Discussion
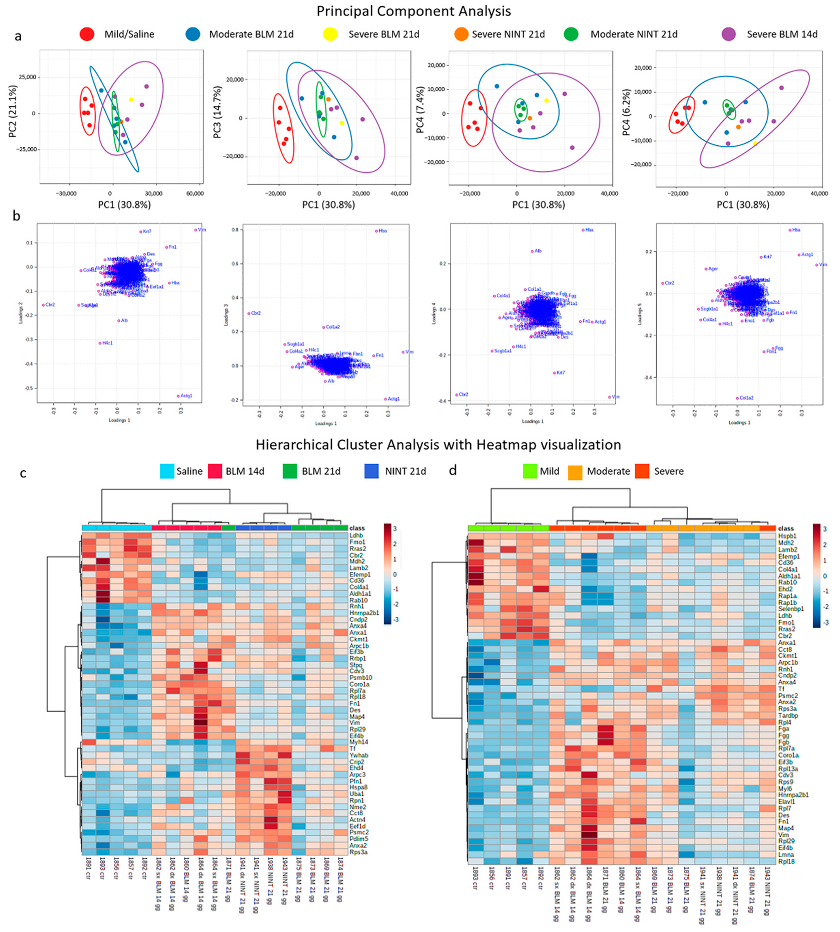
2.1. Multivariate Data Analysis: Principal Component Analysis and Hierarchical Cluster Analysis
2.2. Pulmonary Fibrosis Progression in Bleomycin-Treated Mice: Mild, Moderate, and Severe
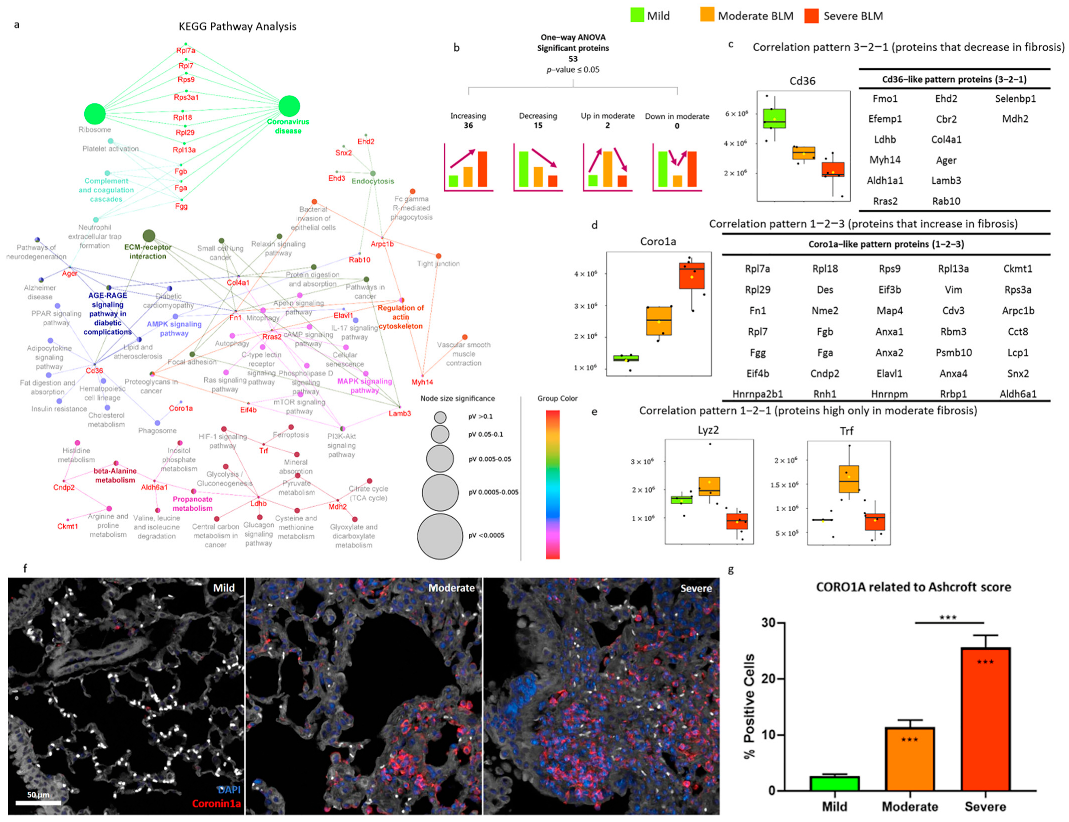
2.3. Proteomic Characterization of the Bleomycin-Induced Mouse Model of Pulmonary Fibrosis
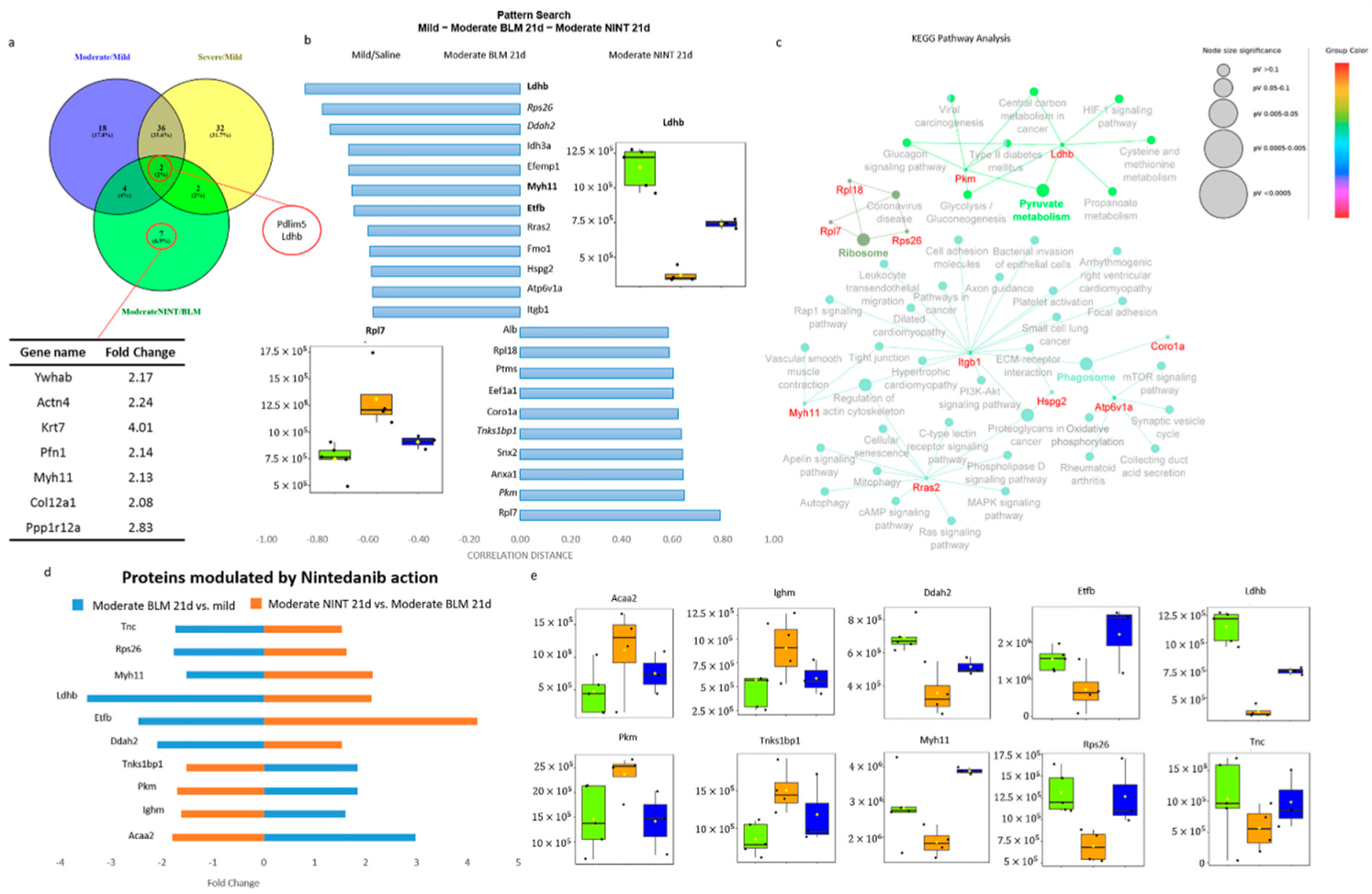
2.4. Proteomic Characterization of Nintedanib-Mediated Fibrosis Slow-Down
3. Materials and Methods
3.1. Reagents and Solutions
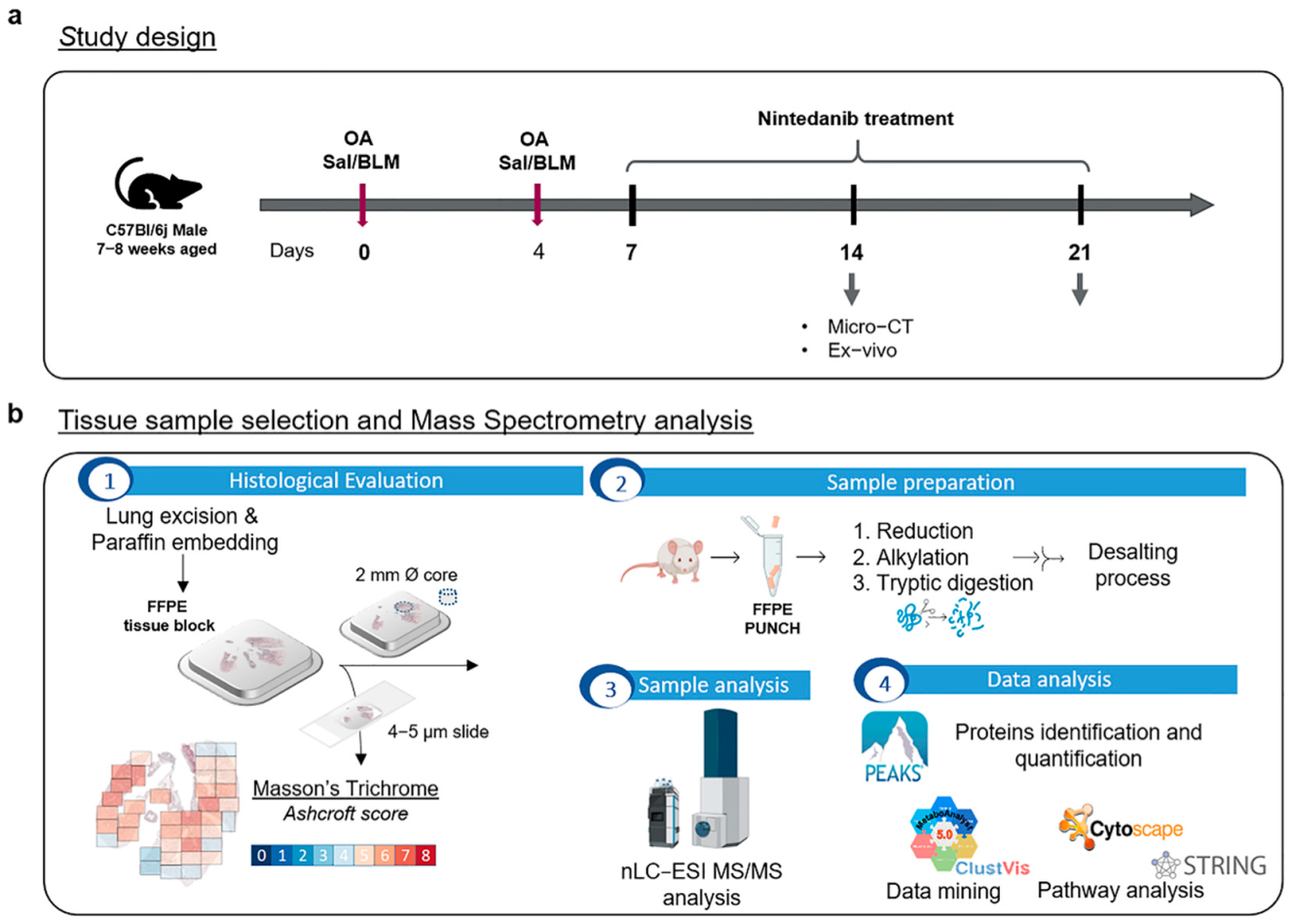
3.2. Experimental Animals
3.3. Bleomycin and Nintedanib Administration
3.4. Micro-CT Lung Imaging
3.5. Histological Assessment of Lung Fibrosis
3.6. Samples
3.7. nLC-ESI-MS/MS Sample Preparation and Analysis
3.8. nLC-ESI-MS/MS Data Elaboration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raghu, G.; Remy-Jardin, M.; Richeldi, L.; Thomson, C.C.; Inoue, Y.; Johkoh, T.; Kreuter, M.; Lynch, D.A.; Maher, T.M.; Martinez, F.J.; et al. Idiopathic Pulmonary Fibrosis (an Update) and Progressive Pulmonary Fibrosis in Adults: An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2022, 205, e18–e47. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Perlman, D.; Tomic, R. Natural history of idiopathic pulmonary fibrosis. Respir. Med. 2015, 109, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Raghu, G.; Remy-Jardin, M.; Myers, J.L.; Richeldi, L.; Ryerson, C.J.; Lederer, D.J.; Behr, J.; Cottin, V.; Danoff, S.K.; Morell, F.; et al. Diagnosis of Idiopathic Pulmonary Fibrosis. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2018, 198, e44–e68. [Google Scholar] [CrossRef] [PubMed]
- Spagnolo, P.; Kropski, J.A.; Jones, M.G.; Lee, J.S.; Rossi, G.; Karampitsakos, T.; Maher, T.M.; Tzouvelekis, A.; Ryerson, C.J. Idiopathic pulmonary fibrosis: Disease mechanisms and drug development. Pharmacol. Ther. 2021, 222, 107798. [Google Scholar] [CrossRef]
- Wollin, L.; Wex, E.; Pautsch, A.; Schnapp, G.; Hostettler, K.E.; Stowasser, S.; Kolb, M. Mode of action of nintedanib in the treatment of idiopathic pulmonary fibrosis. Eur. Respir. J. 2015, 45, 1434–1445. [Google Scholar] [CrossRef]
