Lighting-Up the Far-Red Fluorescence of RNA-Selective Dyes by Switching from Ortho to Para Position
Abstract
:1. Introduction
2. Results
2.1. Photophysical Properties
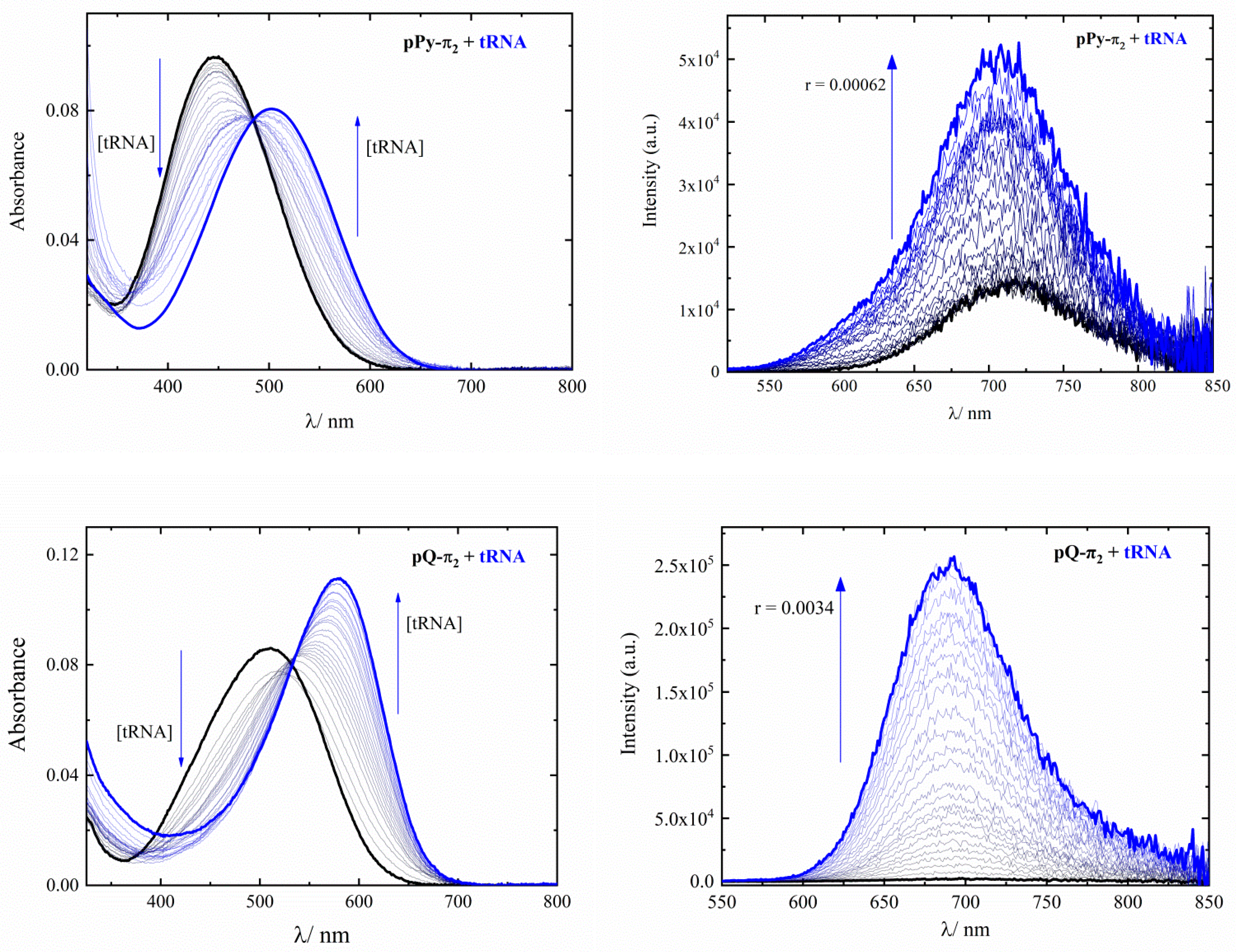
2.2. Interaction with Nucleic Acids
2.3. In Vitro Studies: Antiproliferative Effect and Intracellular Localization
3. Discussion
4. Materials and Methods
4.1. Synthesis
4.2. Characterization
4.3. Materials
4.4. Photophysical Measurements
4.5. Femtosecond Spectroscopy
4.6. Quantum Mechanical Calculations
4.7. Spectroscopic Titrations
4.8. Cell Cultures
4.9. Antiproliferative Assay
4.10. Fluorescence Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Le, P.; Ahmed, N.; Yeo, G.W. Illuminating RNA biology through imaging. Nat. Cell Biol. 2022, 24, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Chilka, P.; Desai, N.; Datta, B. Small Molecule Fluorescent Probes for G-Quadruplex Visualization as Potential Cancer Theranostic Agents. Molecules 2019, 24, 752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunbul, M.; Jäschke, A. SRB-2: A promiscuous rainbow aptamer for live-cell RNA imaging. Nucleic Acids Res. 2018, 46, e110. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, H.J.; Bergqvist, M.H.; Lincoln, P.; Westman, G. Syntheses and DNA-binding studies of a series of unsymmetrical cyanine dyes: Structural influence on the degree of minor groove binding to natural DNA. Bioorganic Med. Chem. 2004, 12, 2369–2384. [Google Scholar] [CrossRef] [PubMed]
- Cesaretti, A.; Mencaroni, L.; Bonaccorso, C.; Botti, V.; Calzoni, E.; Carlotti, B.; Fortuna, C.G.; Montegiove, N.; Spalletti, A.; Elisei, F. Amphiphilicity-Controlled Localization of Red Emitting Bicationic Fluorophores in Tumor Cells Acting as Bio-Probes and Anticancer Drugs. Molecules 2022, 27, 3713. [Google Scholar] [CrossRef] [PubMed]
- Suseela, Y.V.; Narayanaswamy, N.; Pratihar, S.; Govindaraju, T. Far-red fluorescent probes for canonical and non-canonical nucleic acid structures: Current progress and future implications. Chem. Soc. Rev. 2018, 47, 1098–1131. [Google Scholar] [CrossRef]
- Ramesh, J.; Arunkumar, C.; Sujatha, S. Dicationic porphyrins bearing thienyl and pyridinium moieties: Synthesis, characterization, DNA interaction and cancer cell toxicity. Polyhedron 2019, 170, 151–159. [Google Scholar] [CrossRef]
