Alternative Polyadenylation Is a Novel Strategy for the Regulation of Gene Expression in Response to Stresses in Plants
Abstract
1. Introduction
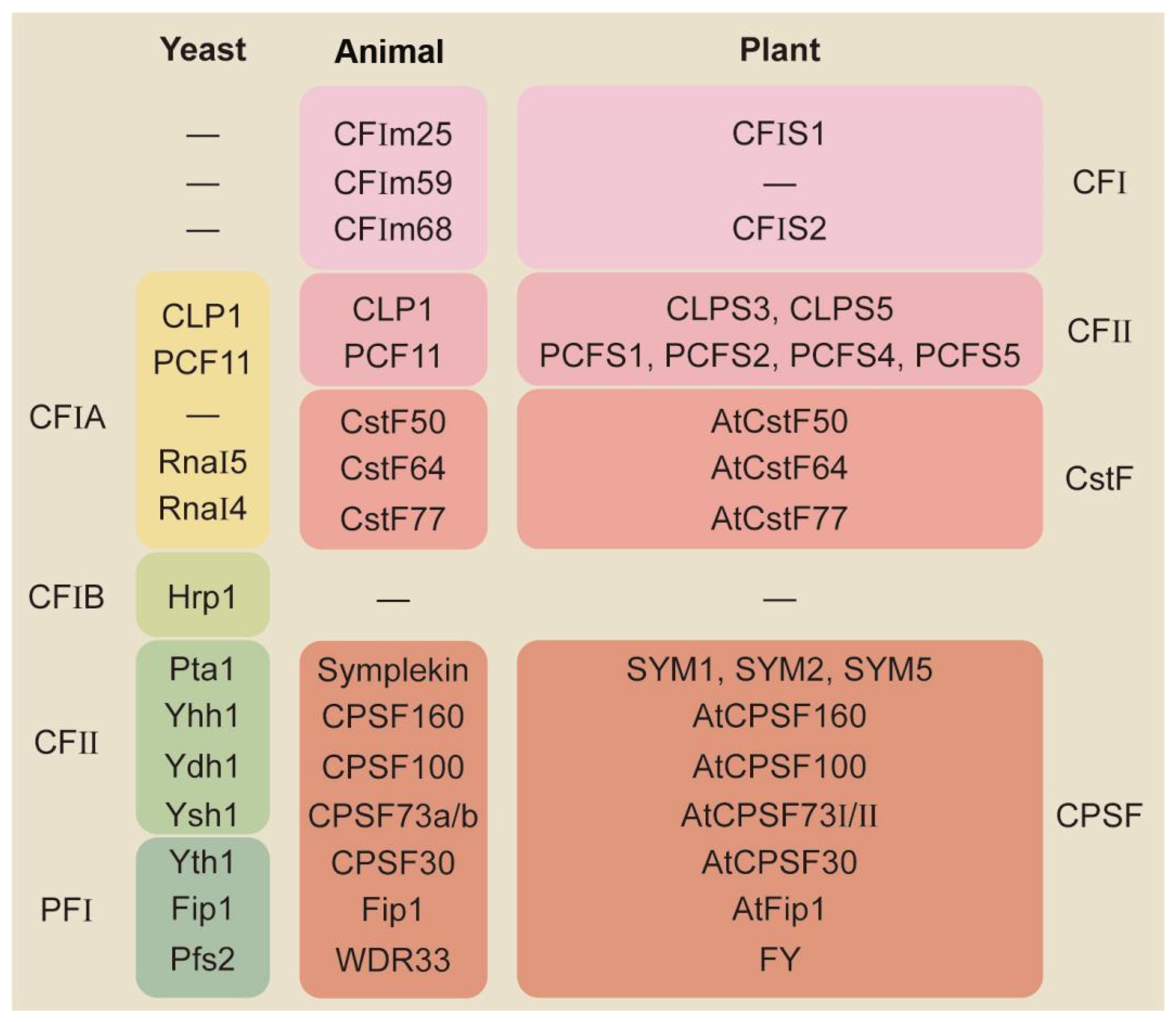
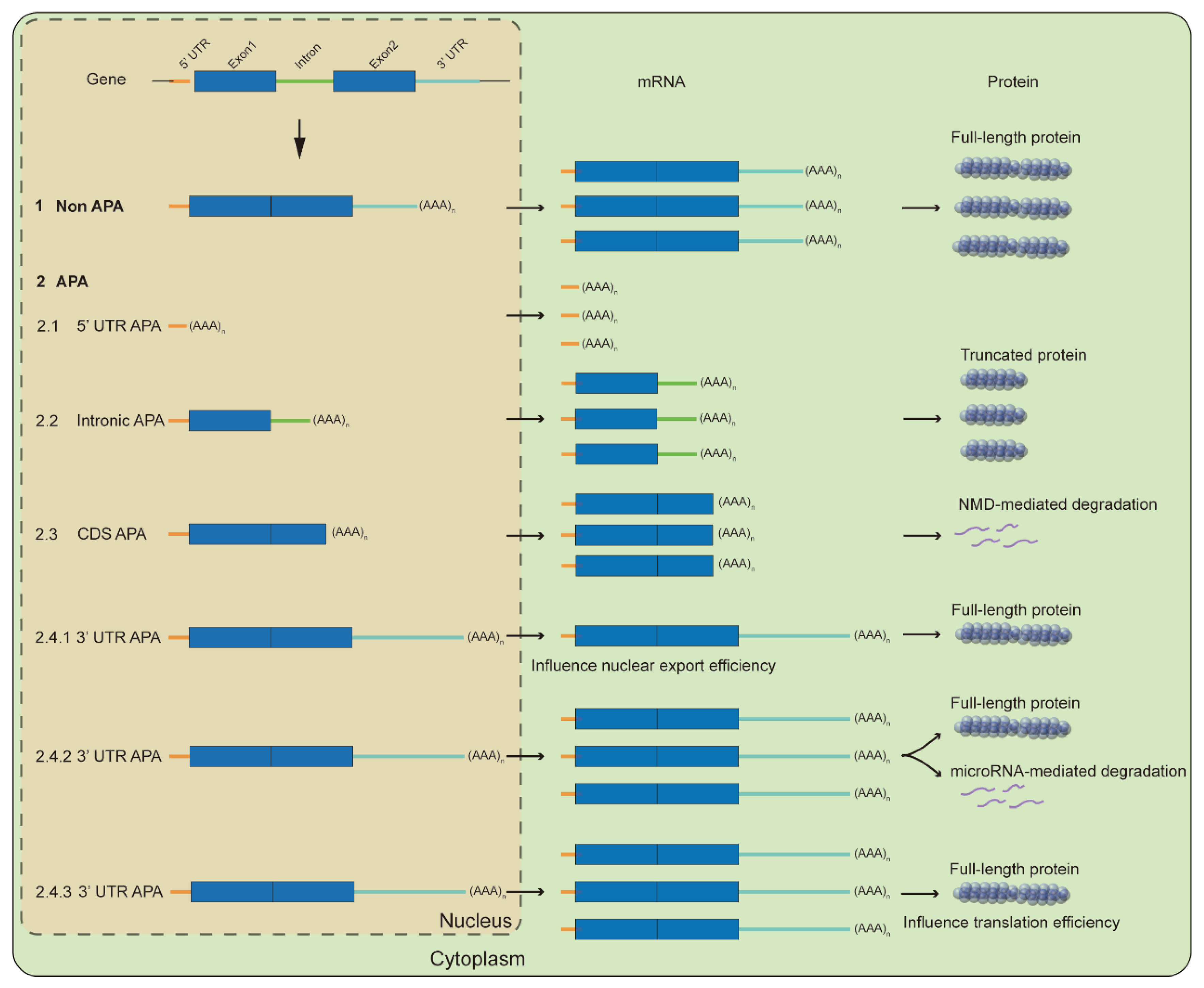
2. Molecular Mechanisms for APA-Mediated Responses
2.1. Influence Full-Length Transcripts
2.2. Influence RNA Fate and Translation Efficiency
2.2.1. RNA Stability
2.2.2. RNA Export
2.2.3. Translation Efficiency
3. An Overview of the Role of APA in Response to Biotic and Abiotic Stresses
3.1. Hypoxic Stress
3.2. Drought Stress
3.3. Salt Stress
3.4. Nitrogen Starvation
3.5. Temperature
3.6. Pathogens
3.7. ROS and ABA
4. Discussion and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, R.; Zheng, D.; Yehia, G.; Tian, B. A compendium of conserved cleavage and polyadenylation events in mammalian genes. Genome Res. 2018, 28, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Passmore, L.A.; Coller, J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell Biol. 2022, 23, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Neve, J.; Burger, K.; Li, W.; Hoque, M.; Patel, R.; Tian, B.; Gullerova, M.; Furger, A. Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation. Genome Res. 2016, 26, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.R.; Kyogoku, C.; Sigurdsson, S.; Vlasova, I.A.; Davies, L.R.L.; Baechler, E.C.; Plenge, R.M.; Koeuth, T.; Ortmann, W.A.; Hom, G.; et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl. Acad. Sci. USA 2007, 104, 6758–6763. [Google Scholar] [CrossRef] [PubMed]
- Hartwick, E.W.; Costantino, D.A.; MacFadden, A.; Nix, J.C.; Tian, S.; Das, R.; Kieft, J.S. Ribosome-induced RNA conformational changes in a viral 3′-UTR sense and regulate translation levels. Nat. Commun. 2018, 9, 5074. [Google Scholar] [CrossRef]
- Berkovits, B.D.; Mayr, C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 2015, 522, 363–367. [Google Scholar] [CrossRef]
- Kumar, A.; Clerici, M.; Muckenfuss, L.M.; Passmore, L.A.; Jinek, M. Mechanistic insights into mRNA 3′-end processing. Curr. Opin. Struct. Biol. 2019, 59, 143–150. [Google Scholar] [CrossRef]
- Hunt, A.G. mRNA 3′ end formation in plants: Novel connections to growth, development and environmental responses. Wiley Interdiscip. Rev. RNA 2020, 11, e1575. [Google Scholar] [CrossRef]
- Clerici, M.; Faini, M.; Muckenfuss, L.M.; Aebersold, R.; Jinek, M. Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex. Nat. Struct. Mol. Biol. 2018, 25, 135–138. [Google Scholar] [CrossRef]
- Yang, W.; Hsu, P.L.; Yang, F.; Song, J.-E.; Varani, G. Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3′-end processing signals. Nucleic Acids Res. 2018, 46, 493–503. [Google Scholar] [CrossRef]
- Yang, Q.; Gilmartin, G.M.; Doublie, S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc. Natl. Acad. Sci. USA 2010, 107, 10062–10067. [Google Scholar] [CrossRef]
- Gross, S.; Moore, C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. USA 2001, 98, 6080–6085. [Google Scholar] [CrossRef]
- Zhao, J.; Kessler, M.M.; Moore, C.L. Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian Cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J. Biol. Chem. 1997, 272, 10831–10838. [Google Scholar] [CrossRef]
