Participants in the Trans-Antarctic Winter Traverse Expedition Showed Increased Bacterial Load and Diversity in Saliva but Maintained Individual Differences within Stool Microbiota and Across Metabolite Fingerprints
Abstract
1. Introduction
2. Results and Discussion
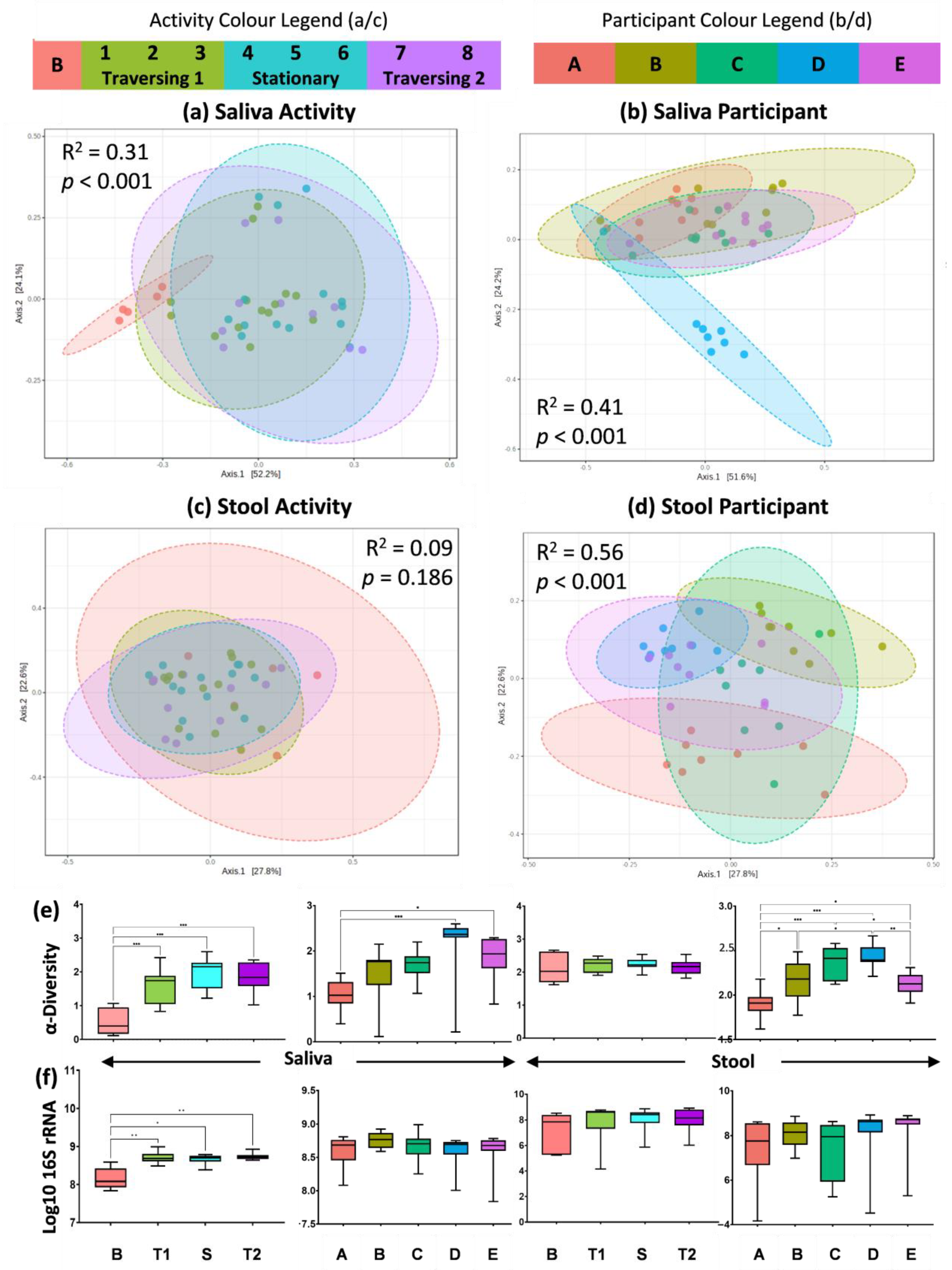
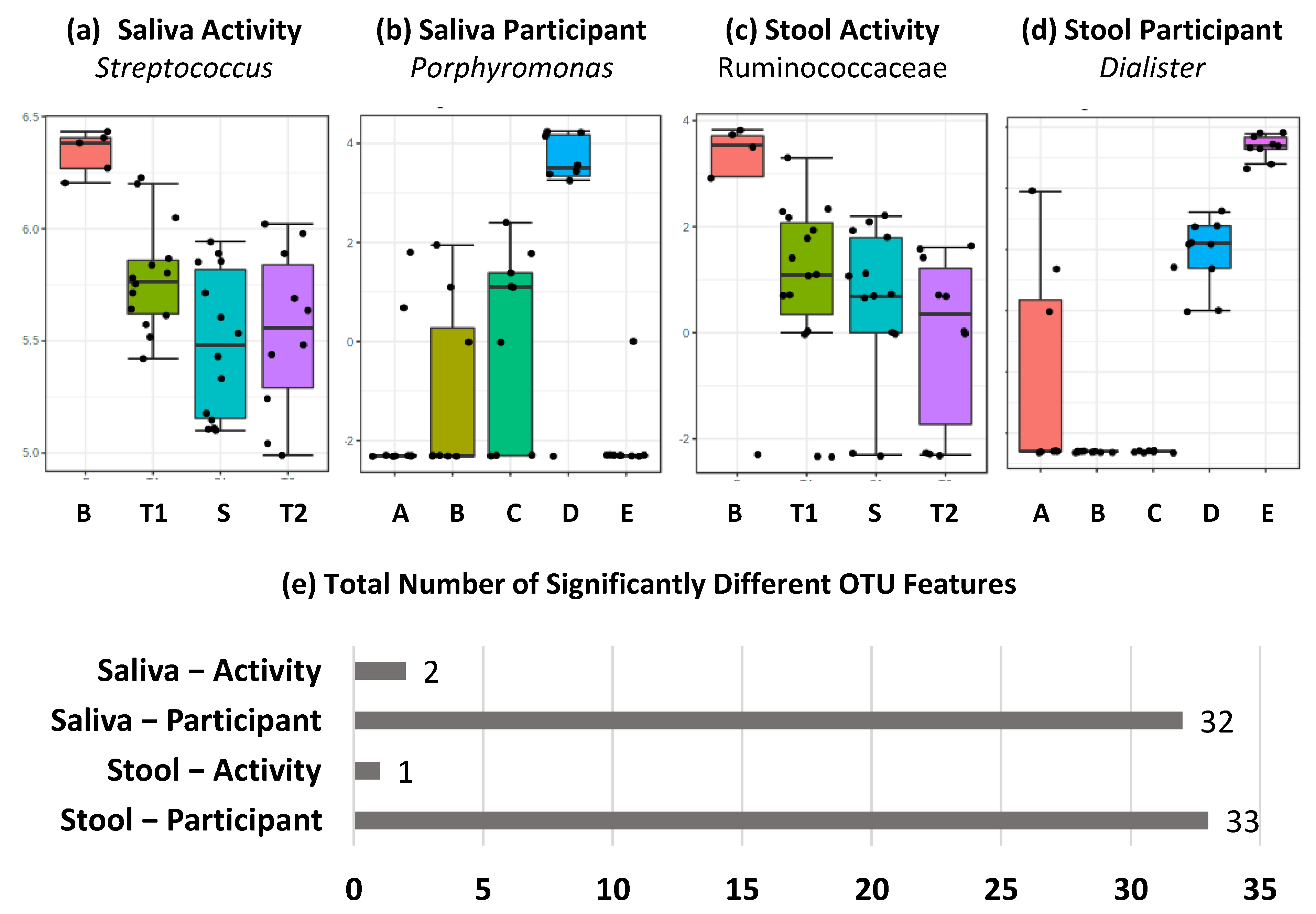
2.1. Amplicon Sequencing of the V3–V4 Region of the 16S rRNA Gene Shows Differential Effect of TAWT Journey on Saliva and Stool Microbiota
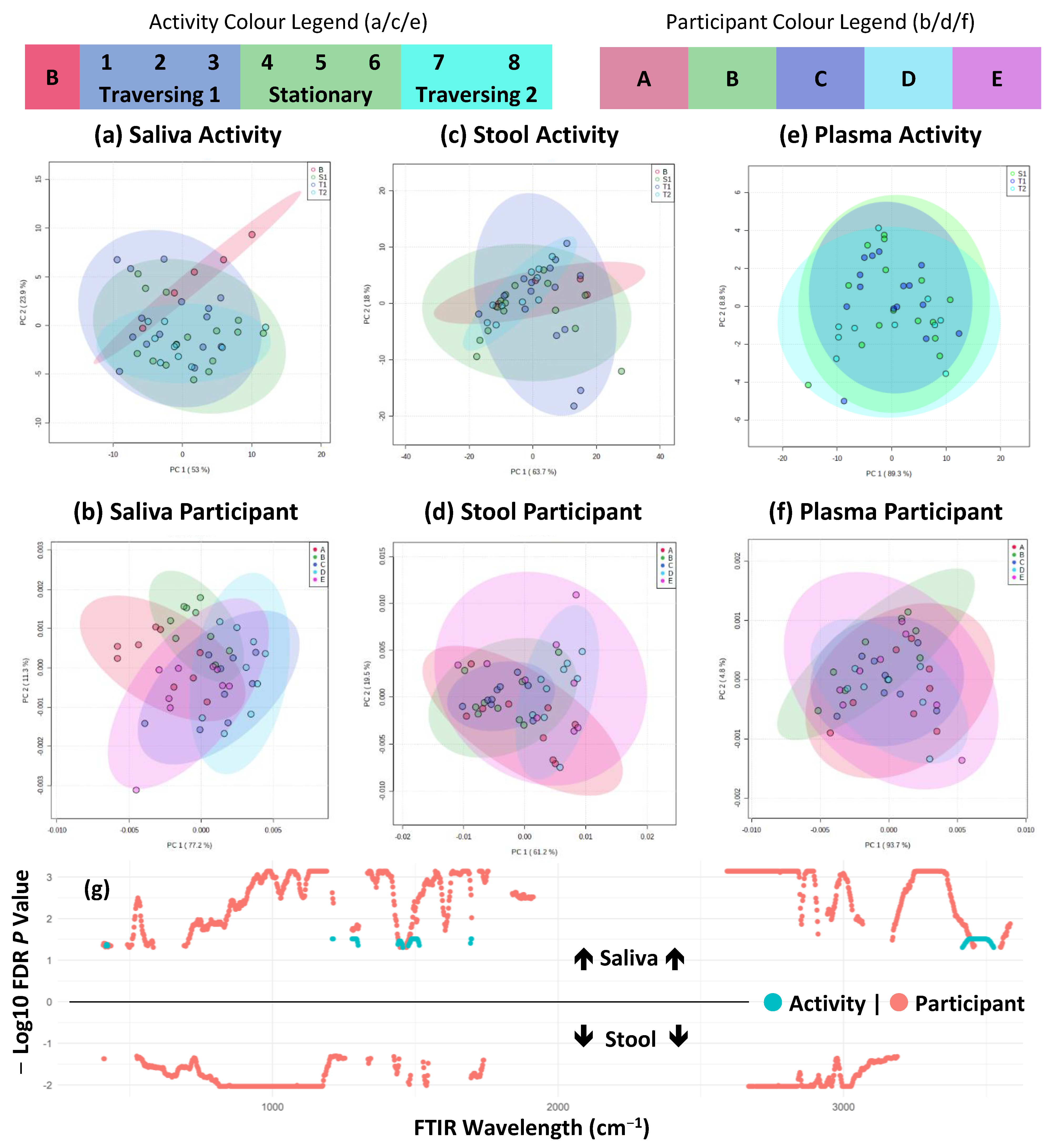
2.2. FTIR Fingerprinting Shows Minimal Impact of Activity during TAWT Expedition
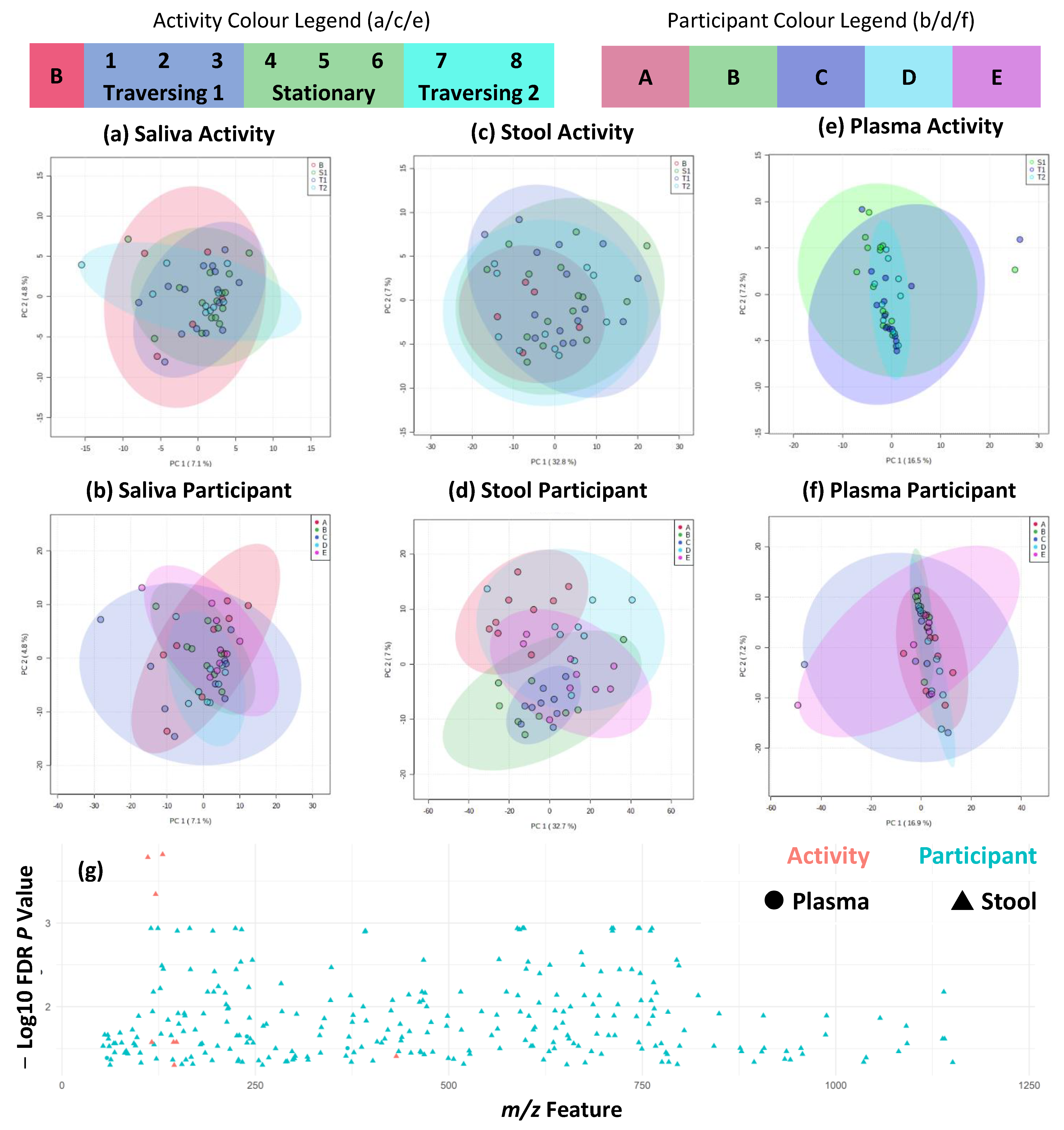
2.3. Metabolite Fingerprinting Shows Maintenance of Individual Differences during TAWT Expedition
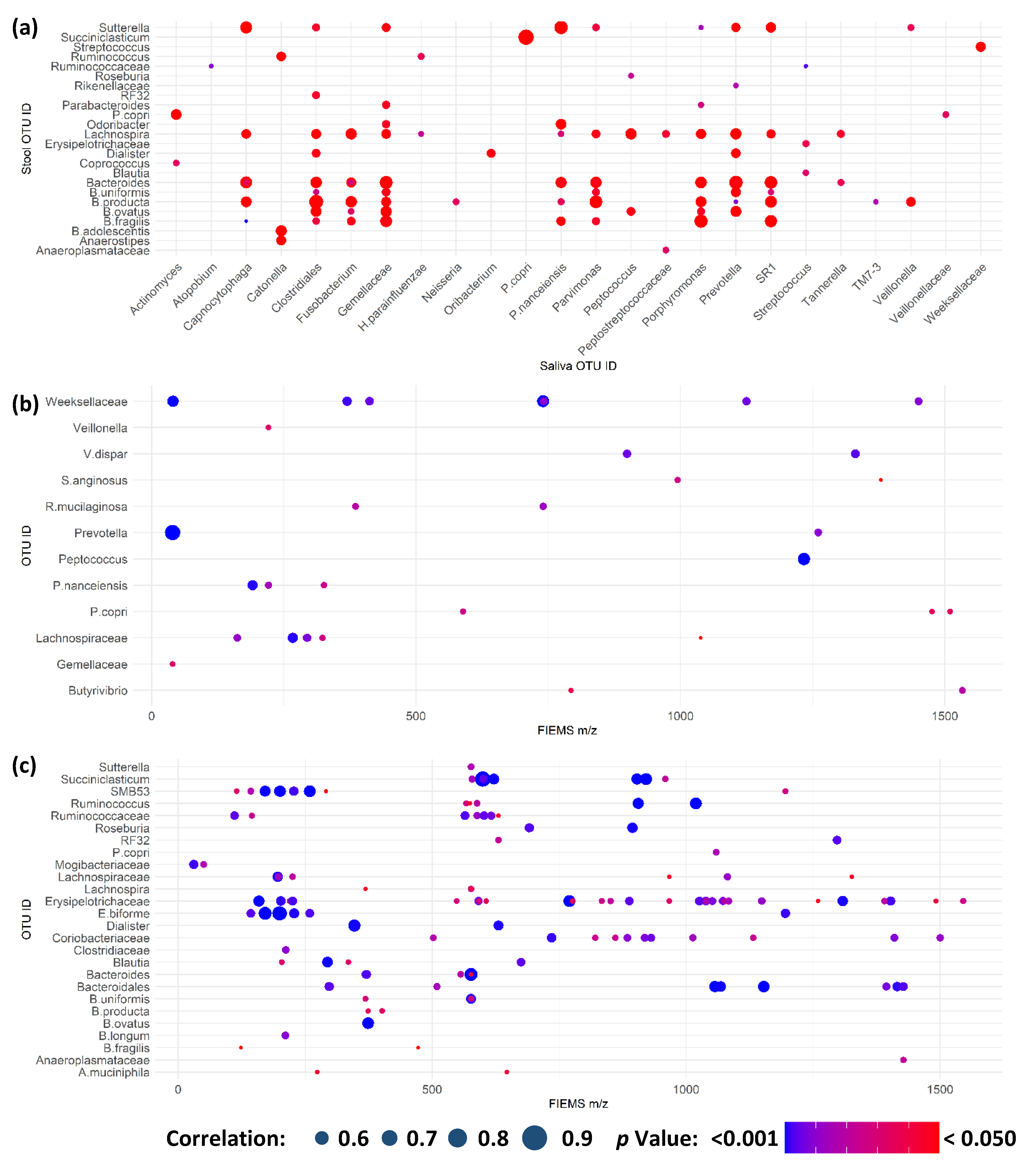
2.4. Multi-Omic Correlation Indicates Potential for Microbiota Modulation in Extreme Stress
3. Materials and Methods
3.1. Ethics Statement and Role of Funding Source
3.2. Participant Recruitment and Sampling
3.3. Sample Processing, DNA Extraction, and pH Measurement
3.4. 16S rRNA Gene Quantitative PCR
3.5. 16S rRNA Gene Amplicon Preparation, Sequencing and Analysis
3.6. Flow Infusion Electrospray Ionisation Mass Spectrometry Metabolite Fingerprinting
3.7. Fourier Transformed Infrared Spectroscopy Fingerprinting
3.8. Statistical and Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kinross, J.M.; Darzi, A.W.; Nicholson, J.K. Gut microbiome-host interactions in health and disease. Genome Med. 2011, 3, 14. [Google Scholar] [CrossRef]