- Tian, Y.; Li, H.; Gao, Y.; Liu, C.; Qiu, T.; Wu, H.; Cao, M.; Zhang, Y.; Ding, H.; Chen, J.; et al. Quantitative Proteomic Characterization of Lung Tissue in Idiopathic Pulmonary Fibrosis. Clin. Proteom. 2019, 16, 6. [Google Scholar] [CrossRef]
- Korfei, M.; Schmitt, S.; Ruppert, C.; Henneke, I.; Markart, P.; Loeh, B.; Mahavadi, P.; Wygrecka, M.; Klepetko, W.; Fink, L.; et al. Comparative Proteomic Analysis of Lung Tissue from Patients with Idiopathic Pulmonary Fibrosis (IPF) and Lung Transplant Donor Lungs. J. Proteome Res. 2011, 10, 2185–2205. [Google Scholar] [CrossRef]
- Korfei, M.; von der Beck, D.; Henneke, I.; Markart, P.; Ruppert, C.; Mahavadi, P.; Ghanim, B.; Klepetko, W.; Fink, L.; Meiners, S.; et al. Comparative proteome analysis of lung tissue from patients with idiopathic pulmonary fibrosis (IPF), non-specific interstitial pneumonia (NSIP) and organ donors. J. Proteom. 2013, 85, 109–128. [Google Scholar] [CrossRef]
- Zheng, P.; Sun, S.; Wang, J.; Cheng, Z.J.; Lei, K.C.; Xue, M.; Zhang, T.; Huang, H.; Zhang, X.D.; Sun, B. Integrative omics analysis identifies biomarkers of idiopathic pulmonary fibrosis. Cell. Mol. Life Sci. 2022, 79, 66. [Google Scholar] [CrossRef]
- Konigsberg, I.R.; Borie, R.; Walts, A.D.; Cardwell, J.; Rojas, M.; Metzger, F.; Hauck, S.M.; Fingerlin, T.E.; Yang, I.V.; Schwartz, D.A. Molecular Signatures of Idiopathic Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2021, 65, 430–441. [Google Scholar] [CrossRef]
- Toren, D.; Yanai, H.; Abu Taha, R.; Bunu, G.; Ursu, E.; Ziesche, R.; Tacutu, R.; Fraifeld, V.E. Systems biology analysis of lung fibrosis-related genes in the bleomycin mouse model. Sci. Rep. 2021, 11, 19269. [Google Scholar] [CrossRef]
- Jenkins, R.G.; Moore, B.B.; Chambers, R.C.; Eickelberg, O.; Königshoff, M.; Kolb, M.; Laurent, G.J.; Nanthakumar, C.B.; Olman, M.A.; Pardo, A.; et al. An Official American Thoracic Society Workshop Report: Use of Animal Models for the Preclinical Assessment of Potential Therapies for Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2017, 56, 667–679. [Google Scholar] [CrossRef]