- Zhao, J.; Kessler, M.; Helmling, S.; O’Connor, J.P.; Moore, C. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol. 1999, 19, 7733–7740. [Google Scholar] [CrossRef][Green Version]
- Preker, P.J.; Ohnacker, M.; Minvielle-Sebastia, L.; Keller, W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997, 16, 4727–4737. [Google Scholar] [CrossRef]
- Ruegsegger, U.; Blank, D.; Keller, W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1998, 1, 243–253. [Google Scholar] [CrossRef]
- de Vries, H.; Rüegsegger, U.; Hübner, W.; Friedlein, A.; Langen, H.; Keller, W. Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000, 19, 5895–5904. [Google Scholar] [CrossRef]
- Takagaki, Y.; Manley, J.L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 2000, 20, 1515–1525. [Google Scholar] [CrossRef]
- Clerici, M.; Faini, M.; Aebersold, R.; Jinek, M. Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex. eLife 2017, 6, e33111. [Google Scholar] [CrossRef]
- Yu, Z.; Hong, L.; Li, Q.Q. Signatures of mRNA alternative polyadenylation in Arabidopsis leaf development. Front. Genet. 2022, 13, 863253. [Google Scholar] [CrossRef]
- Xing, D.; Zhao, H.; Li, Q.Q. Arabidopsis CLP1-SIMILAR PROTEIN3, an ortholog of human polyadenylation factor CLP1, functions in gametophyte, embryo, and postembryonic development. Plant Physiol. 2008, 148, 2059–2069. [Google Scholar] [CrossRef]
- Xing, D.; Zhao, H.; Xu, R.; Li, Q.Q. Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J. 2008, 54, 899–910. [Google Scholar] [CrossRef]
- Yao, Y.L.; Song, L.H.; Katz, Y.; Galili, G. Cloning and characterization of Arabidopsis homologues of the animal CstF complex that regulates 3′ mRNA cleavage and polyadenylation. J. Exp. Bot. 2002, 53, 2277–2278. [Google Scholar] [CrossRef][Green Version]
- Herr, A.J.; Molnar, A.; Jones, A.; Baulcombe, D.C. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 14994–15001. [Google Scholar] [CrossRef]
- Chakrabarti, M.; Hunt, A.G. CPSF30 at the interface of alternative polyadenylation and cellular signaling in plants. Biomolecules 2015, 5, 1151–1168. [Google Scholar] [CrossRef]
- Elliott, B.J.; Dattaroy, T.; Meeks-Midkiff, L.R.; Forbes, K.P.; Hunt, A.G. An interaction between an Arabidopsis poly(A) polymerase and a homologue of the 100 kDa subunit of CPSF. Plant Mol. Biol. 2003, 51, 373–384. [Google Scholar] [CrossRef]
- Xu, R.Q.; Ye, X.F.; Li, Q.S.Q. AtCPSF73-II gene encoding an Arabidopsis homolog of CPSF 73 kDa subunit is critical for early embryo development. Gene 2004, 324, 35–45. [Google Scholar] [CrossRef]
- Li, Z.; Wang, R.; Gao, Y.; Wang, C.; Zhao, L.; Xu, N.; Chen, K.-E.; Qi, S.; Zhang, M.; Tsay, Y.-F.; et al. The Arabidopsis CPSF30-L gene plays an essential role in nitrate signaling and regulates the nitrate transceptor gene NRT1.1. New Phytol. 2017, 216, 1205–1222. [Google Scholar] [CrossRef]
- Forbes, K.P.; Addepalli, B.; Hunt, A.G. An Arabidopsis Fip1 homolog interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J. Biol. Chem. 2006, 281, 176–186. [Google Scholar] [CrossRef]
- Simpson, G.G.; Dijkwel, P.P.; Quesada, V.; Henderson, I.; Dean, C. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 2003, 113, 777–787. [Google Scholar] [CrossRef]
- Vi, S.L.; Trost, G.; Lange, P.; Czesnick, H.; Rao, N.; Lieber, D.; Laux, T.; Gray, W.M.; Manley, J.L.; Groth, D.; et al. Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. Proc. Natl. Acad. Sci. USA 2013, 110, 13994–13999. [Google Scholar] [CrossRef]
