Inter- and Intramolecular RNA–RNA Interactions Modulate the Regulation of Translation Mediated by the 3′ UTR in West Nile Virus
Abstract
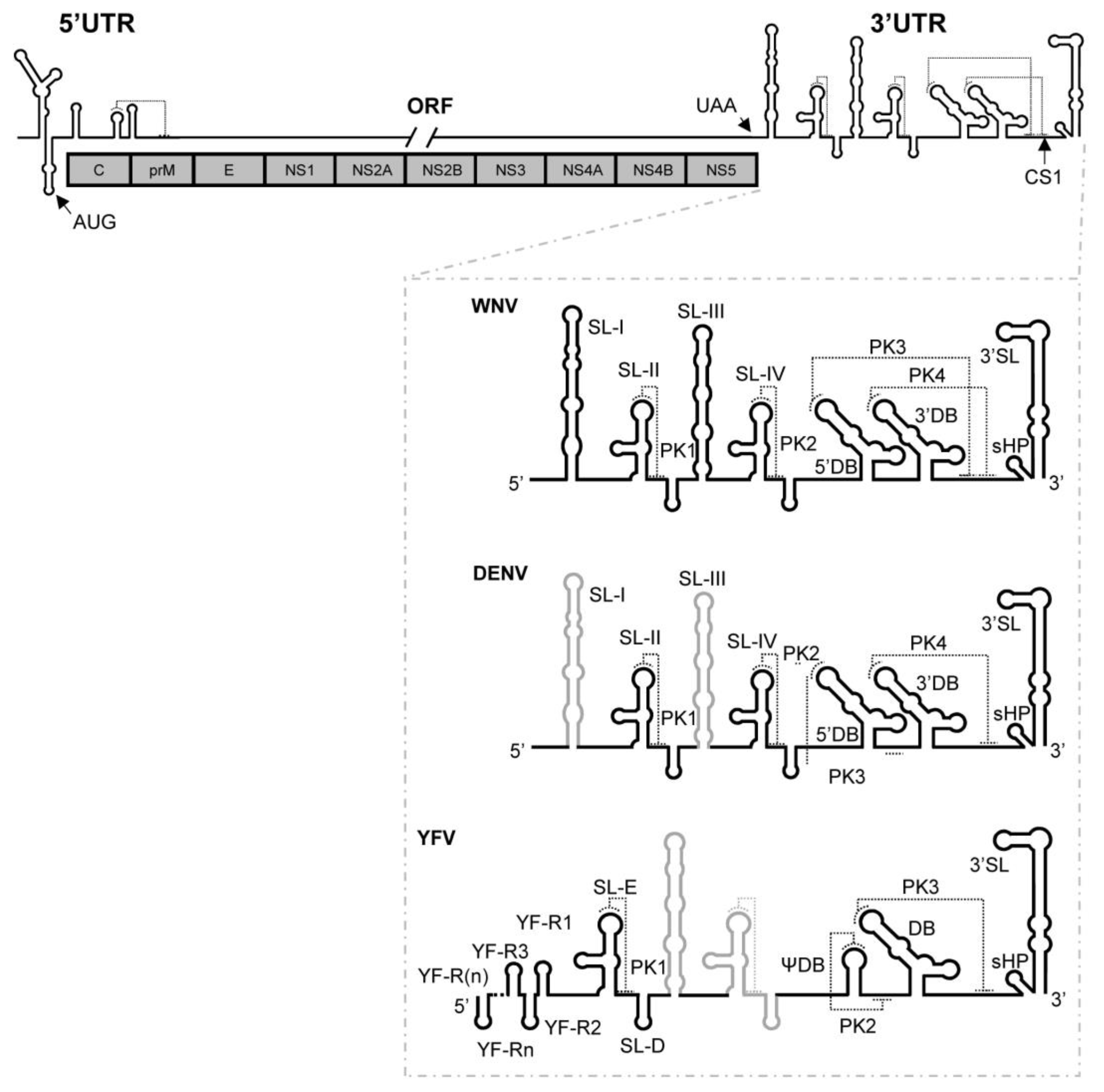
1. Introduction
2. Results
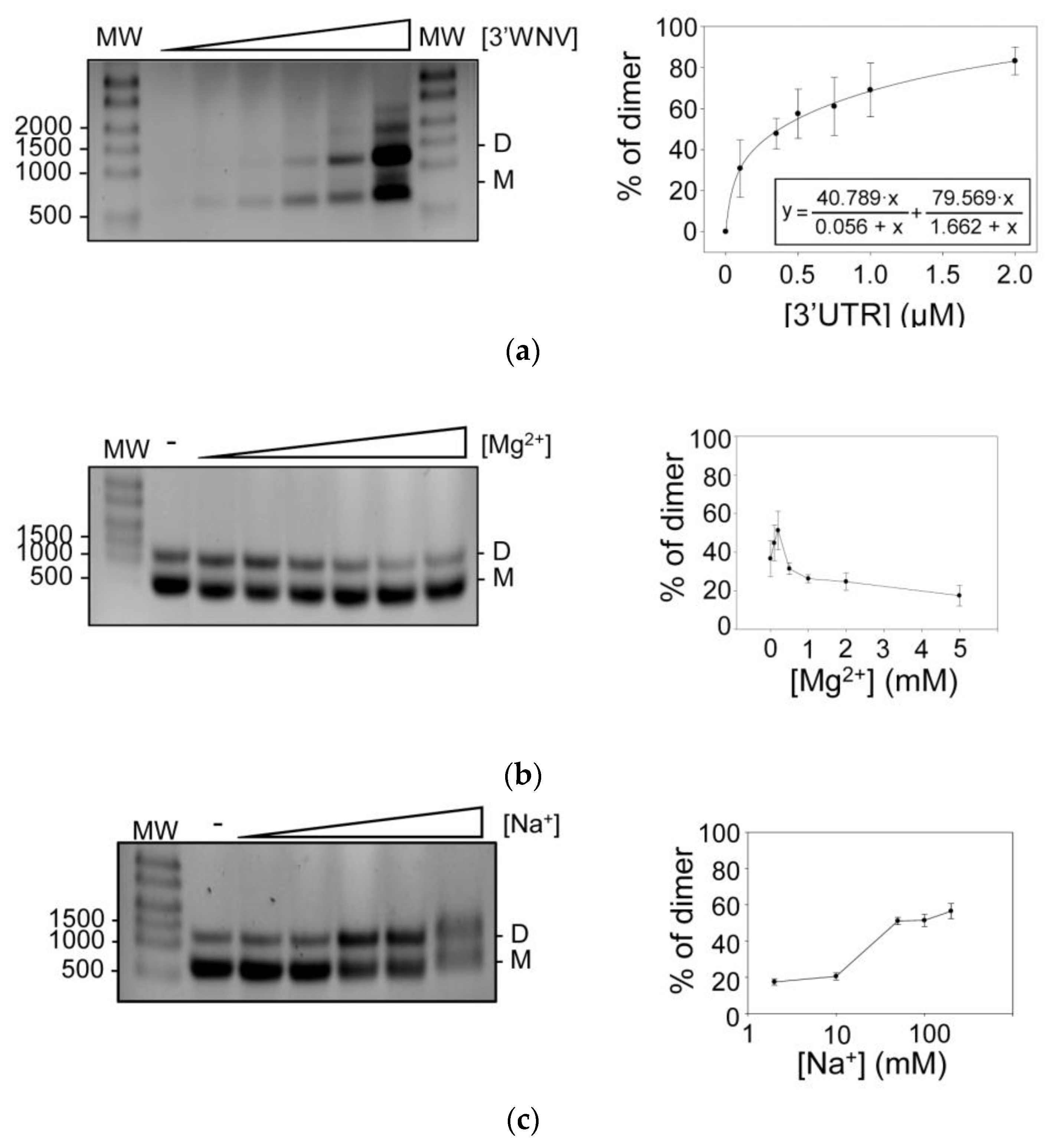
2.1. The 3′ UTR of WNV Dimerizes In Vitro
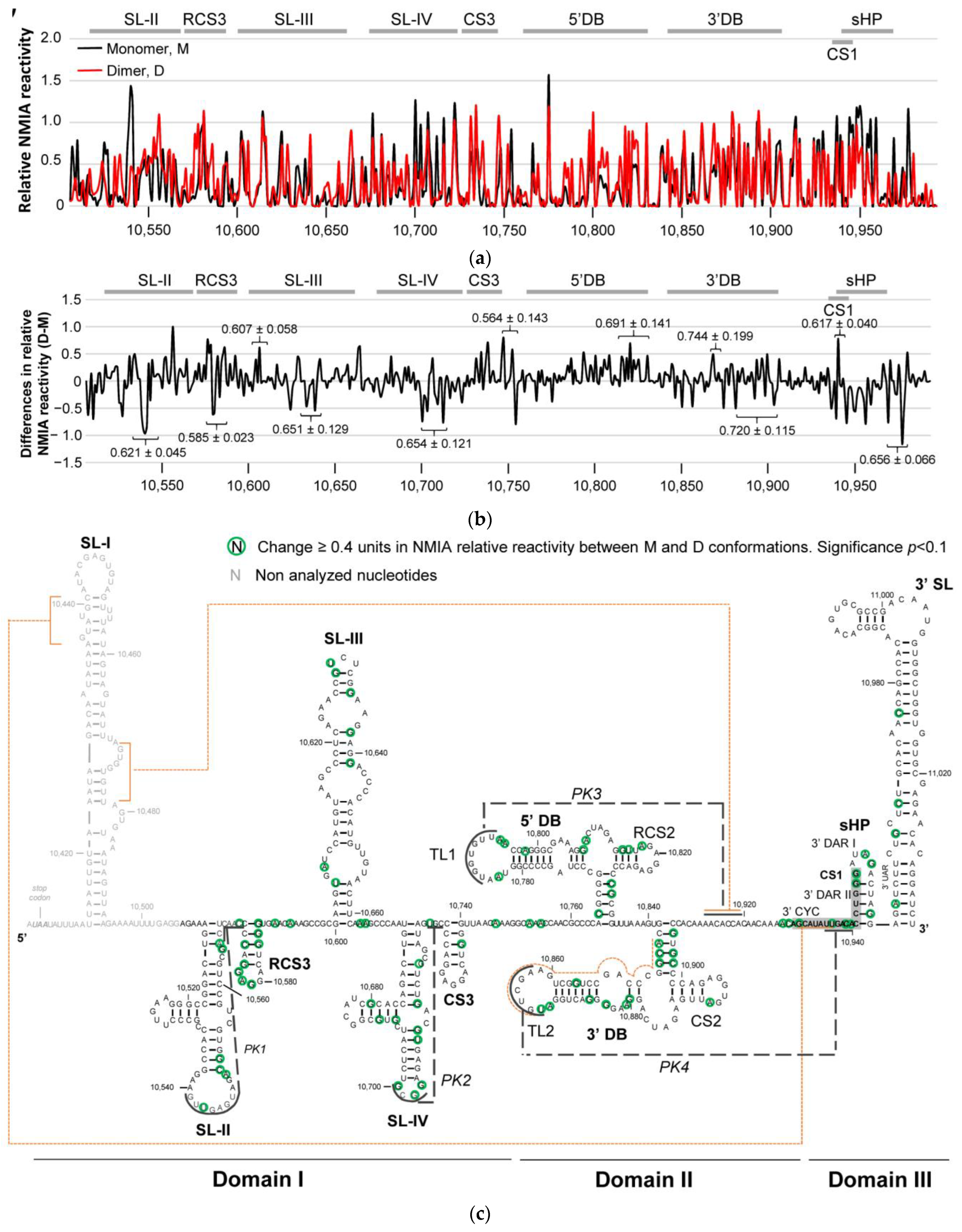
2.2. Structural Elements Essential for the Dimerization of the 3′ UTR, as Revealed by HMX
2.3. The SL-I and 3′DB Elements Are Essential for Efficient 3′ UTR Dimer Formation
2.4. Specific Sequences in SL-I and 3′DB Mediate 3′ UTR Dimerization
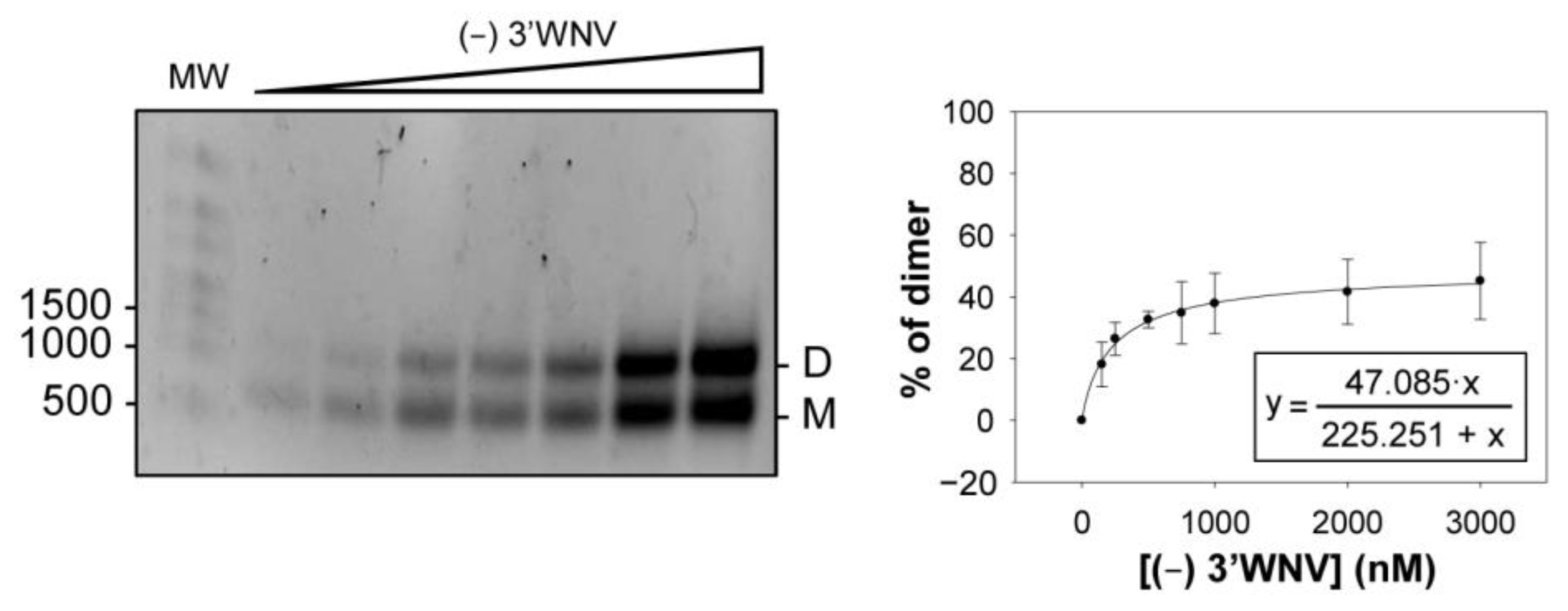
2.5. Dimerization Is also Observed for the (−) Strand of the WNV Genome
2.6. New Roles for the SL-I and 3′DB Elements in the Control of WNV Translation
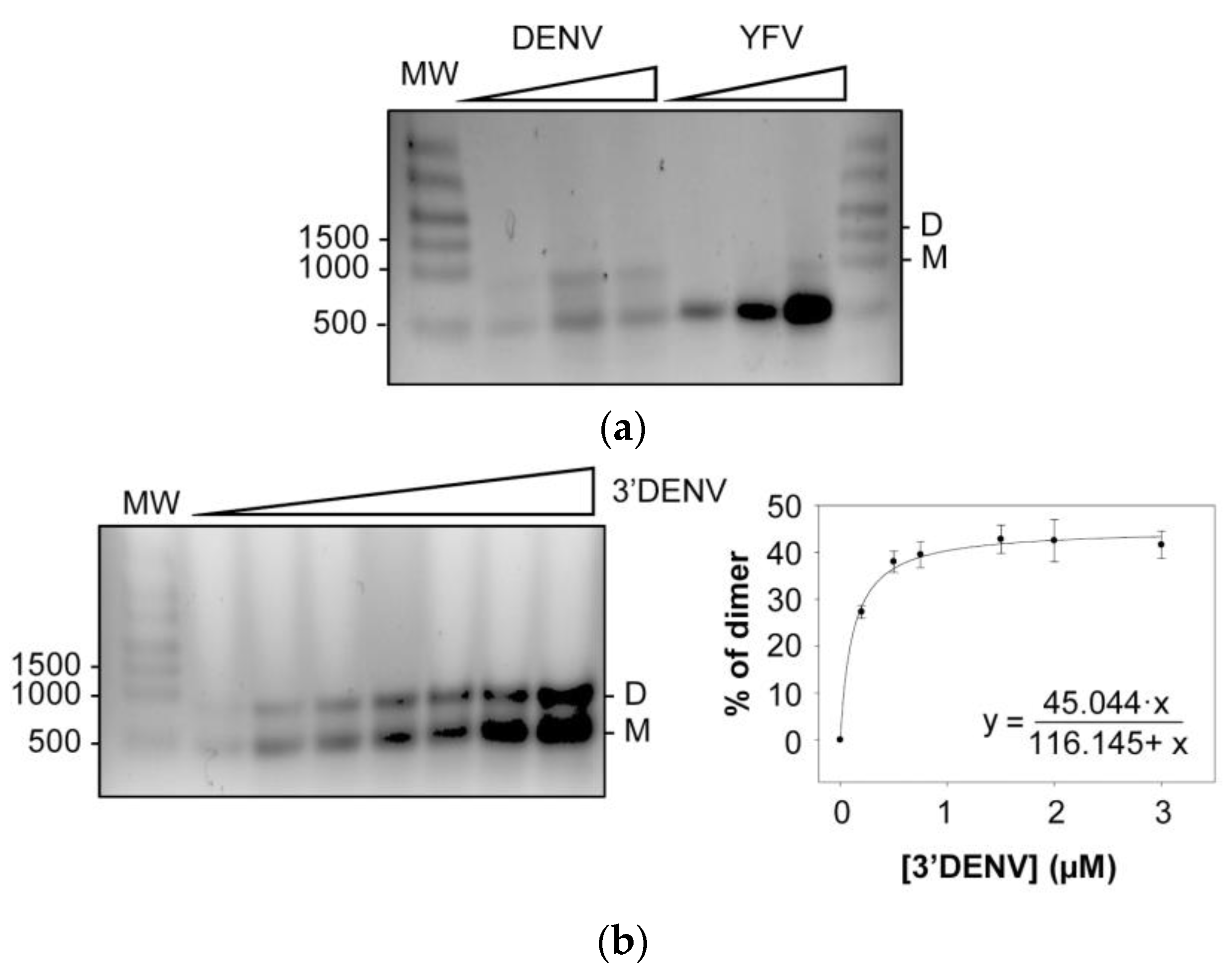
2.7. Genomic Dimerization in Flavivirus: DENV and YFV
3. Discussion
4. Materials and Methods
4.1. DNA Templates and RNA Synthesis
4.2. Dimerization Assays
4.3. HMX (2′ Hydroxyl Molecular Interference) Assays
4.4. Cell Culture
4.5. Cell Transfection and Luciferase Assays
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bou-Nader, C.; Zhang, J. Structural insights into RNA dimerization: Motifs, interfaces and functions. Molecules 2020, 25, 2881. [Google Scholar] [CrossRef] [PubMed]