- Ruscitti, F.; Ravanetti, F.; Bertani, V.; Ragionieri, L.; Mecozzi, L.; Sverzellati, N.; Silva, M.; Ruffini, L.; Menozzi, V.; Civelli, M.; et al. Quantification of Lung Fibrosis in IPF-Like Mouse Model and Pharmacological Response to Treatment by Micro-Computed Tomography. Front. Pharmacol. 2020, 11, 1117. [Google Scholar] [CrossRef]
- Stellari, F.F.; Ruscitti, F.; Pompilio, D.; Ravanetti, F.; Tebaldi, G.; Macchi, F.; Verna, A.E.; Villetti, G.; Donofrio, G. Heterologous Matrix Metalloproteinase Gene Promoter Activity Allows In Vivo Real-time Imaging of Bleomycin-Induced Lung Fibrosis in Transiently Transgenized Mice. Front. Immunol. 2017, 8, 199. [Google Scholar] [CrossRef]
- Ravanetti, F.; Ragionieri, L.; Ciccimarra, R.; Ruscitti, F.; Pompilio, D.; Gazza, F.; Villetti, G.; Cacchioli, A.; Stellari, F.F. Modeling pulmonary fibrosis through bleomycin delivered by osmotic minipump: A new histomorphometric method of evaluation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 318, L376–L385. [Google Scholar] [CrossRef]
- Jonsdottir, H.R.; Arason, A.J.; Palsson, R.; Franzdottir, S.R.; Gudbjartsson, T.; Isaksson, H.J.; Gudmundsson, G.; Gudjonsson, T.; Magnusson, M.K. Basal cells of the human airways acquire mesenchymal traits in idiopathic pulmonary fibrosis and in culture. Lab. Investig. 2015, 95, 1418–1428. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Zhang, Z.; Mukherjee, A.B. Mice lacking uteroglobin are highly susceptible to developing pulmonary fibrosis. FEBS Lett. 2006, 580, 4515–4520. [Google Scholar] [CrossRef]
- Konishi, K.; Gibson, K.F.; Lindell, K.O.; Richards, T.J.; Zhang, Y.; Dhir, R.; Bisceglia, M.; Gilbert, S.; Yousem, S.A.; Song, J.W.; et al. Gene Expression Profiles of Acute Exacerbations of Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2009, 180, 167–175. [Google Scholar] [CrossRef]
- Queisser, M.A.; Kouri, F.M.; Königshoff, M.; Wygrecka, M.; Schubert, U.; Eickelberg, O.; Preissner, K.T. Loss of RAGE in Pulmonary Fibrosis: Molecular Relations to Functional Changes in Pulmonary Cell Types. Am. J. Respir. Cell Mol. Biol. 2008, 39, 337–345. [Google Scholar] [CrossRef]
- Kreus, M.; Lehtonen, S.; Skarp, S.; Kaarteenaho, R. Extracellular matrix proteins produced by stromal cells in idiopathic pulmonary fibrosis and lung adenocarcinoma. PLoS ONE 2021, 16, e0250109. [Google Scholar] [CrossRef]