- Čipor, I.; Kurutos, A.; Dobrikov, G.M.; Kamounah, F.S.; Majhen, D.; Nestić, D.; Piantanida, I. Structure-dependent mitochondria or lysosome-targeting styryl fluorophores bearing remarkable Stokes shift. Dye. Pigment. 2022, 206, 110626. [Google Scholar] [CrossRef]
- Ihmels, H.; Karbasiyoun, M.; Löhl, K.; Stremmel, C. Structural flexibility versus rigidity of the aromatic unit of DNA ligands: Binding of aza- and azoniastilbene derivatives to duplex and quadruplex DNA. Org. Biomol. Chem. 2019, 17, 6404–6413. [Google Scholar] [CrossRef] [Green Version]
- Fudickar, W.; Linker, T. Structural motives controlling the binding affinity of 9,10-bis(methylpyridinium)anthracenes towards DNA. Bioorganic Med. Chem. 2020, 28, 115432. [Google Scholar] [CrossRef]
- Doan, P.H.; Pitter, D.R.G.; Kocher, A.; Wilson, J.N.; Goodson, T.I. Two-Photon Spectroscopy as a New Sensitive Method for Determining the DNA Binding Mode of Fluorescent Nuclear Dyes. J. Am. Chem. Soc. 2015, 137, 9198–9201. [Google Scholar] [CrossRef] [PubMed]
- Tanious, F.A.; Veal, J.M.; Buczak, H.; Ratmeyer, L.S.; Wilson, W.D. DAPI (4’,6-diamidino-2-phenylindole) binds differently to DNA and RNA: Minor-groove binding at AT sites and intercalation at AU sites. Biochemistry 1992, 31, 3103–3112. [Google Scholar] [CrossRef] [PubMed]
- Biver, T. Use of UV-Vis Spectrometry to Gain Information on the Mode of Binding of Small Molecules to DNAs and RNAs. Appl. Spectrosc. Rev. 2012, 47, 272–325. [Google Scholar] [CrossRef]
- Roeland Boer, D.; Canals, A.; Coll, M. DNA-binding drugs caught in action: The latest 3D pictures of drug-DNA complexes. Dalton Trans. 2009, 0, 399–414. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Chi, W.; Qiao, Q.; Tan, D.; Xu, Z.; Liu, X. Twisted intramolecular charge transfer (TICT) and twists beyond TICT: From mechanisms to rational designs of bright and sensitive fluorophores. Chem. Soc. Rev. 2021, 50, 12656–12678. [Google Scholar] [CrossRef]
- Luo, X.; Xue, B.; Feng, G.; Zhang, J.; Lin, B.; Zeng, P.; Li, H.; Yi, H.; Zhang, X.-L.; Zhu, H.; et al. Lighting up the Native Viral RNA Genome with a Fluorogenic Probe for the Live-Cell Visualization of Virus Infection. J. Am. Chem. Soc. 2019, 141, 5182–5191. [Google Scholar] [CrossRef]
- Carlotti, B.; Cesaretti, A.; Fortuna, C.G.; Spalletti, A.; Elisei, F. Experimental evidence of dual emission in a negatively solvatochromic push–pull pyridinium derivative. Phys. Chem. Chem. Phys. 2015, 17, 1877–1882. [Google Scholar] [CrossRef]