- Kuno, G.; Chang, G.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the genus flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Cook, S.; Holmes, E.C. A multigene analysis of the phylogenetic relationships among the flaviviruses (family: Flaviviridae) and the evolution of vector transmission. Arch. Virol. 2006, 151, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Cook, S.; Moureau, G.; Kitchen, A.; Gould, E.A.; de Lamballerie, X.; Holmes, E.C.; Harbach, R.E. Molecular evolution of the insect-specific flaviviruses. J. Gen. Virol. 2012, 93, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solinska, J.; Teramoto, T.; Rausch, J.W.; Shapiro, B.A.; Padmanabhan, R.; Le Grice, S.F. Structural complexity of dengue virus untranslated regions: Cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome. Nucleic Acids Res. 2013, 41, 5075–5089. [Google Scholar] [CrossRef]
- de Borba, L.; Villordo, S.M.; Marsico, F.L.; Carballeda, J.M.; Filomatori, C.V.; Gebhard, L.G.; Pallares, H.M.; Lequime, S.; Lambrechts, L.; Sanchez Vargas, I.; et al. RNA structure duplication in the dengue virus 3’ UTR: Redundancy or host specificity? mBio 2019, 10, e02506-18. [Google Scholar] [CrossRef]
- Berzal-Herranz, A.; Berzal-Herranz, B.; Ramos-Lorente, S.E.; Romero-López, C. The genomic 3’ UTR of flaviviruses is a translation initiation enhancer. Int. J. Mol. Sci. 2022, 23, 8604. [Google Scholar] [CrossRef]
- Wei, Y.; Qin, C.; Jiang, T.; Li, X.; Zhao, H.; Liu, Z.; Deng, Y.; Liu, R.; Chen, S.; Yu, M.; et al. Translational regulation by the 3’ untranslated region of the dengue type 2 virus genome. Am. J. Trop. Med. Hyg. 2009, 81, 817–824. [Google Scholar] [CrossRef]