- Phillips, I.R.; Shephard, E.A. Drug metabolism by flavin-containing monooxygenases of human and mouse. Expert Opin. Drug Metab. Toxicol. 2017, 13, 167–181. [Google Scholar] [CrossRef] [PubMed]
- UniProt. Q08857·CD36_MOUSE. Available online: https://www.uniprot.org/uniprotkb/Q08857/entry (accessed on 12 January 2023).
- Feitsma, L.J.; Brondijk, H.C.; Jarvis, G.E.; Hagemans, D.; Bihan, D.G.; Jerah, N.E.; Versteeg, M.; Farndale, R.W.; Huizinga, E.G. Structural insights into collagen binding by platelet receptor glycoprotein VI. Blood 2022, 139, 3087–3098. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, R.L.; Febbraio, M. CD36, a Scavenger Receptor Involved in Immunity, Metabolism, Angiogenesis, and Behavior. Sci. Signal. 2009, 2, re3. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, J.; Cui, W.; Silverstein, R.L. CD36, a signaling receptor and fatty acid transporter that regulates immune cell metabolism and fate. J. Exp. Med. 2022, 219, e20211314. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Y.; Lv, L.; Chen, J. Silencing CD36 gene expression results in the inhibition of latent-TGF-β1 activation and suppression of silica-induced lung fibrosis in the rat. Respir. Res. 2009, 10, 36. [Google Scholar] [CrossRef]
- Parks, B.W.; Black, L.L.; Zimmerman, K.A.; Metz, A.E.; Steele, C.; Murphy-Ullrich, J.E.; Kabarowski, J.H. CD36, but not G2A, modulates efferocytosis, inflammation, and fibrosis following bleomycin-induced lung injury. J. Lipid Res. 2013, 54, 1114–1123. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, L.; Huang, Y.; Luo, M.; Wang, H.; Jiang, Z.; Zheng, J.; Yang, Z.; Chen, Z.; Zhang, C.; et al. Pharmaceutical targeting of succinate dehydrogenase in fibroblasts controls bleomycin-induced lung fibrosis. Redox Biol. 2021, 46, 102082. [Google Scholar] [CrossRef]
- Pieters, J.; Müller, P.; Jayachandran, R. On guard: Coronin proteins in innate and adaptive immunity. Nat. Rev. Immunol. 2013, 13, 510–518. [Google Scholar] [CrossRef]
- Punwani, D.; Pelz, B.; Yu, J.; Arva, N.C.; Schafernak, K.; Kondratowicz, K.; Makhija, M.; Puck, J.M. Coronin-1A: Immune Deficiency in Humans and Mice. J. Clin. Immunol. 2015, 35, 100–107. [Google Scholar] [CrossRef]
