Characterization and Vaccine Potential of Outer Membrane Vesicles from Photobacterium damselae subsp. piscicida
Abstract
:1. Introduction
2. Results
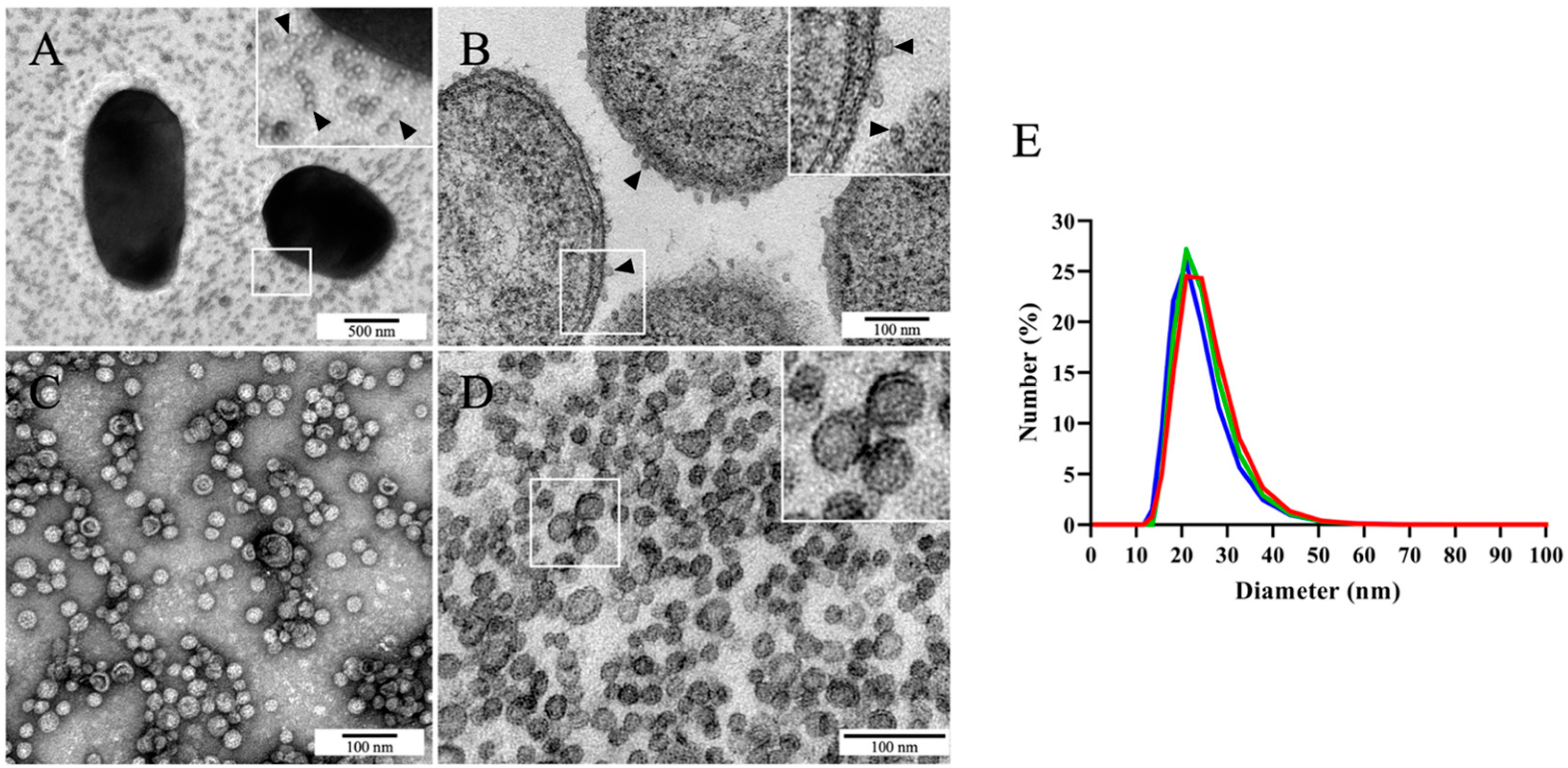
2.1. Photobacterium damselae subsp. piscicida MT1415 Strain Secretes Large Numbers of OMVs In Vitro
2.2. Identification of the Most Abundant Proteins in Phdp MT1415 OMVs
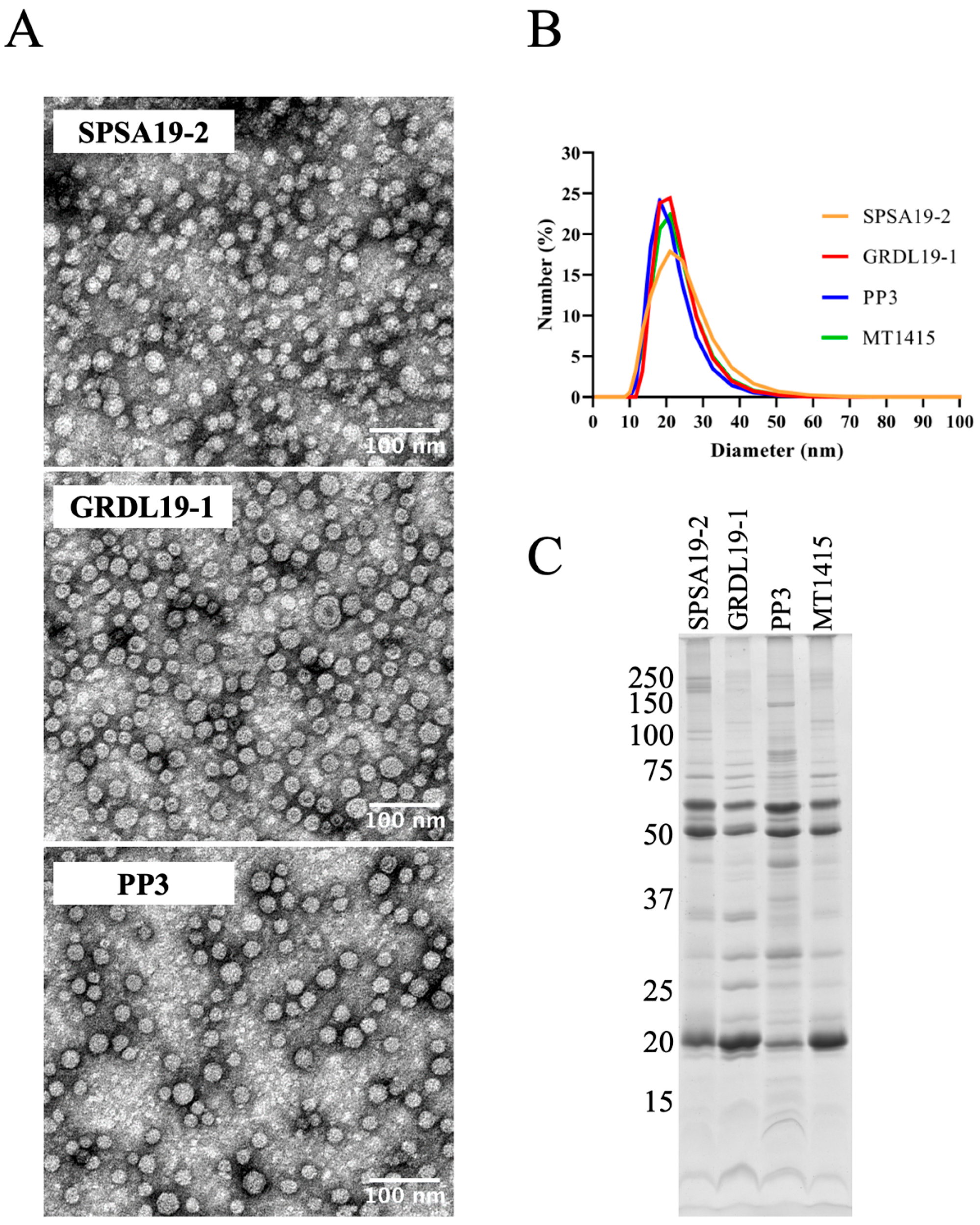
2.3. Large Amounts of OMVs Are Also Secreted In Vitro by the Phdp Strain PP3 and by Two Field Isolates of Phdp
2.4. Phdp Releases OMVs during Infection In Vivo
2.5. OMVs Protect Phdp from the Bactericidal Activity of Fish Antimicrobial Peptides
2.6. Vaccination of Sea Bass with OMVs Confers Partial Protection against a Phdp Challenge
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Generation of a pPHDP10-Negative MT1415 Strain
4.3. Isolation of OMVs
4.4. Density Gradient Separation
4.5. Transmission Electron Microscopy