- Shivaprasad, S.; Sarnow, P. The tale of two flaviviruses: Subversion of host pathways by RNA shapes in dengue and hepatitis C viral RNA genomes. Curr. Opin. Microbiol. 2020, 59, 79–85. [Google Scholar] [CrossRef]
- Fernández-Sanlés, A.; Ríos-Marco, P.; Romero-López, C.; Berzal-Herranz, A. Functional information stored in the conserved structural RNA domains of flavivirus genomes. Front. Microbiol. 2017, 8, 546. [Google Scholar] [CrossRef]
- Zeng, M.; Duan, Y.; Zhang, W.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Zhao, X.; Yang, Q.; Wu, Y.; et al. Universal RNA secondary structure insight into mosquito-borne flavivirus (MBFV) cis-acting RNA biology. Front. Microbiol. 2020, 11, 473. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lorente, S.; Romero-López, C.; Berzal-Herranz, A. Information encoded by the flavivirus genomes beyond the nucleotide sequence. Int. J. Mol. Sci. 2021, 22, 3738. [Google Scholar] [CrossRef] [PubMed]
- Yamada, N.; Tanihara, K.; Takada, A.; Yorihuzi, T.; Tsutsumi, M.; Shimomura, H.; Tsuji, T.; Date, T. Genetic organization and diversity of the 3’ noncoding region of the hepatitis C virus genome. Virology 1996, 223, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Masante, C.; Jaubert, C.; Palau, W.; Plissonneau, J.; Besnard, L.; Ventura, M.; Di Primo, C. Mutations of the SL2 dimerization sequence of the hepatitis C genome abrogate viral replication. Cell. Mol. Life Sci. 2015, 72, 3375–3385. [Google Scholar] [CrossRef]