- Cohen, J.I. Primary Immunodeficiencies Associated with EBV Disease. Curr. Top. Microbiol. Immunol. 2015, 390, 241–265. [Google Scholar] [CrossRef]
- Vignesh, P.; Rawat, A.; Kumrah, R.; Singh, A.; Gummadi, A.; Sharma, M.; Kaur, A.; Nameirakpam, J.; Jindal, A.; Suri, D.; et al. Clinical, Immunological, and Molecular Features of Severe Combined Immune Deficiency: A Multi-Institutional Experience from India. Front. Immunol. 2020, 11, 619146. [Google Scholar] [CrossRef]
- Bao, M.; Huang, Y.; Lang, Z.; Zhao, H.; Saito, Y.; Nagano, T.; Kawagoe, I.; Divisi, D.; Hu, X.; Jiang, G. Proteomic analysis of plasma exosomes in patients with non-small cell lung cancer. Transl. Lung Cancer Res. 2022, 11, 1434–1452. [Google Scholar] [CrossRef]
- He, Y.; Jiang, Z.; Chen, C.; Wang, X. Classification of triple-negative breast cancers based on Immunogenomic profiling. J. Exp. Clin. Cancer Res. 2018, 37, 327. [Google Scholar] [CrossRef]
- Hu, Z.; Liu, Y.; Zhu, Y.; Cui, H.; Pan, J. Identification of key biomarkers and immune infiltration in renal interstitial fibrosis. Ann. Transl. Med. 2022, 10, 190. [Google Scholar] [CrossRef]
- Wu, J.; Cao, J.; Fan, Y.; Li, C.; Hu, X. Comprehensive analysis of miRNA–mRNA regulatory network and potential drugs in chronic chagasic cardiomyopathy across human and mouse. BMC Med. Genom. 2021, 14, 283. [Google Scholar] [CrossRef]
- Moriceau, S.; Kantari, C.; Mocek, J.; Davezac, N.; Gabillet, J.; Guerrera, I.C.; Brouillard, F.; Tondelier, D.; Sermet-Gaudelus, I.; Danel, C.; et al. Coronin-1 Is Associated with Neutrophil Survival and Is Cleaved during Apoptosis: Potential Implication in Neutrophils from Cystic Fibrosis Patients. J. Immunol. 2009, 182, 7254–7263. [Google Scholar] [CrossRef]
- Ganz, T. Antimicrobial Polypeptides in Host Defense of the Respiratory Tract. J. Clin. Investig. 2002, 109, 693–697. [Google Scholar] [CrossRef]
- Joshi, N.; Watanabe, S.; Verma, R.; Jablonski, R.P.; Chen, C.-I.; Cheresh, P.; Markov, N.S.; Reyfman, P.A.; McQuattie-Pimentel, A.C.; Sichizya, L.; et al. A spatially restricted fibrotic niche in pulmonary fibrosis is sustained by M-CSF/M-CSFR signalling in monocyte-derived alveolar macrophages. Eur. Respir. J. 2020, 55, 1900646. [Google Scholar] [CrossRef]