4.6. Dynamic Light Scattering
4.7. SDS-PAGE and Western Blotting
4.8. Protein Quantification
4.9. Proteomic Analysis
4.10. Protease Protection Assay
4.11. Determination of OMVs Concentration
4.12. AMPs Protection Assay
4.13. Fish
4.14. Vaccination Trials
4.15. Experimental Infections
4.16. Assessment of Anti-Phdp Antibody Levels
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andreoni, F.; Magnani, M. Photobacteriosis: Prevention and diagnosis. J. Immunol Res. 2014, 2014, 793817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snieszko, S.F.; Bullock, G.L.; Hollis, E.; Boone, J.G. Pasteurella Sp. from an Epizootic of White Perch (Roccus Americanus) in Chesapeake Bay Tidewater Areas. J. Bacteriol. 1964, 88, 1814–1815. [Google Scholar] [CrossRef] [Green Version]
- Colloca, F.; Cerasi, S. Sparus aurata. Cultured Aquatic Species Information Programme. Available online: https://www.fao.org/fishery/en/culturedspecies/sparus_aurata/en (accessed on 30 December 2022).
- Bagni, M. Dicentrarchus labrax. Cultured Aquatic Species Information Programme. Available online: https://www.fao.org/fishery/en/culturedspecies/dicentrarchus_labrax/en (accessed on 30 December 2022).
- Colen, R.; Ramalho, A.; Rocha, F.; Dinis, M.T. Solea solea. Cultured Aquatic Species Information Programme. Available online: https://www.fao.org/fishery/en/culturedspecies/solea_spp/en (accessed on 30 December 2022).
- Dhirendra, P.T. Seriola quiqueradiata. Cultured Aquatic Species Information Programme. Available online: https://www.fao.org/fishery/en/culturedspecies/seriola_quinqueradiata/en (accessed on 30 December 2022).
- Kaiser, J.B.; Holt, J.G. Rachycentron canadum. Cultured Aquatic Species Information Programme. Available online: https://www.fao.org/fishery/en/culturedspecies/rachycentron_canadum/en (accessed on 30 December 2022).