- Castillo-Martinez, J.; Ovejero, T.; Romero-López, C.; Sanmartin, I.; Berzal-Herranz, B.; Oltra, E.; Berzal-Herranz, A.; Gallego, J. Structure and function analysis of the essential 3’X domain of hepatitis C virus. RNA 2020, 26, 186–198. [Google Scholar] [CrossRef]
- Romero-López, C.; Barroso-delJesus, A.; Berzal-Herranz, A. The chaperone-like activity of the hepatitis C virus IRES and CRE elements regulates genome dimerization. Sci. Rep. 2017, 7, 43415. [Google Scholar] [CrossRef]
- Ellis, R.J. Macromolecular crowding: Obvious but underappreciated. Trends Biochem. Sci. 2001, 26, 597–604. [Google Scholar] [CrossRef]
- Van Treeck, B.; Parker, R. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell 2018, 174, 791–802. [Google Scholar] [CrossRef]
- Cristofari, G.; Ivanyi-Nagy, R.; Gabus, C.; Boulant, S.; Lavergne, J.P.; Penin, F.; Darlix, J.L. The hepatitis C virus core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 2004, 32, 2623–2631. [Google Scholar] [CrossRef]
- Ivanyi-Nagy, R.; Kanevsky, I.; Gabus, C.; Lavergne, J.P.; Ficheux, D.; Penin, F.; Fosse, P.; Darlix, J.L. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res. 2006, 34, 2618–2633. [Google Scholar] [CrossRef]