- Ozsolak, F.; Kapranov, P.; Foissac, S.; Kim, S.W.; Fishilevich, E.; Monaghan, A.P.; John, B.; Milos, P.M. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 2010, 143, 1018–1029. [Google Scholar] [CrossRef]
- Liu, X.; Hoque, M.; Larochelle, M.; Lemay, J.-F.; Yurko, N.; Manley, J.L.; Bachand, F.; Tian, B. Comparative analysis of alternative polyadenylation in S. cerevisiae and S. pombe. Genome Res. 2017, 27, 1685–1695. [Google Scholar] [CrossRef]
- Derti, A.; Garrett-Engele, P.; MacIsaac, K.D.; Stevens, R.C.; Sriram, S.; Chen, R.; Rohl, C.A.; Johnson, J.M.; Babak, T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012, 22, 1173–1183. [Google Scholar] [CrossRef]
- Hoque, M.; Ji, Z.; Zheng, D.; Luo, W.; Li, W.; You, B.; Park, J.Y.; Yehia, G.; Tian, B. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods 2013, 10, 133–139. [Google Scholar] [CrossRef]
- Bell, S.A.; Shen, C.; Brown, A.; Hunt, A.G. Experimental Genome-Wide Determination of RNA Polyadenylation in Chlamydomonas reinhardtii. PLoS ONE 2016, 11, e0146107. [Google Scholar] [CrossRef]
- Wu, X.; Liu, M.; Downie, B.; Liang, C.; Ji, G.; Li, Q.Q.; Hunt, A.G. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA 2011, 108, 12533–12538. [Google Scholar] [CrossRef]
- Shen, Y.; Ji, G.; Haas, B.J.; Wu, X.; Zheng, J.; Reese, G.J.; Li, Q.Q. Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation. Nucleic Acids Res. 2008, 36, 3150–3161. [Google Scholar] [CrossRef]
- Elkon, R.; Ugalde, A.P.; Agami, R. Alternative cleavage and polyadenylation: Extent, regulation and function. Nat. Rev. Genet. 2013, 14, 496–506. [Google Scholar] [CrossRef]
- Jabre, I.; Reddy, A.S.; Kalyna, M.; Chaudhary, S.; Khokhar, W.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Does co-transcriptional regulation of alternative splicing mediate plant stress responses? Nucleic Acids Res. 2019, 47, 2716–2726. [Google Scholar] [CrossRef]
- Shang, X.; Cao, Y.; Ma, L. Alternative splicing in plant genes: A means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef]
- Tsuchiya, T.; Eulgem, T. An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. USA 2013, 110, E3535–E3543. [Google Scholar] [CrossRef]
- Ma, X.; Cheng, S.; Ding, R.; Zhao, Z.; Zou, X.; Guang, S.; Wang, Q.; Jing, H.; Yu, C.; Ni, T.; et al. ipaQTL-atlas: An atlas of intronic polyadenylation quantitative trait loci across human tissues. Nucleic Acids Res. 2022, 51, 1046–1052. [Google Scholar] [CrossRef]
- Zhao, Z.; Xu, Q.; Wei, R.; Wang, W.; Ding, D.; Yang, Y.; Yao, J.; Zhang, L.; Hu, Y.-Q.; Wei, G.; et al. Cancer-associated dynamics and potential regulators of intronic polyadenylation revealed by IPAFinder using standard RNA-seq data. Genome Res. 2021, 31, 2095–2106. [Google Scholar] [CrossRef]
- Yang, J.; Cao, Y.; Ma, L. Co-Transcriptional RNA Processing in Plants: Exploring from the Perspective of Polyadenylation. Int. J. Mol. Sci. 2021, 22, 3300. [Google Scholar] [CrossRef]
- Kyburz, A.; Friedlein, A.; Langen, H.; Keller, W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol. Cell 2006, 23, 195–205. [Google Scholar] [CrossRef]