- FAO. FAO Yearbook. Fishery and Aquaculture Statistics 2019/FAO Annuaire. Statistiques des Pêches et de l’Aquaculture 2019/FAO Anuario. Estadísticas de Pesca y Acuicultura 2019; FAO: Rome, Italy, 2021; 110p. [Google Scholar]
- Magariños, B.; Toranzo, A.E.; Romalde, J.L. Phenotypic and pathobiological characteristics of Pasteurella piscicida. Annu. Rev. Fish. Dis. 1996, 6, 41–64. [Google Scholar] [CrossRef]
- Hawke, J.P.; Plakas, S.M.; Minton, R.V.; McPhearson, R.M.; Snider, T.G.; Guarino, A.M. Fish pasteurellosis of cultured striped bass (Morone saxatilis) in coastal Alabama. Aquaculture 1987, 65, 193–204. [Google Scholar] [CrossRef]
- Do Vale, A.; Costa-Ramos, C.; Silva, A.; Silva, D.S.; Gartner, F.; dos Santos, N.M.; Silva, M.T. Systemic macrophage and neutrophil destruction by secondary necrosis induced by a bacterial exotoxin in a Gram-negative septicaemia. Cell Microbiol. 2007, 9, 988–1003. [Google Scholar] [CrossRef]
- Tung, M.-C.; Tsai, S.-S.; Ho, L.-F.; Huang, S.T.; Chen, S.C. An acute septicemic infection of Pasteurella organism in pond-cultured Formosa snake-head fish (Channa maculata Lacepeda) in Taiwan. Fish. Pathol. 1985, 20, 143–148. [Google Scholar] [CrossRef] [Green Version]
- Noya, M.; Magarinos, B.; Toranzo, A.E.; Lamas, J. Sequential pathology of experimental pasteurellosis in gilthead seabream Sparus aurata. A light- and electron-microscopic study. Dis. Aquat. Org. 1995, 21, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Steele, J.C.; Meng, X.-Z. Usage, residue, and human health risk of antibiotics in Chinese aquaculture: A review. Environ. Pollut. 2017, 223, 161–169. [Google Scholar] [CrossRef]
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Pepi, M.; Focardi, S. Antibiotic-Resistant Bacteria in Aquaculture and Climate Change: A Challenge for Health in the Mediterranean Area. Int. J. Environ. Res. Public Health 2021, 18, 5723. [Google Scholar] [CrossRef]
- Ma, J.; Bruce, T.J.; Jones, E.M.; Cain, K.D. A Review of Fish Vaccine Development Strategies: Conventional Methods and Modern Biotechnological Approaches. Microorganisms 2019, 7, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lattos, A.; Giantsis, I.A.; Tsavea, E.; Kolygas, M.; Athanassopoulou, F.; Bitchava, K. Virulence Genes and In Vitro Antibiotic Profile of Photobacterium damselae Strains, Isolated from Fish Reared in Greek Aquaculture Facilities. Animals 2022, 12, 3133. [Google Scholar] [CrossRef]
- Baseggio, L.; Silayeva, O.; Buller, N.; Landos, M.; Englestädter, J.; Barnes, A.C. Complete, closed and curated genome sequences of Photobacterium damselae subsp. piscicida isolates from Australia indicate mobilome-driven localized evolution and novel pathogenicity determinants. Microb. Genom. 2021, 7, 000562. [Google Scholar] [CrossRef]
- Pham, T.H.; Cheng, T.-C.; Wang, P.-C.; Chen, S.-C. Genotypic diversity, and molecular and pathogenic characterization of Photobacterium damselae subsp. piscicida isolated from different fish species in Taiwan. J. Fish. Dis. 2020, 43, 757–774. [Google Scholar] [CrossRef]