- Yang, T.; Jia, Y.; Ma, Y.; Cao, L.; Chen, X.; Qiao, B. Comparative Proteomic Analysis of Bleomycin-induced Pulmonary Fibrosis Based on Isobaric Tag for Quantitation. Am. J. Med. Sci. 2017, 353, 49–58. [Google Scholar] [CrossRef]
- Ogger, P.P.; Byrne, A.J. Lung fibrosis enters the iron age†. J. Pathol. 2020, 252, 1–3. [Google Scholar] [CrossRef]
- Xu, X.; Xiong, X.; Sun, Y. The role of ribosomal proteins in the regulation of cell proliferation, tumorigenesis, and genomic integrity. Sci. China Life Sci. 2016, 59, 656–672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xin, Q.; Wu, Z.; Wang, C.; Wang, Y.; Wu, Q.; Niu, R. Application of Isobaric Tags for Relative and Absolute Quantification (iTRAQ) Coupled with Two-Dimensional Liquid Chromatography/Tandem Mass Spectrometry in Quantitative Proteomic Analysis for Discovery of Serum Biomarkers for Idiopathic Pulmonary Fibrosis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 4146–4153. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chen, J.; Wang, S.; Tian, Z.; Fan, Y.; Wang, M.; Zhao, J.; Tang, K.; Xie, J. Identification of Genetic Signature Associated with Aging in Pulmonary Fibrosis. Front. Med. 2021, 8, 744239. [Google Scholar] [CrossRef] [PubMed]
- Colunga Biancatelli, R.M.L.; Solopov, P.; Catravas, J. HSP90 Inhibitors for IPF/COVID-19. Sch. Community Encycl. 2020, 1–7. [Google Scholar] [CrossRef]
- Sibinska, Z.; Tian, X.; Korfei, M.; Kojonazarov, B.; Kolb, J.S.; Klepetko, W.; Kosanovic, D.; Wygrecka, M.; Ghofrani, H.A.; Weissmann, N.; et al. Amplified canonical transforming growth factor-β signalling via heat shock protein 90 in pulmonary fibrosis. Eur. Respir. J. 2017, 49, 1501941. [Google Scholar] [CrossRef]
- Chakraborty, A.; Mastalerz, M.; Ansari, M.; Schiller, H.B.; Staab-Weijnitz, C.A. Emerging Roles of Airway Epithelial Cells in Idiopathic Pulmonary Fibrosis. Cells 2022, 11, 1050. [Google Scholar] [CrossRef]
- MYH11 Myosin Heavy Chain 11 [Homo Sapiens (Human)]-Gene-NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/4629#summary (accessed on 12 January 2023).