- Li, X.; Kan, E.M.; Lu, J.; Cao, Y.; Wong, R.K.; Keshavarzian, A.; Wilder-Smith, C.H. Combat-training increases intestinal permeability, immune activation and gastrointestinal symptoms in soldiers. Aliment. Pharmacol. Ther. 2013, 37, 799–809. [Google Scholar] [CrossRef]
- Groeneweg, F.L.; Karst, H.; de Kloet, R.; Joels, M. Rapid non-genomic effects of corticosteroids and their role in the central stress response. J. Endocrinol. 2011, 209, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Dinan, T.G.; Cryan, J.F. Regulation of the stress response by the gut microbiota: Implications for psychoneuroendocrinology. Psychoneuroendocrinology 2012, 37, 1369–1378. [Google Scholar] [CrossRef] [PubMed]
- Gareau, M.G.; Jury, J.; MacQueen, G.; Sherman, P.M.; Perdue, M.H. Probiotic treatment of rat pups normalises corticosterone release and ameliorates colonic dysfunction in-duced by maternal separation. Gut 2007, 56, 1522–1528. [Google Scholar] [CrossRef] [PubMed]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Repáraz, J.; Mielcarz, D.W.; Haque, S.B.; Kasper, L.H. Gut, bugs, and brain: Role of commensal bacteria in the control of central nervous system disease. Ann. Neurol. 2010, 69, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kasper, L.H. The role of microbiome in central nervous system disorders. Brain Behav. Immun. 2013, 38, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Messaoudi, M.; Lalonde, R.; Violle, N.; Javelot, H.; Desor, D.; Nejdi, A.; Bisson, J.F.; Rougeot, C.; Pichelin, M.; Cazaubiel, M.; et al. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br. J. Nutr. 2011, 105, 755–764. [Google Scholar] [CrossRef]
- Saei, A.A.; Barzegari, A. The microbiome: The forgotten organ of the astronaut’s body—Probiotics beyond terrestrial limits. Future Microbiol. 2012, 7, 1037–1046. [Google Scholar] [CrossRef]