- Fischer, N.M.; Poleto, M.D.; Steuer, J.; van der Spoel, D. Influence of Na+ and Mg2+ ions on RNA structures studied with molecular dynamics simulations. Nucleic Acids Res. 2018, 46, 4872–4882. [Google Scholar] [CrossRef] [PubMed]
- Homan, P.J.; Tandon, A.; Rice, G.M.; Ding, F.; Dokholyan, N.V.; Weeks, K.M. RNA tertiary structure analysis by 2’-hydroxyl molecular interference. Biochemistry 2014, 53, 6825–6833. [Google Scholar] [CrossRef]
- Bellaousov, S.; Reuter, J.S.; Seetin, M.G.; Mathews, D.H. RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013, 41, W471–W474. [Google Scholar] [CrossRef] [PubMed]
- Hofacker, I.L. RNA secondary structure analysis using the Vienna RNA package. Curr. Protoc. Bioinform. 2004, 26, 12. [Google Scholar] [CrossRef]
- Schwartz, M.; Chen, J.; Janda, M.; Sullivan, M.; den Boon, J.; Ahlquist, P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 2002, 9, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Guil, S.; Esteller, M. RNA-RNA interactions in gene regulation: The coding and noncoding players. Trends Biochem. Sci. 2015, 40, 248–256. [Google Scholar] [CrossRef]
- Langdon, E.M.; Qiu, Y.; Ghanbari Niaki, A.; McLaughlin, G.A.; Weidmann, C.A.; Gerbich, T.M.; Smith, J.A.; Crutchley, J.M.; Termini, C.M.; Weeks, K.M.; et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 2018, 360, 922–927. [Google Scholar] [CrossRef]
- Dubois, N.; Marquet, R.; Paillart, J.C.; Bernacchi, S. Retroviral RNA dimerization: From structure to functions. Front. Microbiol. 2018, 9, 527. [Google Scholar] [CrossRef]