- Cesaretti, A.; Spalletti, A.; Elisei, F.; Foggi, P.; Germani, R.; Fortuna, C.G.; Carlotti, B. The role of twisting in driving excited-state symmetry breaking and enhanced two-photon absorption in quadrupolar cationic pyridinium derivatives. Phys. Chem. Chem. Phys. 2021, 23, 16739–16753. [Google Scholar] [CrossRef]
- Cesaretti, A.; Carlotti, B.; Germani, R.; Spalletti, A.; Elisei, F. Inclusion of push–pull N -methylpyridinium salts within surfactant hydrogels: Is their excited state intramolecular charge transfer mediated by twisting? Phys. Chem. Chem. Phys. 2015, 17, 17214–17220. [Google Scholar] [CrossRef]
- Viviano-Posadas, A.O.; Romero-Mendoza, U.; Bazany-Rodríguez, I.J.; Velázquez-Castillo, R.V.; Martínez-Otero, D.; Bautista-Renedo, J.M.; González-Rivas, N.; Galindo-Murillo, R.; Salomón-Flores, M.K.; Dorazco-González, A. Efficient fluorescent recognition of ATP/GTP by a water-soluble bisquinolinium pyridine-2,6-dicarboxamide compound. Crystal structures, spectroscopic studies and interaction mode with DNA. RSC Adv. 2022, 12, 27826–27838. [Google Scholar] [CrossRef]
- Zhu, H.; Fan, J.; Du, J.; Peng, X. Fluorescent probes for sensing and imaging within specific cellular organelles. Acc. Chem. Res. 2016, 49, 2115–2126. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, N.; Arya, D.P. Linker dependent intercalation of bisbenzimidazole-aminosugars in an RNA duplex; selectivity in RNA vs. DNA binding. Bioorganic Med. Chem. Lett. 2016, 26, 5989–5994. [Google Scholar] [CrossRef] [PubMed]
- Weißenstein, A.; Vysotsky, M.O.; Piantanida, I.; Würthner, F. Naphthalene diimide–amino acid conjugates as novel fluorimetric and CD probes for differentiation between ds-DNA and ds-RNA. Beilstein J. Org. Chem. 2020, 16, 2032–2045. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Wei, P.; Li, R.; Zhong, Y.; Li, X.; Xue, F.; Shi, Y.; Yi, T. Ribosomal RNA-Selective Light-Up Fluorescent Probe for Rapidly Imaging the Nucleolus in Live Cells. ACS Sens. 2019, 4, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, Y.; Sato, Y.; Nishizawa, S. Deep-Red Light-up Signaling of Benzo[c,d]indole–Quinoline Monomethine Cyanine for Imaging of Nucleolar RNA in Living Cells and for Sequence-Selective RNA Analysis. Anal. Chem. 2019, 91, 14254–14260. [Google Scholar] [CrossRef]