- Magarinos, B.; Santos, Y.; Romalde, J.L.; Rivas, C.; Barja, J.L.; Toranzo, A.E. Pathogenic activities of live cells and extracellular products of the fish pathogen Pasteurella piscicida. J. Gen. Microbiol. 1992, 138, 2491–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, D.S.; Pereira, L.M.G.; Moreira, A.R.; Ferreira-da-Silva, F.; Brito, R.M.; Faria, T.Q.; Zornetta, I.; Montecucco, C.; Oliveira, P.; Azevedo, J.E.; et al. The Apoptogenic Toxin AIP56 Is a Metalloprotease A-B Toxin that Cleaves NF-κb P65. PLoS Pathog. 2013, 9, e1003128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Do Vale, A.; Silva, M.T.; dos Santos, N.M.S.; Nascimento, D.S.; Reis-Rodrigues, P.; Costa-Ramos, C.; Ellis, A.E.; Azevedo, J.E. AIP56, a novel plasmid-encoded virulence factor of Photobacterium damselae subsp. piscicida with apoptogenic activity against sea bass macrophages and neutrophils. Mol. Microbiol. 2005, 58, 1025–1038. [Google Scholar] [CrossRef]
- Lisboa, J.; Pereira, C.; Rifflet, A.; Ayala, J.; Terceti, M.S.; Barca, A.V.; Rodrigues, I.; Pereira, P.J.B.; Osorio, C.R.; García-del Portillo, F.; et al. A Secreted NlpC/P60 Endopeptidase from Photobacterium damselae subsp. piscicida Cleaves the Peptidoglycan of Potentially Competing Bacteria. mSphere 2021, 6, e00736-20. [Google Scholar] [CrossRef]
- Do Vale, A.; Pereira, C.; Osorio, C.R.; dos Santos, N.M.S. The Apoptogenic Toxin AIP56 Is Secreted by the Type II Secretion System of Photobacterium damselae subsp. piscicida. Toxins 2017, 9, 368. [Google Scholar] [CrossRef] [Green Version]
- Abushattal, S.; Vences, A.; Osorio, C.R. A virulence gene typing scheme for Photobacterium damselae subsp. piscicida, the causative agent of fish photobacteriosis, reveals a high prevalence of plasmid-encoded virulence factors and of type III secretion system genes. Aquaculture 2020, 521, 735057. [Google Scholar] [CrossRef]
- Abushattal, S.; Vences, A.; Dos Santos, N.M.S.; do Vale, A.; Osorio, C.R. Draft Genome Sequences of Photobacterium damselae subsp. piscicida SNW-8.1 and PP3, Two Fish-Isolated Strains Containing a Type III Secretion System. Microbiol. Resour. Announc. 2019, 8, e00426-19. [Google Scholar] [CrossRef] [Green Version]
- Abushattal, S.; Vences, A.; Osorio, C.R. A Highly Unstable and Elusive Plasmid That Encodes the Type III Secretion System Is Necessary for Full Virulence in the Marine Fish Pathogen Photobacterium damselae subsp. piscicida. Int. J. Mol. Sci. 2022, 23, 4729. [Google Scholar] [CrossRef] [PubMed]
- Sartorio, M.G.; Pardue, E.J.; Feldman, M.F.; Haurat, M.F. Bacterial Outer Membrane Vesicles: From Discovery to Applications. Annu. Rev. Microbiol. 2021, 75, 609–630. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiku, V.; Tan, M.-W. Host immunity and cellular responses to bacterial outer membrane vesicles. Trends Immunol. 2021, 42, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Prior, J.T.; Davitt, C.; Kurtz, J.; Gellings, P.; McLachlan, J.B.; Morici, L.A. Bacterial-Derived Outer Membrane Vesicles are Potent Adjuvants that Drive Humoral and Cellular Immune Responses. Pharmaceutics 2021, 13, 131. [Google Scholar] [CrossRef]
- Anand, D.; Chaudhuri, A. Bacterial outer membrane vesicles: New insights and applications. Mol. Membr. Biol. 2016, 33, 125–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micoli, F.; MacLennan, C.A. Outer membrane vesicle vaccines. Semin. Immunol. 2020, 50, 101433. [Google Scholar] [CrossRef] [PubMed]
- Feavers, I.M.; Maiden, M.C.J. Recent Progress in the Prevention of Serogroup B Meningococcal Disease. Clin. Vaccine Immunol. CVI 2017, 24, e00566-16. [Google Scholar] [CrossRef] [Green Version]