- Ishimaru, D.; Plant, E.P.; Sims, A.C.; Yount, B.L., Jr.; Roth, B.M.; Eldho, N.V.; Perez-Alvarado, G.C.; Armbruster, D.W.; Baric, R.S.; Dinman, J.D.; et al. RNA dimerization plays a role in ribosomal frameshifting of the SARS coronavirus. Nucleic Acids Res. 2013, 41, 2594–2608. [Google Scholar] [CrossRef]
- Manzano, M.; Reichert, E.D.; Polo, S.; Falgout, B.; Kasprzak, W.; Shapiro, B.A.; Padmanabhan, R. Identification of cis-acting elements in the 3’-untranslated region of the Dengue virus type 2 RNA that modulate translation and replication. J. Biol. Chem. 2011, 286, 22521–22534. [Google Scholar] [CrossRef]
- Schneider, A.B.; Wolfinger, M.T. Musashi binding elements in Zika and related flavivirus 3’UTRs: A comparative study in silico. Sci. Rep. 2019, 9, 6911. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Gupta, M.; Li, W.; Miller, I.; Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999, 147, 1431–1442. [Google Scholar] [CrossRef]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.; Lloyd, R.E. Poliovirus unlinks TIA1 aggregation and mRNA stress granule formation. J. Virol. 2011, 85, 12442–12454. [Google Scholar] [CrossRef] [PubMed]