- Calzoni, E.; Argentati, C.; Cesaretti, A.; Montegiove, N.; Tortorella, I.; Bazzucchi, M.; Morena, F.; Martino, S.; Emiliani, C. RNA Modifications in Neurodegenerations. In Epitranscriptomics; Jurga, S., Barciszewski, J., Eds.; RNA Technologies; Springer: Cham, Switzerland, 2021; pp. 23–77. ISBN 978-3-030-71612-7. [Google Scholar]
- Lu, Y.-J.; Deng, Q.; Hu, D.-P.; Wang, Z.-Y.; Huang, B.-H.; Du, Z.-Y.; Fang, Y.-X.; Wong, W.-L.; Zhang, K.; Chow, C.-F. A molecular fluorescent dye for specific staining and imaging of RNA in live cells: A novel ligand integration from classical thiazole orange and styryl compounds. Chem. Commun. 2015, 51, 15241–15244. [Google Scholar] [CrossRef]
- Song, G.; Sun, Y.; Liu, Y.; Wang, X.; Chen, M.; Miao, F.; Zhang, W.; Yu, X.; Jin, J. Low molecular weight fluorescent probes with good photostability for imaging RNA-rich nucleolus and RNA in cytoplasm in living cells. Biomaterials 2014, 35, 2103–2112. [Google Scholar] [CrossRef]
- Zhou, B.; Liu, W.; Zhang, H.; Wu, J.; Liu, S.; Xu, H.; Wang, P. Imaging of nucleolar RNA in living cells using a highly photostable deep-red fluorescent probe. Biosens. Bioelectron. 2015, 68, 189–196. [Google Scholar] [CrossRef]
- Higuchi, K.; Sato, Y.; Togashi, N.; Suzuki, M.; Yoshino, Y.; Nishizawa, S. Bright and Light-Up Sensing of Benzo[c,d]indole-oxazolopyridine Cyanine Dye for RNA and Its Application to Highly Sensitive Imaging of Nucleolar RNA in Living Cells. ACS Omega 2022, 7, 23744–23748. [Google Scholar] [CrossRef]
- Sato, Y.; Igarashi, Y.; Suzuki, M.; Higuchi, K.; Nishizawa, S. Deep-red fluorogenic cyanine dyes carrying an amino group-terminated side chain for improved RNA detection and nucleolar RNA imaging. RSC Adv. 2021, 11, 35436–35439. [Google Scholar] [CrossRef]
- Mazzoli, A.; Carlotti, B.; Bonaccorso, C.; Fortuna, C.G.; Mazzucato, U.; Miolo, G.; Spalletti, A. Photochemistry and DNA-affinity of some pyrimidine-substituted styryl-azinium iodides. Photochem. Photobiol. Sci. 2011, 10, 1830–1836. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, A.; Carlotti, B.; Consiglio, G.; Fortuna, C.G.; Miolo, G.; Spalletti, A. Photobehaviour of methyl-pyridinium and quinolinium iodide derivatives, free and complexed with DNA. A case of bisintercalation. Photochem. Photobiol. Sci. 2014, 13, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, A.; Spalletti, A.; Carlotti, B.; Emiliani, C.; Fortuna, C.G.; Urbanelli, L.; Tarpani, L.; Germani, R. Spectroscopic Investigation of Interactions of New Potential Anticancer Drugs with DNA and Non-Ionic Micelles. J. Phys. Chem. B 2015, 119, 1483–1495. [Google Scholar] [CrossRef] [PubMed]