- Müller-McNicoll, M.; Botti, V.; de Jesus Domingues, A.M.; Brandl, H.; Schwich, O.D.; Steiner, M.C.; Curk, T.; Poser, I.; Zarnack, K.; Neugebauer, K.M. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016, 30, 553–566. [Google Scholar] [CrossRef]
- Dubbury, S.J.; Boutz, P.L.; Sharp, P.A. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 2018, 564, 141–145. [Google Scholar] [CrossRef]
- Vorlová, S.; Rocco, G.; LeFave, C.V.; Jodelka, F.M.; Hess, K.; Hastings, M.L.; Henke, E.; Cartegni, L. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol. Cell 2011, 43, 927–939. [Google Scholar] [CrossRef]
- Adam, S.; Polo, S.E.; Almouzni, G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 2013, 155, 963. [Google Scholar] [CrossRef]
- Devany, E.; Park, J.Y.; Murphy, M.R.; Zakusilo, G.; Baquero, J.; Zhang, X.; Hoque, M.; Tian, B.; Kleiman, F.E. Intronic cleavage and polyadenylation regulates gene expression during DNA damage response through U1 snRNA. Cell Discov. 2016, 2, 16013. [Google Scholar] [CrossRef]
- Krajewska, M.; Dries, R.; Grassetti, A.V.; Dust, S.; Gao, Y.; Huang, H.; Sharma, B.; Day, D.S.; Kwiatkowski, N.; Pomaville, M.; et al. CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat. Commun. 2019, 10, 1757. [Google Scholar] [CrossRef]
- Mayr, C. What are 3′ UTRs doing? Cold Spring Harb. Perspect. Biol. 2019, 11, a034728. [Google Scholar] [CrossRef]
- Geisberg, J.V.; Moqtaderi, Z.; Fan, X.; Ozsolak, F.; Struhl, K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 2014, 156, 812–824. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Lu, Y.; Zinta, G.; Lang, Z.; Zhu, J.-K. UTR-dependent control of gene expression in plants. Trends Plant Sci. 2018, 23, 248–259. [Google Scholar] [CrossRef]
- Axtell, M.J.; Meyers, B.C. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef]
- Deka, K.; Saha, S. Heat stress induced arginylation of HuR promotes alternative polyadenylation of Hsp70.3 by regulating HuR stability and RNA binding. Cell Death Differ. 2021, 28, 730–747. [Google Scholar] [CrossRef]
- Zheng, T.; Tan, Y.; Qiu, J.; Xie, Z.; Hu, X.; Zhang, J.; Na, N. Alternative polyadenylation trans-factor FIP1 exacerbates UUO/IRI-induced kidney injury and contributes to AKI-CKD transition via ROS-NLRP3 axis. Cell Death Dis. 2021, 12, 512. [Google Scholar] [CrossRef]
- Ripin, N.; Boudet, J.; Duszczyk, M.M.; Hinniger, A.; Faller, M.; Krepl, M.; Gadi, A.; Schneider, R.J.; Šponer, J.; Meisner-Kober, N.C.; et al. Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM. Proc. Natl. Acad. Sci. USA 2019, 116, 2935–2944. [Google Scholar] [CrossRef]
- Zheng, D.; Wang, R.; Ding, Q.; Wang, T.; Xie, B.; Wei, L.; Zhong, Z.; Tian, B. Cellular stress alters 3′ UTR landscape through alternative polyadenylation and isoform-specific degradation. Nature Commun. 2018, 9, 2268. [Google Scholar] [CrossRef]
- Wolf, E.J.; Miles, A.; Lee, E.S.; Nabeel-Shah, S.; Greenblatt, J.F.; Palazzo, A.F.; Tropepe, V.; Emili, A. MKRN2 Physically Interacts with GLE1 to Regulate mRNA Export and Zebrafish Retinal Development. Cell Rep. 2020, 31, 107693. [Google Scholar] [CrossRef]
- Rosa-Mercado, N.A.; Steitz, J.A. Who let the DoGs out?—Biogenesis of stress-induced readthrough transcripts. Trends Biochem. Sci. 2022, 47, 206–217. [Google Scholar] [CrossRef]