- Wang, R.-J.; Wu, P.; Cai, G.-X.; Wang, Z.-M.; Xu, Y.; Peng, J.-J.; Sheng, W.-Q.; Lu, H.-F.; Cai, S.-J. Down-regulated MYH11 Expression Correlates with Poor Prognosis in Stage II and III Colorectal Cancer. Asian Pac. J. Cancer Prev. 2014, 15, 7223–7228. [Google Scholar] [CrossRef]
- Nie, M.; Pan, X.; Tao, H.; Xu, M.; Liu, S.; Sun, W.; Wu, J.; Zou, X. Clinical and prognostic significance of MYH11 in lung cancer. Oncol. Lett. 2020, 19, 3899–3906. [Google Scholar] [CrossRef]
- Seitz, S.; Korsching, E.; Weimer, J.; Jacobsen, A.; Arnold, N.; Meindl, A.; Arnold, W.; Gustavus, D.; Klebig, C.; Petersen, I.; et al. Genetic background of different cancer cell lines influences the gene set involved in chromosome 8 mediated breast tumor suppression. Genes Chromosom. Cancer 2006, 45, 612–627. [Google Scholar] [CrossRef]
- Le, T.-T.T.; Berg, N.K.; Harting, M.T.; Li, X.; Eltzschig, H.K.; Yuan, X. Purinergic Signaling in Pulmonary Inflammation. Front. Immunol. 2019, 10, 1633. [Google Scholar] [CrossRef]
- Kottmann, R.M.; Kulkarni, A.A.; Smolnycki, K.A.; Lyda, E.; Dahanayake, T.; Salibi, R.; Honnons, S.; Jones, C.; Isern, N.G.; Hu, J.Z.; et al. Lactic Acid Is Elevated in Idiopathic Pulmonary Fibrosis and Induces Myofibroblast Differentiation via pH-Dependent Activation of Transforming Growth Factor-β. Am. J. Respir. Crit. Care Med. 2012, 186, 740–751. [Google Scholar] [CrossRef]
- Newton, D.A.; Lottes, R.G.; Ryan, R.M.; Spyropoulos, D.D.; Baatz, J.E. Dysfunctional lactate metabolism in human alveolar type II cells from idiopathic pulmonary fibrosis lung explant tissue. Respir. Res. 2021, 22, 278. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. We need to talk about the Warburg effect. Nat. Metab. 2020, 2, 127–129. [Google Scholar] [CrossRef]
- Maher, T.M. Aerobic Glycolysis and the Warburg Effect. An Unexplored Realm in the Search for Fibrosis Therapies? Am. J. Respir. Crit. Care Med. 2015, 192, 1407–1409. [Google Scholar] [CrossRef]
- Institute for Laboratory Animal Research; Commission on Life Sciences; Division on Earth and Life Studies; National Research Council. Guide for the Care and Use of Laboratory Animals; National Academies Press: Washington, DC, USA, 1996. [Google Scholar]
- Ferrini, E.; Mecozzi, L.; Corsi, L.; Ragionieri, L.; Donofrio, G.; Stellari, F.F. Alfaxalone and Dexmedetomidine as an Alternative to Gas Anesthesia for Micro-CT Lung Imaging in a Bleomycin-Induced Pulmonary Fibrosis Murine Model. Front. Vet. Sci. 2020, 7, 588592. [Google Scholar] [CrossRef]
- Mambrini, M.; Mecozzi, L.; Ferrini, E.; Leo, L.; Bernardi, D.; Grandi, A.; Sverzellati, N.; Ruffini, L.; Silva, M.; Stellari, F.F. The importance of routine quality control for reproducible pulmonary measurements by in vivo micro-CT. Sci. Rep. 2022, 12, 9695. [Google Scholar] [CrossRef]
- Vincenzi, E.; Fantazzini, A.; Basso, C.; Barla, A.; Odone, F.; Leo, L.; Mecozzi, L.; Mambrini, M.; Ferrini, E.; Sverzellati, N.; et al. A fully automated deep learning pipeline for micro-CT-imaging-based densitometry of lung fibrosis murine models. Respir. Res. 2022, 23, 308. [Google Scholar] [CrossRef]
- Mecozzi, L.; Mambrini, M.; Ruscitti, F.; Ferrini, E.; Ciccimarra, R.; Ravanetti, F.; Sverzellati, N.; Silva, M.; Ruffini, L.; Belenkov, S.; et al. In-Vivo lung fibrosis staging in a bleomycin-mouse model: A new micro-CT guided densitometric approach. Sci. Rep. 2020, 10, 18735. [Google Scholar] [CrossRef]
- Ciccimarra, R.; Bolognesi, M.M.; Zoboli, M.; Cattoretti, G.; Stellari, F.F.; Ravanetti, F. The normal and fibrotic mouse lung classified by spatial proteomic analysis. Sci. Rep. 2022, 12, 8742. [Google Scholar] [CrossRef]
- Ashcroft, T.; Simpson, J.M.; Timbrell, V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J. Clin. Pathol. 1988, 41, 467–470. [Google Scholar] [CrossRef]