- Botti, V.; Cesaretti, A.; Ban, Ž.; Crnolatac, I.; Consiglio, G.; Elisei, F.; Piantanida, I. Fine structural tuning of styryl-based dyes for fluorescence and CD-based sensing of various ds-DNA/RNA sequences. Org. Biomol. Chem. 2019, 17, 8243–8258. [Google Scholar] [CrossRef] [PubMed]
- Botti, V.; Urbanelli, L.; Sagini, K.; Tarpani, L.; Cesaretti, A.; Fortuna, C.G.; Elisei, F. Quaternized styryl-azinium fluorophores as cellular RNA-binders. Photochem. Photobiol. Sci. 2020, 19, 362–370. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; Hu, W.; Liu, Z. A Mitochondrial-Targeting Near-Infrared Fluorescent Probe for Visualizing and Monitoring Viscosity in Live Cells and Tissues. Anal. Chem. 2019, 91, 10302–10309. [Google Scholar] [CrossRef]
- Cesaretti, A.; Foggi, P.; Fortuna, C.G.; Elisei, F.; Spalletti, A.; Carlotti, B. Uncovering Structure-Property Relationships in Push-Pull Chromophores: A Promising Route to Large Hyperpolarizability and Two-Photon Absorption. J. Phys. Chem. C 2020, 124, 15739–15748. [Google Scholar] [CrossRef]
- Reichardt, C. Solvatochromic dyes as solvent polarity indicators. Chem. Rev. 1994, 94, 2319–2358. [Google Scholar] [CrossRef]
- Oudar, J.L.; Chemla, D.S. Hyperpolarizabilities of the nitroanilines and their relations to the excited state dipole moment. J. Chem. Phys. 1977, 66, 2664–2668. [Google Scholar] [CrossRef]
- Horng, M.L.; Gardecki, J.A.; Papazyan, A.; Maroncelli, M. Subpicosecond Measurements of Polar Solvation Dynamics: Coumarin 153 Revisited. J. Phys. Chem. 1995, 99, 17311–17337. [Google Scholar] [CrossRef]
- Park, M.; Im, D.; Rhee, Y.H.; Joo, T. Coherent and Homogeneous Intramolecular Charge-Transfer Dynamics of 1-tert-Butyl-6-cyano-1,2,3,4-tetrahydroquinoline (NTC6), a Rigid Analogue of DMABN. J. Phys. Chem. A 2014, 118, 5125–5134. [Google Scholar] [CrossRef] [PubMed]
- Carlotti, B.; Flamini, R.; Kikaš, I.; Mazzucato, U.; Spalletti, A. Intramolecular charge transfer, solvatochromism and hyperpolarizability of compounds bearing ethenylene or ethynylene bridges. Chem. Phys. 2012, 407, 9–19. [Google Scholar] [CrossRef]