- Hennig, T.; Michalski, M.; Rutkowski, A.J.; Djakovic, L.; Whisnant, A.W.; Friedl, M.-S.; Jha, B.A.; Baptista, M.A.P.; L’Hernault, A.; Erhard, F.; et al. HSV-1-induced disruption of transcription termination resembles a cellular stress response but selectively increases chromatin accessibility downstream of genes. PLoS Pathog. 2018, 14, e1006954. [Google Scholar] [CrossRef]
- Vilborg, A.; Passarelli, M.; Yario, T.A.; Tycowski, K.T.; Steitz, J.A. Widespread inducible transcription downstream of human genes. Mol. Cell 2015, 59, 449–461. [Google Scholar] [CrossRef]
- Mayr, C. Regulation by 3′-Untranslated regions. Annu. Rev. Genet. 2017, 51, 171–194. [Google Scholar] [CrossRef]
- Sandberg, R.; Neilson, J.R.; Sarma, A.; Sharp, P.A.; Burge, C.B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 2008, 320, 1643–1647. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, L.; Chen, C.; Ge, Y.; Kang, M.; Song, Z.; Li, J.; Feng, Y.; Huo, Z.; He, G.; et al. Crosstalk between alternative polyadenylation and miRNAs in the regulation of protein translational efficiency. Genome Res. 2018, 28, 1656–1663. [Google Scholar] [CrossRef]
- Huggins, C.J.; Mayekar, M.K.; Martin, N.; Saylor, K.L.; Gonit, M.; Jailwala, P.; Kasoji, M.; Haines, D.C.; Quiñones, O.A.; Johnson, P.F. C/EBPγ is a critical regulator of cellular stress response networks through heterodimerization with ATF4. Mol. Cell. Biol. 2015, 36, 693–713. [Google Scholar] [CrossRef]
- Hunt, A.G. Review: Mechanisms underlying alternative polyadenylation in plants—Looking in the right places. Plant Sci. 2022, 324, 111430. [Google Scholar] [CrossRef]
- Deng, X.; Cao, X. Roles of pre-mRNA splicing and polyadenylation in plant development. Curr. Opin. Plant Biol. 2017, 35, 45–53. [Google Scholar] [CrossRef]
- Hornyik, C.; Duc, C.; Rataj, K.; Terzi, L.C.; Simpson, G.G. Alternative polyadenylation of antisense RNAs and flowering time control. Biochem. Soc. Trans. 2010, 38, 1077–1081. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.P.I. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef]
- VanWallendael, A.; Soltani, A.; Emery, N.C.; Peixoto, M.M.; Olsen, J.; Lowry, D.B. A molecular view of plant local adaptation: Incorporating stress-response networks. Annu. Rev. Plant Biol. 2019, 70, 559–583. [Google Scholar] [CrossRef]
- de Lorenzo, L.; Sorenson, R.; Bailey-Serres, J.; Hunt, A.G. Noncanonical alternative polyadenylation contributes to gene regulation in response to hypoxia. Plant Cell 2017, 29, 1262–1277. [Google Scholar] [CrossRef]
- Klauer, A.A.; van Hoof, A. Degradation of mRNAs that lack a stop codon: A decade of nonstop progress. Wiley Interdiscip. Rev. RNA 2012, 3, 649–660. [Google Scholar] [CrossRef]
- Singh, I.; Lee, S.-H.; Sperling, A.S.; Samur, M.K.; Tai, Y.-T.; Fulciniti, M.; Munshi, N.C.; Mayr, C.; Leslie, C.S. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nature Commun. 2018, 9, 1716. [Google Scholar] [CrossRef]
- Chew, G.-L.; Pauli, A.; Rinn, J.; Regev, A.; Schier, A.F.; Valen, E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5’ leaders of coding RNAs. Development 2013, 140, 2828–2834. [Google Scholar] [CrossRef]
- Sun, H.-X.; Li, Y.; Niu, Q.-W.; Chua, N.-H. Dehydration stress extends mRNA 3′ untranslated regions with noncoding RNA functions in Arabidopsis. Genome Res. 2017, 27, 1427–1436. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef]