- Tan, K.; Li, R.; Huang, X.; Liu, Q. Outer Membrane Vesicles: Current Status and Future Direction of These Novel Vaccine Adjuvants. Front. Microbiol. 2018, 9, 783. [Google Scholar] [CrossRef] [Green Version]
- Amanda Izeli, P.; Elizabeth De, G. Outer Membrane Vesicles: A Challenging Yet Promising Platform for COVID-19 Vaccines. In COVID-19 Vaccines—Current State and Perspectives; Ibrokhim, Y.A., Ed.; IntechOpen: Rijeka, Croatia, 2022; p. Ch. 7. [Google Scholar]
- Irene, C.; Fantappiè, L.; Caproni, E.; Zerbini, F.; Anesi, A.; Tomasi, M.; Zanella, I.; Stupia, S.; Prete, S.; Valensin, S.; et al. Bacterial outer membrane vesicles engineered with lipidated antigens as a platform for Staphylococcus aureus vaccine. Proc. Natl. Acad. Sci. USA 2019, 116, 21780–21788. [Google Scholar] [CrossRef] [Green Version]
- Gerritzen, M.J.H.; Martens, D.E.; Wijffels, R.H.; van der Pol, L.; Stork, M. Bioengineering bacterial outer membrane vesicles as vaccine platform. Biotechnol. Adv. 2017, 35, 565–574. [Google Scholar] [CrossRef]
- Park, S.B.; Jang, H.B.; Nho, S.W.; Cha, I.S.; Hikima, J.; Ohtani, M.; Aoki, T.; Jung, T.S. Outer membrane vesicles as a candidate vaccine against edwardsiellosis. PLoS ONE 2011, 6, e17629. [Google Scholar] [CrossRef]
- Hacker, C.; Asadi, J.; Pliotas, C.; Ferguson, S.; Sherry, L.; Marius, P.; Tello, J.; Jackson, D.; Naismith, J.; Lucocq, J.M. Nanoparticle suspensions enclosed in methylcellulose: A new approach for quantifying nanoparticles in transmission electron microscopy. Sci. Rep. 2016, 6, 25275. [Google Scholar] [CrossRef] [Green Version]
- Toranzo, A.E.; Barreiro, S.; Casal, J.F.; Figueras, A.; Magarinos, B.; Barja, J.L. Pasteurellosis in cultured gilthead seabream (Sparus aurata): First report in Spain. Aquaculture 1991, 99, 1–15. [Google Scholar] [CrossRef]
- Manning, A.J.; Kuehn, M.J. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011, 11, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Robles, T.; Dillard, R.S.; Cairns, L.S.; Silva-Valenzuela, C.A.; Housman, M.; Ali, A.; Wright, E.R.; Camilli, A. Vibrio cholerae Outer Membrane Vesicles Inhibit Bacteriophage Infection. J. Bacteriol. 2018, 200, e00792-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balhuizen, M.D.; Dijk, A.v.; Jansen, J.W.A.; Lest, C.H.A.v.d.; Veldhuizen, E.J.A.; Haagsman, H.P.; Dunman, P. Outer Membrane Vesicles Protect Gram-Negative Bacteria against Host Defense Peptides. mSphere 2021, 6, e00523-21. [Google Scholar] [CrossRef]
- Kulkarni, H.M.; Nagaraj, R.; Jagannadham, M.V. Protective role of E. coli outer membrane vesicles against antibiotics. Microbiol. Res. 2015, 181, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Duperthuy, M.; Sjöström, A.E.; Sabharwal, D.; Damghani, F.; Uhlin, B.E.; Wai, S.N. Role of the Vibrio cholerae Matrix Protein Bap1 in Cross-Resistance to Antimicrobial Peptides. PLoS Pathog. 2013, 9, e10036-20. [Google Scholar] [CrossRef] [Green Version]
- Valero, Y.; Saraiva-Fraga, M.; Costas, B.; Guardiola, F.A. Antimicrobial peptides from fish: Beyond the fight against pathogens. Rev. Aquac. 2020, 12, 224–253. [Google Scholar] [CrossRef]
- Shabir, U.; Ali, S.; Magray, A.R.; Ganai, B.A.; Firdous, P.; Hassan, T.; Nazir, R. Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: A review. Microb. Pathog. 2018, 114, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Barroso, C.; Carvalho, P.; Carvalho, C.; Santarém, N.; Gonçalves, J.F.M.; Rodrigues, P.N.S.; Neves, J.V. The Diverse Piscidin Repertoire of the European Sea Bass (Dicentrarchus labrax): Molecular Characterization and Antimicrobial Activities. Int. J. Mol. Sci. 2020, 21, 4613. [Google Scholar] [CrossRef] [PubMed]