- Seki, H.; Onishi, S.; Asamura, N.; Suzuki, Y.; Kawamata, J.; Kaneno, D.; Hadano, S.; Watanabe, S.; Niko, Y. Bright and two-photon active red fluorescent dyes that selectively move back and forth between the mitochondria and nucleus upon changing the mitochondrial membrane potential. J. Mater. Chem. B 2018, 6, 7396–7401. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Yang, L.; Ma, B.; Kong, Q.; Li, G.; Wang, Y.; Tang, B.Z. Multifunctional Two-Photon AIE Luminogens for Highly Mitochondria-Specific Bioimaging and Efficient Photodynamic Therapy. ACS Appl. Mater. Interfaces 2019, 11, 20715–20724. [Google Scholar] [CrossRef] [PubMed]
- Zinchuk, V.; Wu, Y.; Grossenbacher-Zinchuk, O. Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Sci. Rep. 2013, 3, 1365. [Google Scholar] [CrossRef] [Green Version]
- Zinchuk, V.; Grossenbacher-Zinchuk, O. Quantitative Colocalization Analysis of Confocal Fluorescence Microscopy Images. Curr. Protoc. Cell Biol. 2011, 52, 4.16.1–4.16.19. [Google Scholar] [CrossRef]
- Giordano, A.; Romano, G. Cell Cycle Control and Dysregulation Protocols; Springer: Berlin/Heidelberg, Germany, 2008; ISBN 978-1-59259-822-9. [Google Scholar]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]
- Anderson, G.R.; Wardell, S.E.; Cakir, M.; Yip, C.; Ahn, Y.; Ali, M.; Yllanes, A.P.; Chao, C.A.; McDonnell, D.P.; Wood, K.C. Dysregulation of mitochondrial dynamics proteins are a targetable feature of human tumors. Nat. Commun. 2018, 9, 1677. [Google Scholar] [CrossRef] [Green Version]
- Haugland, R.P. Assays for cell viability, proliferation and function. In The Handbook, A Guide to Fluorescent Probes and Labeling Technologies, 11th ed.; Life Technologies: Carlsbad, CA, USA, 2010; pp. 664–665. [Google Scholar]
- Birks, J.B. Photophysics of Aromatic Molecules; Wiley Monographs in Chemical Physics; Wiley-Interscience: London, UK, 1970; ISBN 978-0-471-07420-5. [Google Scholar]
- Montalti, M.; Credi, A.; Prodi, L.; Gandolfi, M.T. Handbook of Photochemistry; CRC Press: Boca Raton, FL, USA, 2006; ISBN 978-1-4200-1519-5. [Google Scholar]
- Mencaroni, L.; Bonaccorso, C.; Botti, V.; Carlotti, B.; Consiglio, G.; Elisei, F.; Fortuna, C.G.; Spalletti, A.; Cesaretti, A. Nonlinear optical properties of a new panchromatic series of water-soluble bicationic push-pull fluorophores. Dye. Pigment. 2021, 194, 109620. [Google Scholar] [CrossRef]