- Reineke, L.C.; Lloyd, R.E. Diversion of stress granules and P-bodies during viral infection. Virology 2013, 436, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, B.M.; Graham, M.E.; O′ Donoghue, Z.; Beckham, J.D.; Kieft, J.S. Three-dimensional structure of a flavivirus dumbbell RNA reveals molecular details of an RNA regulator of replication. Nucleic Acids Res. 2021, 49, 7122–7138. [Google Scholar] [CrossRef]
- Martin-Acebes, M.A.; Blazquez, A.B.; Jimenez de Oya, N.; Escribano-Romero, E.; Saiz, J.C. West Nile virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PLoS ONE 2011, 6, e24970. [Google Scholar] [CrossRef]
- Romero-López, C.; Berzal-Herranz, A. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell. Mol. Life Sci. 2012, 69, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Díaz-González, R.; Berzal-Herranz, A. Inhibition of hepatitis C virus internal ribosome entry site-mediated translation by an RNA targeting the conserved IIIf domain. Cell. Mol. Life Sci. 2007, 64, 2994–3006. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Barroso-delJesus, A.; García-Sacristán, A.; Briones, C.; Berzal-Herranz, A. The folding of the hepatitis C virus internal ribosome entry site depends on the 3’-end of the viral genome. Nucleic Acids Res. 2012, 40, 11697–11713. [Google Scholar] [CrossRef]
- Romero-López, C.; Berzal-Herranz, A. A long-range RNA-RNA interaction between the 5’ and 3’ ends of the HCV genome. RNA 2009, 15, 1740–1752. [Google Scholar] [CrossRef] [PubMed]