- Hübner, R.-H.; Gitter, W.; El Mokhtari, N.E.; Mathiak, M.; Both, M.; Bolte, H.; Freitag-Wolf, S.; Bewig, B. Standardized quantification of pulmonary fibrosis in histological samples. BioTechniques 2008, 44, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Stella, M.; Chinello, C.; Cazzaniga, A.; Smith, A.; Galli, M.; Piga, I.; Grasso, A.; Grasso, M.; Del Puppo, M.; Varallo, M.; et al. Histology-guided proteomic analysis to investigate the molecular profiles of clear cell Renal Cell Carcinoma grades. J. Proteom. 2019, 191, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Chinello, C.; Stella, M.; Piga, I.; Smith, A.J.; Bovo, G.; Varallo, M.; Ivanova, M.; Denti, V.; Grasso, M.; Grasso, A.; et al. Proteomics of liquid biopsies: Depicting RCC infiltration into the renal vein by MS analysis of urine and plasma. J. Proteom. 2019, 191, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]



| Animal ID and Treatment | Average Ashcroft | Ashcroft Frequencies | Fibrosis Severity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||
| 1856 (SAL) | 1.19 | 0 | 80.6 | 19.4 | 0 | 0 | 0 | 0 | 0 | 0 | Mild |
| 1857 (SAL) | 1.24 | 0 | 78 | 20 | 2 | 0 | 0 | 0 | 0 | 0 | Mild |
| 1860 (BLM 14d) | 5.03 | 0 | 0 | 2.8 | 8.6 | 20 | 34.3 | 20 | 14.3 | 0 | Severe |
| * 1862 (BLM 14d) | 4.95 | 0 | 0 | 0 | 4.9 | 31.7 | 31.7 | 26.8 | 4.9 | 0 | Severe |
| * 1864 (BLM 14d) | 5.94 | 0 | 0 | 0 | 0 | 12.1 | 18.2 | 33.3 | 36.4 | 0 | Severe |
| 1869 (BLM 21d) | 4.33 | 0 | 0 | 0 | 16.3 | 44.9 | 28.6 | 10.2 | 0 | 0 | Moderate |
| 1871 (BLM 21d) | 5.4 | 0 | 0 | 0 | 2.1 | 27.7 | 19.1 | 29.8 | 21.3 | 0 | Severe |
| 1873 (BLM 21d) | 3.94 | 0 | 0 | 0 | 34.3 | 48.6 | 5.7 | 11.4 | 0 | 0 | Moderate |
| 1874 (BLM 21d) | 3.98 | 0 | 0 | 0 | 34.6 | 42.3 | 15.4 | 5.8 | 1.9 | 0 | Moderate |
| 1875 (BLM 21d) | 3.92 | 0 | 0 | 0 | 41.7 | 36.1 | 11.1 | 11.1 | 0 | 0 | Moderate |
| 1891 (SAL) | 1.68 | 0 | 41.2 | 50 | 8.8 | 0 | 0 | 0 | 0 | 0 | Mild |
| 1892 (SAL) | 1.25 | 6.2 | 62.5 | 31.3 | 0 | 0 | 0 | 0 | 0 | 0 | Mild |
| 1893 (SAL) | 0.85 | 21.7 | 71.7 | 6.6 | 0 | 0 | 0 | 0 | 0 | 0 | Mild |
| 1938 (NINT 21d) | 3.77 | 0 | 0 | 0 | 40.9 | 43.2 | 13.6 | 2.3 | 0 | 0 | Moderate |
| * 1941 (NINT 21d) | 3.61 | 0 | 0 | 0 | 44.7 | 50 | 5.3 | 0 | 0 | 0 | Moderate |
| * 1943 (NINT 21d) | 4.66 | 0 | 0 | 3.1 | 12.5 | 21.9 | 46.9 | 9.4 | 6.2 | 0 | Severe |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Principi, L.; Ferrini, E.; Ciccimarra, R.; Pagani, L.; Chinello, C.; Previtali, P.; Smith, A.; Villetti, G.; Zoboli, M.; Ravanetti, F.; et al. Proteomic Fingerprint of Lung Fibrosis Progression and Response to Therapy in Bleomycin-Induced Mouse Model. Int. J. Mol. Sci. 2023, 24, 4410. https://doi.org/10.3390/ijms24054410
Principi L, Ferrini E, Ciccimarra R, Pagani L, Chinello C, Previtali P, Smith A, Villetti G, Zoboli M, Ravanetti F, et al. Proteomic Fingerprint of Lung Fibrosis Progression and Response to Therapy in Bleomycin-Induced Mouse Model. International Journal of Molecular Sciences. 2023; 24(5):4410. https://doi.org/10.3390/ijms24054410
Chicago/Turabian StylePrincipi, Lucrezia, Erica Ferrini, Roberta Ciccimarra, Lisa Pagani, Clizia Chinello, Paolo Previtali, Andrew Smith, Gino Villetti, Matteo Zoboli, Francesca Ravanetti, and et al. 2023. "Proteomic Fingerprint of Lung Fibrosis Progression and Response to Therapy in Bleomycin-Induced Mouse Model" International Journal of Molecular Sciences 24, no. 5: 4410. https://doi.org/10.3390/ijms24054410
APA StylePrincipi, L., Ferrini, E., Ciccimarra, R., Pagani, L., Chinello, C., Previtali, P., Smith, A., Villetti, G., Zoboli, M., Ravanetti, F., Stellari, F. F., Magni, F., & Piga, I. (2023). Proteomic Fingerprint of Lung Fibrosis Progression and Response to Therapy in Bleomycin-Induced Mouse Model. International Journal of Molecular Sciences, 24(5), 4410. https://doi.org/10.3390/ijms24054410