- Bonaccorso, C.; Cesaretti, A.; Elisei, F.; Mencaroni, L.; Spalletti, A.; Fortuna, C.G. New Styryl Phenanthroline Derivatives as Model D−π−A−π−D Materials for Non-Linear Optics. Chem. Phys. Chem. 2018, 19, 1917–1929. [Google Scholar] [CrossRef]
- Cesaretti, A.; Carlotti, B.; Elisei, F.; Fortuna, C.G.; Spalletti, A. Photoinduced ICT vs. excited rotamer intercoversion in two quadrupolar polyaromatic N -methylpyridinium cations. Phys. Chem. Chem. Phys. 2018, 20, 2851–2864. [Google Scholar] [CrossRef] [PubMed]
- Golub, G.H.; Loan, C.F.V. Matrix Computations; JHU Press: Baltimore, MD, USA, 2013; ISBN 978-1-4214-0859-0. [Google Scholar]
- Strang, G.; Strang, G.; Strang, G.; Strang, G. Introduction to Linear Algebra; Wellesley-Cambridge Press: Wellesley, MA, USA, 1993; Volume 3. [Google Scholar]
- Snellenburg, J.J.; Laptenok, S.; Seger, R.; Mullen, K.M.; Stokkum, I.H.M. van Glotaran: A Java-Based Graphical User Interface for the R Package TIMP. J. Stat. Softw. 2012, 49, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H. Gaussian 16 revision a. 03. 2016; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Becke, A.D. Density-functional thermochemistry. I. The effect of the exchange-only gradient correction. J. Chem. Phys. 1992, 96, 2155–2160. [Google Scholar] [CrossRef] [Green Version]
- Barone, V.; Cossi, M. Quantum Calculation of Molecular Energies and Energy Gradients in Solution by a Conductor Solvent Model. J. Phys. Chem. A 1998, 102, 1995–2001. [Google Scholar] [CrossRef]
- McGhee, J.D.; von Hippel, P.H. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
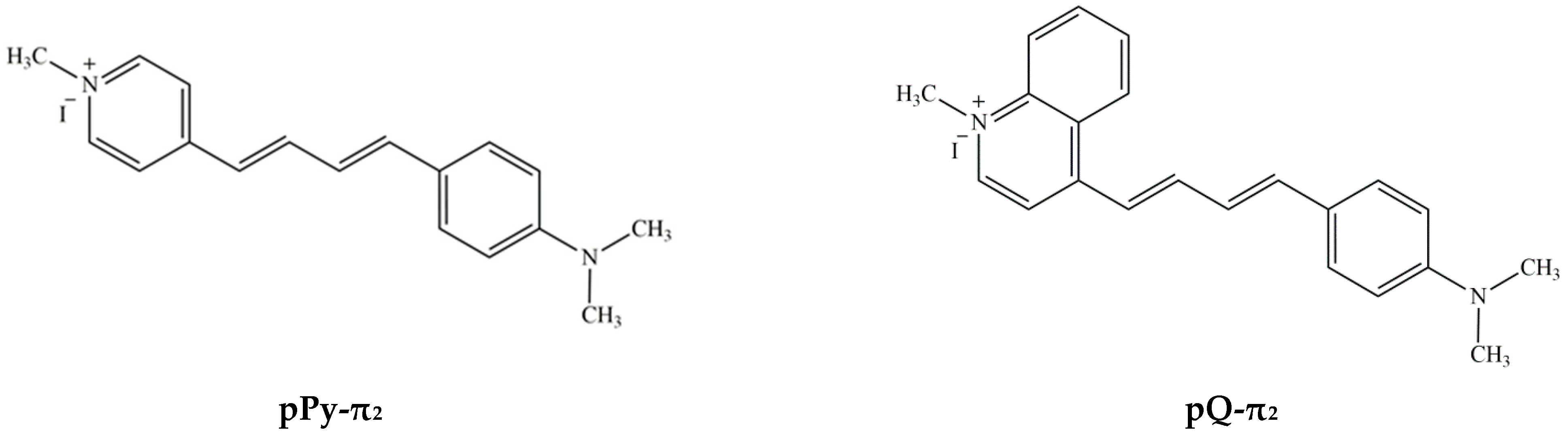






| pPy-π2 a | pQ-π2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Solvent | λabs/nm | λem/nm | Δν/cm−1 | ΦF | λabs/nm | λem/nm | Δν/cm−1 | ΦF | |
| DCM | 0.321 | 557 | 705 | 3770 | 0.25 | 591 | 679 | 2200 | 0.053 |
| DCE | 0.346 | 552 | 710 | 4030 | 0.19 | 584 | 687 | 2600 | 0.042 |
| 2–PrOH/DCM 50:50 | 0.428 | 523 | 705 | 4900 | 0.14 | 569 | 678 | 2800 | 0.032 |
| 2–PrOH | 0.552 | 504 | 707 | 5700 | 0.071 | 552 | 674 | 3300 | 0.026 |
| EtOH | 0.654 | 502 | 709 | 5820 | 0.049 | 548 | 678 | 3500 | 0.016 |
| MeOH | 0.765 | 492 | 712 | 6280 | 0.027 | 542 | 686 | 3900 | 0.006 |
| W/EtOH 50:50 | 0.827 | 484 | 710 | 6580 | 0.038 | ||||
| W/EtOH 70:30 | 0.8962 | 473 | 708 | 7020 | 0.019 | 538 | 684 | 4000 | 0.006 |
| W | 1 | 447 | 709 | 8270 | 0.0056 | 511 | 688 | 5000 | 0.002 |