- Karabiber, F.; McGinnis, J.L.; Favorov, O.V.; Weeks, K.M. Qushape: Rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 2013, 19, 63–73. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero-López, C.; Roda-Herreros, M.; Berzal-Herranz, B.; Ramos-Lorente, S.E.; Berzal-Herranz, A. Inter- and Intramolecular RNA–RNA Interactions Modulate the Regulation of Translation Mediated by the 3′ UTR in West Nile Virus. Int. J. Mol. Sci. 2023, 24, 5337. https://doi.org/10.3390/ijms24065337
Romero-López C, Roda-Herreros M, Berzal-Herranz B, Ramos-Lorente SE, Berzal-Herranz A. Inter- and Intramolecular RNA–RNA Interactions Modulate the Regulation of Translation Mediated by the 3′ UTR in West Nile Virus. International Journal of Molecular Sciences. 2023; 24(6):5337. https://doi.org/10.3390/ijms24065337
Chicago/Turabian StyleRomero-López, Cristina, Margarita Roda-Herreros, Beatriz Berzal-Herranz, Sara Esther Ramos-Lorente, and Alfredo Berzal-Herranz. 2023. "Inter- and Intramolecular RNA–RNA Interactions Modulate the Regulation of Translation Mediated by the 3′ UTR in West Nile Virus" International Journal of Molecular Sciences 24, no. 6: 5337. https://doi.org/10.3390/ijms24065337
APA StyleRomero-López, C., Roda-Herreros, M., Berzal-Herranz, B., Ramos-Lorente, S. E., & Berzal-Herranz, A. (2023). Inter- and Intramolecular RNA–RNA Interactions Modulate the Regulation of Translation Mediated by the 3′ UTR in West Nile Virus. International Journal of Molecular Sciences, 24(6), 5337. https://doi.org/10.3390/ijms24065337