- Singh, N.K.; Wood, J.M.; Karouia, F.; Venkateswaran, K. Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces. Microbiome 2018, 6, 204. [Google Scholar] [CrossRef]
- Checinska Sielaff, A.; Urbaniak, C.; Mohan, G.B.M.; Stepanov, V.G.; Tran, Q.; Wood, J.M.; Minich, J.; McDonald, D.; Mayer, T.; Knight, R.; et al. Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces. Microbiome 2019, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, W.A.; Fazelinia, H.; Rosenthal, S.B.; Laiakis, E.C.; Kim, M.S.; Meydan, C.; Kidane, Y.; Rathi, K.S.; Smith, S.M.; Stear, B.; et al. Comprehensive multi-omics analysis reveals mitochondrial stress as a central biological hub for space-flight impact. Cell 2020, 183, 1185–1201.e20. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Babykin, M.M.; Beletsky, A.V.; Grigoriev, A.I.; Zinchenko, V.; Kadnikov, V.V.; Kirpichnikov, M.P.; Mazur, A.M.; Nedoluzhko, A.; Novikova, N.D.; et al. Metagenomic Analysis of the Dynamic Changes in the Gut Microbiome of the Participants of the MARS-500 Experiment, Simulating Long Term Space Flight. Acta Nat. 2013, 5, 116–125. [Google Scholar] [CrossRef]
- Douglas, G.; Voorhies, A. Evidence based selection of probiotic strains to promote astronaut health or alleviate symptoms of illness on long duration spaceflight missions. Benef. Microbes 2017, 8, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Cahill, T.; Hardiman, G. Nutritional Challenges and Countermeasures for Space Travel; Wiley Online Library: Hoboken, NJ, USA, 2020. [Google Scholar]