- Bell, O.; Tiwari, V.K.; Thoma, N.H.; Schubeler, D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011, 12, 554–564. [Google Scholar] [CrossRef]
- Zhao, Z.; Wu, X.; Ji, G.; Liang, C.; Li, Q.Q. Genome-wide comparative analyses of polyadenylation signals in eukaryotes suggest a possible origin of the AAUAAA signal. Int. J. Mol. Sci. 2019, 20, 958. [Google Scholar] [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Téllez-Robledo, B.; Manzano, C.; Saez, A.; Navarro-Neila, S.; Silva-Navas, J.; de Lorenzo, L.; González-García, M.; Toribio, R.; Hunt, A.G.; Baigorri, R.; et al. The polyadenylation factor FIP1 is important for plant development and root responses to abiotic stresses. Plant J. 2019, 99, 1203–1219. [Google Scholar] [CrossRef]
- Yu, Z.; Lin, J.; Li, Q.Q. Transcriptome analyses of FY mutants reveal its role in mRNA alternative polyadenylation. Plant Cell 2019, 31, 2332–2352. [Google Scholar] [CrossRef]
- Chakrabarti, M.; Lorenzo, L.; Abdel-Ghany, S.E.; Reddy, A.S.N.; Hunt, A.G. Wide-ranging transcriptome remodelling mediated by alternative polyadenylation in response to abiotic stresses in Sorghum. Plant J. 2020, 102, 916–930. [Google Scholar] [CrossRef]
- Ma, H.; Cai, L.; Lin, J.; Zhou, K.; Li, Q.Q. Divergence in the regulation of the salt tolerant response between Arabidopsis thaliana and its halophytic relative Eutrema Salsugineum by mRNA alternative polyadenylation. Front. Plant Sci. 2022, 13, 866054. [Google Scholar] [CrossRef]
- Conesa, C.M.; Saez, A.; Navarro-Neila, S.; de Lorenzo, L.; Hunt, A.G.; Sepúlveda, E.B.; Baigorri, R.; Garcia-Mina, J.M.; Zamarreño, A.M.; Sacristán, S.; et al. Alternative Polyadenylation and Salicylic Acid Modulate Root Responses to Low Nitrogen Availability. Plants 2020, 9, 251. [Google Scholar] [CrossRef]
- Wang, R.; Xing, X.; Wang, Y.; Tran, A.; Crawford, N.M. A Genetic Screen for Nitrate Regulatory Mutants Captures the Nitrate Transporter Gene NRT1.1. Plant Physiol. 2009, 151, 472–478. [Google Scholar] [CrossRef]
- Higuera, J.J.; Fernandez, E.; Galvan, A. Chlamydomonas NZF1, a tandem-repeated zinc finger factor involved in nitrate signalling by controlling the regulatory gene NIT2. Plant Cell Environ. 2014, 37, 2139–2150. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, W.; Li, Z.; Li, Z.; Bi, Y.; Crawford, N.M.; Wang, Y. FIP1 Plays an Important Role in Nitrate Signaling and Regulates CIPK8 and CIPK23 Expression in Arabidopsis. Front. Plant Sci. 2018, 9, 593. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.-K. Mutational Evidence for the Critical Role of CBF Transcription Factors in Cold Acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar] [CrossRef] [PubMed]
- Kindgren, P.; Ard, R.; Ivanov, M.; Marquardt, S. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat. Commun. 2018, 9, 4561. [Google Scholar] [CrossRef]
- Banerjee, A.K.; Blanco, M.R.; Bruce, E.A.; Honson, D.D.; Chen, L.M.; Chow, A.; Bhat, P.; Ollikainen, N.; Quinodoz, S.A.; Loney, C.; et al. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell 2020, 183, 1325–1339. [Google Scholar] [CrossRef]
- Zhang, K.; Xie, Y.; Muñoz-Moreno, R.; Wang, J.; Zhang, L.; Esparza, M.; García-Sastre, A.; Fontoura, B.M.A.; Ren, Y. Structural basis for influenza virus NS1 protein block of mRNA nuclear export. Nat. Microbiol. 2019, 4, 1671–1679. [Google Scholar] [CrossRef]