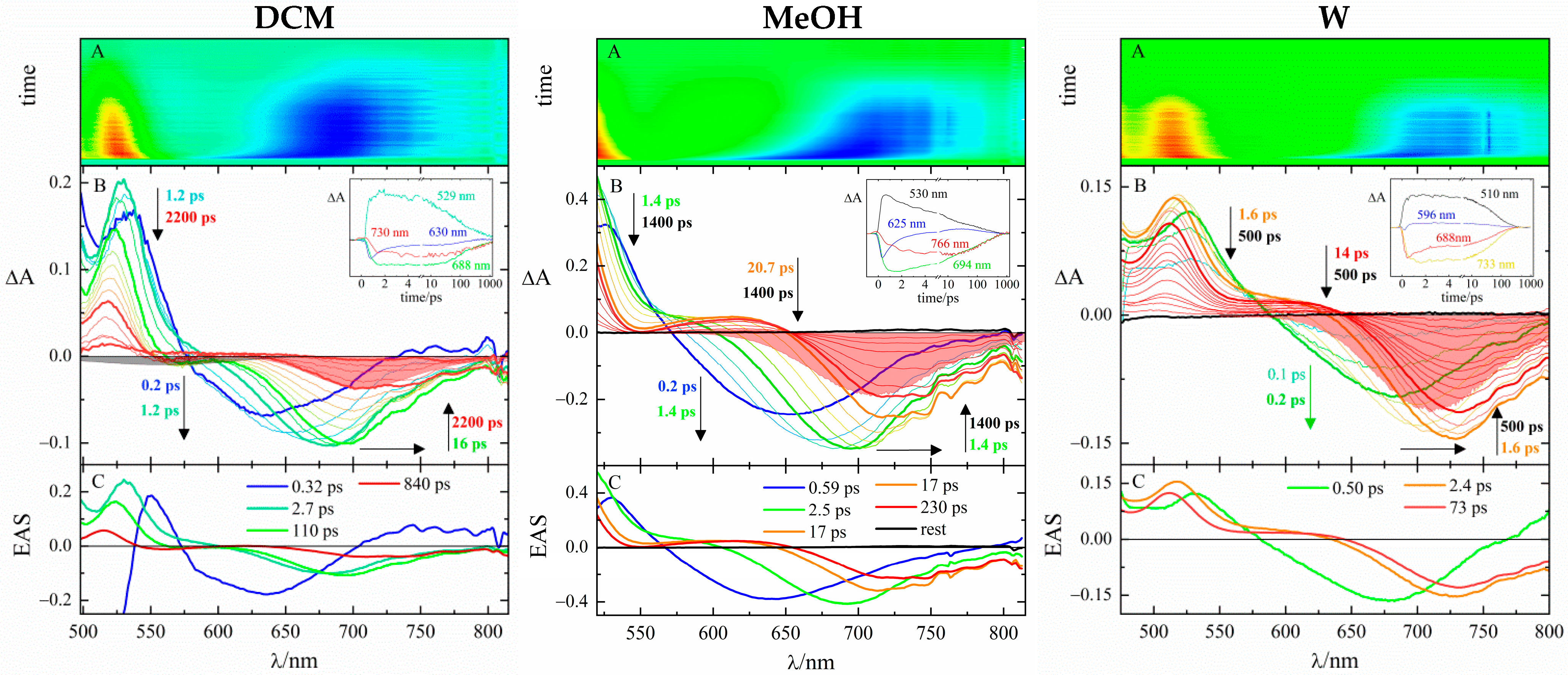
| pPy-π2 | pQ-π2 | Assignment | ||||
|---|---|---|---|---|---|---|
| DCM | MeOH | W | DCM | MeOH | W | |
| τ (ps) | τ (ps) | |||||
| 0.32 | 0.59 | 0.14 | 0.46 | Solv./LE | ||
| 2.7 | Solv. | |||||
| 110 | 2.5 | 0.50 | 4.1 | 1.9 | 0.39 | ICT |
| 17 | 2.4 | 4.8 | 1.4 | Solv. | ||
| 840 | 230 | 73 | 82 | 15 | 5.6 | TICT |
| rest | rest | rest | rest | rest | rest | |
| n Fixed = 0.1 | Kass | |
|---|---|---|
| pPy-π2 | pQ-π2 | |
| tRNA | (4 ± 2) × 103 | (2.4 ± 0.3) × 104 |
| ct-DNA | (3.0 ± 0.2) × 104 | (1.8 ± 0.2) × 105 |
| pPy-π2 | pQ-π2 | Assignment | ||||
|---|---|---|---|---|---|---|
| +tRNA | +ct-DNA | +tRNA | +ct-DNA | |||
| τ (ps) | τ (ps) | |||||
| 0.50 | 0.55 | 1.3 | 0.39 | 0.76 | 0.50 | ICT |
| 2.4 | 3.4 | 6.0 | 1.4 | 8.8 | 8.7 | Solv. |
| 73 | 88 | 82 | 5.6 | TICT | ||
| 1100 | 1300 | 500 | 320 | Complex | ||
| rest | rest | rest | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cesaretti, A.; Calzoni, E.; Montegiove, N.; Bianconi, T.; Alebardi, M.; La Serra, M.A.; Consiglio, G.; Fortuna, C.G.; Elisei, F.; Spalletti, A. Lighting-Up the Far-Red Fluorescence of RNA-Selective Dyes by Switching from Ortho to Para Position. Int. J. Mol. Sci. 2023, 24, 4812. https://doi.org/10.3390/ijms24054812
Cesaretti A, Calzoni E, Montegiove N, Bianconi T, Alebardi M, La Serra MA, Consiglio G, Fortuna CG, Elisei F, Spalletti A. Lighting-Up the Far-Red Fluorescence of RNA-Selective Dyes by Switching from Ortho to Para Position. International Journal of Molecular Sciences. 2023; 24(5):4812. https://doi.org/10.3390/ijms24054812
Chicago/Turabian StyleCesaretti, Alessio, Eleonora Calzoni, Nicolò Montegiove, Tommaso Bianconi, Martina Alebardi, Maria Antonietta La Serra, Giuseppe Consiglio, Cosimo Gianluca Fortuna, Fausto Elisei, and Anna Spalletti. 2023. "Lighting-Up the Far-Red Fluorescence of RNA-Selective Dyes by Switching from Ortho to Para Position" International Journal of Molecular Sciences 24, no. 5: 4812. https://doi.org/10.3390/ijms24054812
APA StyleCesaretti, A., Calzoni, E., Montegiove, N., Bianconi, T., Alebardi, M., La Serra, M. A., Consiglio, G., Fortuna, C. G., Elisei, F., & Spalletti, A. (2023). Lighting-Up the Far-Red Fluorescence of RNA-Selective Dyes by Switching from Ortho to Para Position. International Journal of Molecular Sciences, 24(5), 4812. https://doi.org/10.3390/ijms24054812