- Beheshti, A.; Chakravarty, K.; Fogle, H.; Fazelinia, H.; da Silveira, W.A.; Boyko, V.; Polo, S.-H.L.; Saravia-Butler, A.M.; Hardiman, G.; Taylor, D.; et al. Multi-omics analysis of multiple missions to space reveal a theme of lipid dysregulation in mouse liver. Sci. Rep. 2019, 9, 19195. [Google Scholar] [CrossRef] [PubMed]
- Morgan, F.; Barker, G.; Briggs, C.; Price, R.; Keys, H.; Zealand, A.N. Environmental Domains of Antarctica Version 2.0 Final Report; Landcare Research New Zealand Ltd.: Lincoln, New Zealand, 2007; pp. 1–89. [Google Scholar]
- Jones, A.W.; Cameron, S.; Thatcher, R.; Beecroft, M.S.; Mur, L.; Davison, G. Effects of bovine colostrum supplementation on upper respiratory illness in active males. Brain Behav. Immun. 2014, 39, 194–203. [Google Scholar] [CrossRef]
- Cameron, S.; Huws, S.; Hegarty, M.J.; Smith, D.; Mur, L.A.J. The human salivary microbiome exhibits temporal stability in bacterial diversity. FEMS Microbiol. Ecol. 2015, 91, fiv091. [Google Scholar] [CrossRef]
- Trochimiak, T.; Hübner-Woźniak, E. Effect of exercise on the level of immunoglobulin A in saliva. Biol. Sport 2012, 29, 255–261. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, N.; Wang, Z.; Li, L.; Zhang, J.; Ma, R.; Nie, H.; Li, Z. Influences of pH and Iron Concentration on the Salivary Microbiome in Individual Humans with and without Caries. Appl. Environ. Microbiol. 2017, 83, e02412-16. [Google Scholar] [CrossRef]
- Bacci, G.; Mengoni, A.; Emiliani, G.; Chiellini, C.; Cipriani, E.G.; Bianconi, G.; Canganella, F.; Fani, R. Defining the resilience of the human salivary microbiota by a 520-day longitudinal study in a confined envi-ronment: The Mars500 mission. Microbiome 2021, 9, 152. [Google Scholar] [CrossRef] [PubMed]