- Jia, X.; Yuan, S.; Wang, Y.; Fu, Y.; Ge, Y.; Ge, Y.; Lan, X.; Feng, Y.; Qiu, F.; Li, P.; et al. The role of alternative polyadenylation in the antiviral innate immune response. Nat. Commun. 2017, 8, 14605. [Google Scholar] [CrossRef]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef]
- van Wersch, S.; Li, X. Stronger when together: Clustering of plant NLR disease resistance genes. Trends Plant Sci. 2019, 24, 688–699. [Google Scholar] [CrossRef]
- Zhou, Q.; Fu, H.; Yang, D.; Ye, C.; Zhu, S.; Lin, J.; Ye, W.; Ji, G.; Ye, X.; Wu, X.; et al. Differential alternative polyadenylation contributes to the developmental divergence between two rice subspecies, japonica and indica. Plant J. 2019, 98, 260–276. [Google Scholar] [CrossRef]
- Ji, C.; Ji, Z.; Liu, B.; Cheng, H.; Liu, H.; Liu, S.; Yang, B.; Chen, G. Xa1 allelic R genes activate rice blight resistance suppressed by interfering TAL effectors. Plant Commun. 2020, 1, 100087. [Google Scholar] [CrossRef]
- Fitzgerald, H.A.; Canlas, P.E.; Chern, M.S.; Ronald, P.C. Alteration of TGA factor activity in rice results in enhanced tolerance to Xanthomonas oryzae pv. oryzae. Plant J. 2005, 43, 335–347. [Google Scholar] [CrossRef]
- Grant, M.R.; Godiard, L.; Straube, E.; Ashfield, T.; Lewald, J.; Sattler, A.; Innes, R.W.; Dangl, J.L. Structure of the Arabidopsis RPM1 Gene Enabling Dual Specificity Disease Resistance. Science 1995, 269, 843–846. [Google Scholar] [CrossRef]
- Boch, J.; Verbsky, M.L.; Robertson, T.L.; Larkin, J.C.; Kunkel, B.N. Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in CPR5. Mol. Plant-Microbe Interact. 1998, 11, 1196–1206. [Google Scholar] [CrossRef]
- Peng, S.; Guo, D.; Guo, Y.; Zhao, H.; Mei, J.; Han, Y.; Guan, R.; Wang, T.; Song, T.; Sun, K.; et al. Constitutive expresser of pathogenesis-related genes 5 is an RNA-binding protein controlling plant immunity via an RNA processing complex. Plant Cell 2022, 34, 1724–1744. [Google Scholar] [CrossRef]
- Donahue, J.L.; Alford, S.R.; Torabinejad, J.; Kerwin, R.E.; Nourbakhsh, A.; Ray, W.K.; Hernick, M.; Huang, X.; Lyons, B.M.; Hein, P.P.; et al. The Arabidopsis thaliana myo-inositol 1-phosphate synthase1 gene is required for myo-inositol synthesis and suppression of cell death. Plant Cell 2010, 22, 888–903. [Google Scholar] [CrossRef]
- Bruggeman, Q.; Garmier, M.; de Bont, L.; Soubigou-Taconnat, L.; Mazubert, C.; Benhamed, M.; Raynaud, C.; Bergounioux, C.; Delarue, M.; Wang, G.; et al. The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: A key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol. 2014, 165, 732–746. [Google Scholar] [CrossRef]
- Sun, T.; Zhang, Y.; Li, Y.; Zhang, Q.; Ding, Y.; Zhang, Y. ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 2015, 6, 10159. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- Cao, J.; Ye, C.; Hao, G.; Dabney-Smith, C.; Hunt, A.G.; Li, Q.Q. Root Hair Single Cell Type Specific Profiles of Gene Expression and Alternative Polyadenylation Under Cadmium Stress. Front. Plant Sci. 2019, 10, 589. [Google Scholar] [CrossRef]
- Qi, J.; Wang, J.; Gong, Z.; Zhou, J.-M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 2017, 38, 92–100. [Google Scholar] [CrossRef]