- Freitas, I.L.; Teixeira, A.; Loureiro, I.; Lisboa, J.; Saraiva, A.; dos Santos, N.M.S.; do Vale, A. Susceptibility of Sea Bream (Sparus aurata) to AIP56, an AB-Type Toxin Secreted by Photobacterium damselae subsp. piscicida. Toxins 2022, 14, 119. [Google Scholar] [CrossRef]
- Rivas, A.J.; Lemos, M.L.; Osorio, C.R. Photobacterium damselae subsp. damselae, a bacterium pathogenic for marine animals and humans. Front. Microbiol. 2013, 4, 283. [Google Scholar] [CrossRef] [Green Version]
- Confer, A.W.; Ayalew, S. The OmpA family of proteins: Roles in bacterial pathogenesis and immunity. Vet. Microbiol. 2013, 163, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Prasadarao, N.V. Outer membrane protein A and OprF: Versatile roles in Gram-negative bacterial infections. FEBS J. 2012, 279, 919–931. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Basak, A.J.; Nair, A.V.; Duraivelan, K.; Samanta, D. Immunoglobulin-fold containing bacterial adhesins: Molecular and structural perspectives in host tissue colonization and infection. FEMS Microbiol. Lett. 2020, 368, fnaa220. [Google Scholar] [CrossRef]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miccoli, A.; Manni, M.; Picchietti, S.; Scapigliati, G. State-of-the-Art Vaccine Research for Aquaculture Use: The Case of Three Economically Relevant Fish Species. Vaccines 2021, 9, 140. [Google Scholar] [CrossRef]
- Villumsen, K.R.; Koppang, E.O.; Christensen, D.; Bojesen, A.M. Alternatives to mineral oil adjuvants in vaccines against Aeromonas salmonicida subsp. salmonicida in rainbow trout offer reductions in adverse effects. Sci. Rep. 2017, 7, 5930. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Tang, L.; Li, S.; Li, G.; Mo, Z. The efficacy and side-effects of oil-based adjuvants emulsified Vibrio anguillarum bivalent inactivated vaccine in turbot (Scophthalmus maximus) under production mode. Aquaculture 2020, 524, 735259. [Google Scholar] [CrossRef]
- Tziouvas, H.; Varvarigos, P. Intensity scale of side effects in European sea bass (Dicentrarchus labrax) post intraperitoneal injection with commercial oil-adjuvanted vaccines. Bull. Eur. Assoc. Fish Pathol. 2021, 41, 103–110. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Morton, R.E.; Evans, T.A. Modification of the bicinchoninic acid protein assay to eliminate lipid interference in determining lipoprotein protein content. Anal. Biochem. 1992, 204, 332–334. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.; Almeida, A.; Ferreira, J.A.; Silva, L.; Santos-Sousa, H.; Pinto-de-Sousa, J.; Santos, L.L.; Amado, F.; Schwientek, T.; Levery, S.B.; et al. Glycoproteomic Analysis of Serum from Patients with Gastric Precancerous Lesions. J. Proteome Res. 2013, 12, 1454–1466. [Google Scholar] [CrossRef]
- Ferreirinha, P.; Correia, A.; Teixeira-Coelho, M.; Osório, H.; Teixeira, L.; Rocha, A.; Vilanova, M. Mucosal immunization confers long-term protection against intragastrically established Neospora caninum infection. Vaccine 2016, 34, 6250–6258. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, G.; McDowall, A.; Back, R.; Dubochet, J. On the preparation of cryosections for immunocytochemistry. J. Ultrastruct. Res. 1984, 89, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, N.M.S.; Taverne, N.; Taverne-Thiele, A.J.; de Sousa, M.; Rombout, J.H.W.M. Characterisation of monoclonal antibodies specific for sea bass (Dicentrarchus labrax L.) IgM indicates the existence of B cell subpopulations. Fish. Shellfish Immunol. 1997, 7, 175–191. [Google Scholar] [CrossRef]