- Turroni, S.; Rampelli, S.; Biagi, E.; Consolandi, C.; Severgnini, M.; Peano, C.; Quercia, S.; Soverini, M.; Carbonero, F.G.; Bianconi, G.; et al. Temporal dynamics of the gut microbiota in people sharing a confined environment, a 520-day ground-based space simulation, MARS500. Microbiome 2017, 5, 1–11. [Google Scholar] [CrossRef]
- Cui, J.; Lian, Y.; Zhao, C.; Du, H.; Han, Y.; Gao, W.; Xiao, H.; Zheng, J. Dietary fibers from fruits and vegetables and their health benefits via modulation of gut microbiota. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1514–1532. [Google Scholar] [CrossRef]
- Vandeputte, D.; Falony, G.; Vieira-Silva, S.; Tito, R.Y.; Joossens, M.; Raes, J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016, 65, 57–62. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Gajjar, K.B.; Theophilou, G.; Martin, F.L.; Martin-Hirsch, P.L. Vibrational spectroscopy of biofluids for disease screening or diagnosis: Translation from the laboratory to a clinical setting. J. Biophotonics 2014, 7, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Nieman, D.C.; Groen, A.; Pugachev, A.; Vacca, G. Detection of Functional Overreaching in Endurance Athletes Using Proteomics. Proteomes 2018, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Tascher, G.; Brioche, T.; Maes, P.; Chopard, A.; O’Gorman, D.; Gauquelin-Koch, G.; Blanc, S.; Bertile, F. Proteome-wide Adaptations of Mouse Skeletal Muscles during a Full Month in Space. J. Proteome Res. 2017, 16, 2623–2638. [Google Scholar] [CrossRef] [PubMed]