- Lyons, R.; Iwase, A.; Gänsewig, T.; Sherstnev, A.; Duc, C.; Barton, G.J.; Hanada, K.; Higuchi-Takeuchi, M.; Matsui, M.; Sugimoto, K.; et al. The RNA-binding protein FPA regulates flg22-triggered defense responses and transcription factor activity by alternative polyadenylation. Sci. Rep. 2013, 3, 2866. [Google Scholar] [CrossRef]
- Zhang, J.; Addepalli, B.; Yun, K.-Y.; Hunt, A.G.; Xu, R.; Rao, S.; Li, Q.Q.; Falcone, D.L. A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS ONE 2008, 3, e2410. [Google Scholar] [CrossRef]
- Song, P.; Yang, J.; Wang, C.; Lu, Q.; Shi, L.; Tayier, S.; Jia, G. Arabidopsis N6-methyladenosine reader CPSF30-L recognizes FUE signals to control polyadenylation site choice in liquid-like nuclear bodies. Mol. Plant 2021, 14, 571–587. [Google Scholar] [CrossRef]
- Shi, Y.; Manley, J.L. The end of the message: Multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 2015, 29, 889–897. [Google Scholar] [CrossRef]
- Zhang, Z.Q.; Fu, J.H.; Gilmour, D.S. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 2005, 19, 1572–1580. [Google Scholar] [CrossRef]
- Nejat, N.; Ramalingam, A.; Mantri, N. Advances in transcriptomics of plants. Adv. Biochem. Eng. Biotechnol. 2018, 164, 161–185. [Google Scholar]
- Zhao, M.; Liu, B.; Wu, K.; Ye, Y.; Huang, S.; Wang, S.; Wang, Y.; Han, R.; Liu, Q.; Fu, X.; et al. Regulation of osmir156h through alternative polyadenylation improves grain yield in rice. PLoS ONE 2015, 10, e0126154. [Google Scholar] [CrossRef]
| Stress | Species | Target Genes | APA Types | Associated Polyadenylation Factors | References |
|---|---|---|---|---|---|
| Hypoxic | Arabidopsis thaliana | 5′UTR APA CDS APA Intronic APA 3′UTR APA | [74] | ||
| Drought | Arabidopsis thaliana | 3′UTR extension | FPA | [78] | |
| Salt | Arabidopsis thaliana Eutrema salsugineum Sorghum | AKR2 AT3G47610 CIPK21 MAP3Kδ4 | 3′UTR APA | FIP1 CPSF30 | [83,84,85,86] |
| N starvation | Arabidopsis thaliana Chlamydomonas | NRT1.1 CPSF30 | 3′UTR APA Intronic APA | FIP1 CPSF30 | [28,87,89,90] |
| Temperature | Arabidopsis thaliana | SVK | 3′UTR extension | [95] | |
| Pathogens | Rice Arabidopsis thaliana | Xa1 rTGA2.1 CBP60g | CDS APA 3′UTR APA | FIP1 CPSF30 | [101,106,108] |
| ROS | Arabidopsis thaliana | ERF4 | CDS APA | FPA CPSF30 | [111,113,114] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Ma, L.; Cao, Y. Alternative Polyadenylation Is a Novel Strategy for the Regulation of Gene Expression in Response to Stresses in Plants. Int. J. Mol. Sci. 2023, 24, 4727. https://doi.org/10.3390/ijms24054727
Wu J, Ma L, Cao Y. Alternative Polyadenylation Is a Novel Strategy for the Regulation of Gene Expression in Response to Stresses in Plants. International Journal of Molecular Sciences. 2023; 24(5):4727. https://doi.org/10.3390/ijms24054727
Chicago/Turabian StyleWu, Jing, Ligeng Ma, and Ying Cao. 2023. "Alternative Polyadenylation Is a Novel Strategy for the Regulation of Gene Expression in Response to Stresses in Plants" International Journal of Molecular Sciences 24, no. 5: 4727. https://doi.org/10.3390/ijms24054727
APA StyleWu, J., Ma, L., & Cao, Y. (2023). Alternative Polyadenylation Is a Novel Strategy for the Regulation of Gene Expression in Response to Stresses in Plants. International Journal of Molecular Sciences, 24(5), 4727. https://doi.org/10.3390/ijms24054727