| Band | NCBI Accession Number | Relative Abundance 1 | MW (kDa) 2 | Signal Peptide 3 | Description | Present/Absent in subsp. damselae 4 |
|---|---|---|---|---|---|---|
| A | WP_012954632.1 | 93.0% | 56.2 | Sec/SPI | Apoptosis-inducing protein (AIP56) | Absent |
| B | WP_044175038.1 | 79.7% | 82.7 | Sec/SPII | Lipase | Present |
| C | WP_094461548.1 | 81.0% | 83.1 | Sec/SPII | Ig-like domain containing protein | Present |
| D | WP_044175643.1 | na | 70.8 | Sec/SPI | DUF3466 family protein | Present |
| E | WP_044179093.1 | na | 64.4 | Sec/SPI | TonB-dependent receptor | Present |
| F | WP_012954632.1 | na | 56.2 | Sec/SPI | Apoptosis-inducing protein (AIP56) | Absent |
| G | WP_044178512.1 WP_044176547.1 | 69.6% 28.8% | 55.1 60.2 | Sec/SPI Sec/SPI | NlpC/P60 endopeptidase (PnpA) Insecticidal delta-endotoxin Cry8Ea1 family protein | Present Present |
| H | WP_044174887.1 | 92.0% | 32.7 | Sec/SPI | OmpA family protein | Present |
| I | WP_044174816.1 | 99.8% | 33.7 | Sec/SPI | OmpA family protein | Present |
| J | WP_044176214.1 WP_044174887.1 | 33.3% 33.2% | 42.3 32.7 | Sec/SPI Sec/SPI | Omp transport protein OmpA family protein | Present Present |
| K | WP_044179308.1 WP_094461570.1 | 61.4% 35.4% | 20.5 19.6 | Sec/SPI Sec/SPI | TIGR04219 family outer membrane beta-barrel protein Outer membrane beta-barrel protein | Present Present |
| L | WP_094461570.1 | 98.0% | 19.6 | Sec/SPI | Outer membrane beta-barrel protein | Present |
| M | WP_044178211.1 | 87.8% | 18.8 | Sec/SPI | Outer membrane beta-barrel protein | Present |
| N | WP_081282903.1 | 64.3% | 13.6 | Sec/SPII | Glycine zipper 2TM domain-containing protein | Present |
| O | WP_044177989.1 | na | 7.0 | Sec/SPII | Lpp/OprI family alanine-zipper lipoprotein | Present |
| Strain | Species of Isolation | Geographical Origin | Year |
|---|---|---|---|
| MT1415 | Dicentrarchus labrax | Italy | Unknown |
| PP3 | Seriola quinqueradiata | Japan | Unknown |
| GRDL19-1 | Dicentrarchus labrax | Greece | 2019 |
| SPSA19-2 | Sparus aurata | Spain | 2019 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teixeira, A.; Loureiro, I.; Lisboa, J.; Oliveira, P.N.; Azevedo, J.E.; dos Santos, N.M.S.; do Vale, A. Characterization and Vaccine Potential of Outer Membrane Vesicles from Photobacterium damselae subsp. piscicida. Int. J. Mol. Sci. 2023, 24, 5138. https://doi.org/10.3390/ijms24065138
Teixeira A, Loureiro I, Lisboa J, Oliveira PN, Azevedo JE, dos Santos NMS, do Vale A. Characterization and Vaccine Potential of Outer Membrane Vesicles from Photobacterium damselae subsp. piscicida. International Journal of Molecular Sciences. 2023; 24(6):5138. https://doi.org/10.3390/ijms24065138
Chicago/Turabian StyleTeixeira, Alexandra, Inês Loureiro, Johnny Lisboa, Pedro N. Oliveira, Jorge E. Azevedo, Nuno M. S. dos Santos, and Ana do Vale. 2023. "Characterization and Vaccine Potential of Outer Membrane Vesicles from Photobacterium damselae subsp. piscicida" International Journal of Molecular Sciences 24, no. 6: 5138. https://doi.org/10.3390/ijms24065138