- Brzhozovskiy, A.G.; Kononikhin, A.S.; Pastushkova, L.C.; Kashirina, D.N.; Indeykina, M.I.; Popov, I.A.; Custaud, M.-A.; Larina, I.M.; Nikolaev, E.N. The Effects of Spaceflight Factors on the Human Plasma Proteome, Including Both Real Space Missions and Ground-Based Experiments. Int. J. Mol. Sci. 2019, 20, 3194. [Google Scholar] [CrossRef]
- Gao, X.; Pujos-Guillot, E.; Martin, J.-F.; Galan, P.; Juste, C.; Jia, W.; Sebedio, J.-L. Metabolite analysis of human fecal water by gas chromatography/mass spectrometry with ethyl chloroformate derivatization. Anal. Biochem. 2009, 393, 163–175. [Google Scholar] [CrossRef]
- Sternberg, D.E.; Heninger, G.R.; Both, R.H. Plasma homovanillic acid as an index of brain dopamine metabolism: Enhancement with debrisoquin. Life Sci. 1983, 32, 2447–2452. [Google Scholar] [CrossRef]
- Barrios, C.; Spector, T.D.; Menni, C. Blood, urine and faecal metabolite profiles in the study of adult renal disease. Arch. Biochem. Biophys. 2016, 589, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Dall’Asta, M.; Calani, L.; Tedeschi, M.; Jechiu, L.; Brighenti, F.; Del Rio, D. Identification of microbial metabolites derived from in vitro fecal fermentation of different polyphenolic food sources. Nutrition 2012, 28, 197–203. [Google Scholar] [CrossRef]
- Heaney, L.M.; Deighton, K.; Suzuki, T. Non-targeted metabolomics in sport and exercise science. J. Sports Sci. 2017, 37, 959–967. [Google Scholar] [CrossRef] [PubMed]
- San-Millán, I.; Stefanoni, D.; Martinez, J.L.; Hansen, K.C.; D’Alessandro, A.; Nemkov, T. Metabolomics of Endurance Capacity in World Tour Professional Cyclists. Front. Physiol. 2020, 11, 578. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Chen, C.Y.; Hayes, R.B. Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control 2012, 23, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I.; Yamazaki, K. Can oral bacteria affect the microbiome of the gut? J. Oral Microbiol. 2019, 11, 1586422. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, S.; Takeshita, T.; Asakawa, M.; Shibata, Y.; Takeuchi, K.; Yamanaka, W.; Yamashita, Y. Relative abundance of total subgingival plaque-specific bacteria in salivary microbiota reflects the overall periodontal condition in patients with periodontitis. PLoS ONE 2017, 12, e0174782. [Google Scholar] [CrossRef]
- Weng, Y.J.; Gan, H.Y.; Li, X.; Huang, Y.; Li, Z.C.; Deng, H.M.; Chen, S.Z.; Zhou, Y.; Wang, L.S.; Han, Y.P.; et al. Correlation of diet, microbiota and metabolite networks in inflammatory bowel disease. J. Dig. Dis. 2019, 20, 447–459. [Google Scholar] [CrossRef]
- Tuohy, K.M.; Probert, H.M.; Smejkal, C.W.; Gibson, G.R. Using probiotics and prebiotics to improve gut health. Drug Discov. Today 2003, 8, 692–700. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Caporaso, J.G. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ 2018. [Google Scholar] [CrossRef]
- Shenker, N.S.; Perdones-Montero, A.; Burke, A.; Stickland, S.; McDonald, J.A.; Alexander-Hardiman, K.; Flanagan, J.; Takats, Z.; Cameron, S.J. Metabolomic and Metataxonomic Fingerprinting of Human Milk Suggests Compositional Stability over a Natural Term of Breastfeeding to 24 Months. Nutrients 2020, 12, 3450. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Cameron, S.J.; Lewis, K.E.; Beckmann, M.; Allison, G.G.; Ghosal, R.; Lewis, P.D.; Mur, L.A. The metabolomic detection of lung cancer biomarkers in sputum. Lung Cancer 2016, 94, 88–95. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0: The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cameron, S.J.S.; Edwards, A.; Lambert, R.J.; Stroud, M.; Mur, L.A.J. Participants in the Trans-Antarctic Winter Traverse Expedition Showed Increased Bacterial Load and Diversity in Saliva but Maintained Individual Differences within Stool Microbiota and Across Metabolite Fingerprints. Int. J. Mol. Sci. 2023, 24, 4850. https://doi.org/10.3390/ijms24054850
Cameron SJS, Edwards A, Lambert RJ, Stroud M, Mur LAJ. Participants in the Trans-Antarctic Winter Traverse Expedition Showed Increased Bacterial Load and Diversity in Saliva but Maintained Individual Differences within Stool Microbiota and Across Metabolite Fingerprints. International Journal of Molecular Sciences. 2023; 24(5):4850. https://doi.org/10.3390/ijms24054850
Chicago/Turabian StyleCameron, Simon J. S., Arwyn Edwards, Robert J. Lambert, Mike Stroud, and Luis A. J. Mur. 2023. "Participants in the Trans-Antarctic Winter Traverse Expedition Showed Increased Bacterial Load and Diversity in Saliva but Maintained Individual Differences within Stool Microbiota and Across Metabolite Fingerprints" International Journal of Molecular Sciences 24, no. 5: 4850. https://doi.org/10.3390/ijms24054850
APA StyleCameron, S. J. S., Edwards, A., Lambert, R. J., Stroud, M., & Mur, L. A. J. (2023). Participants in the Trans-Antarctic Winter Traverse Expedition Showed Increased Bacterial Load and Diversity in Saliva but Maintained Individual Differences within Stool Microbiota and Across Metabolite Fingerprints. International Journal of Molecular Sciences, 24(5), 4850. https://doi.org/10.3390/ijms24054850






