The Role of Regulated Programmed Cell Death in Osteoarthritis: From Pathogenesis to Therapy
Abstract
1. Introduction
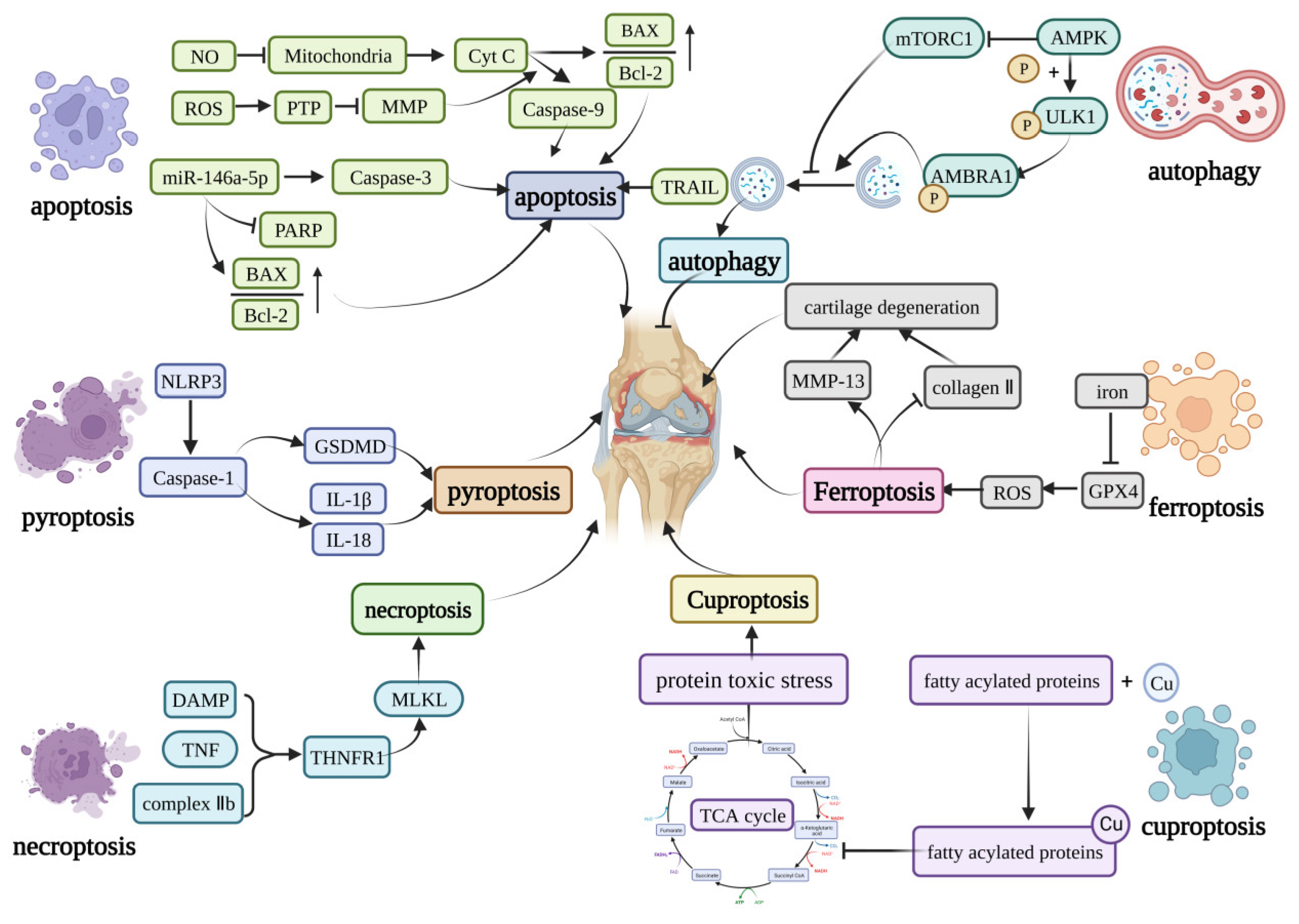
2. Mechanisms of Cell Death in Osteoarthritis
2.1. Apoptosis
2.2. Pyroptosis
2.3. Necroptosis
2.4. Ferroptosis
2.5. Autophagy
2.6. Cuproptosis
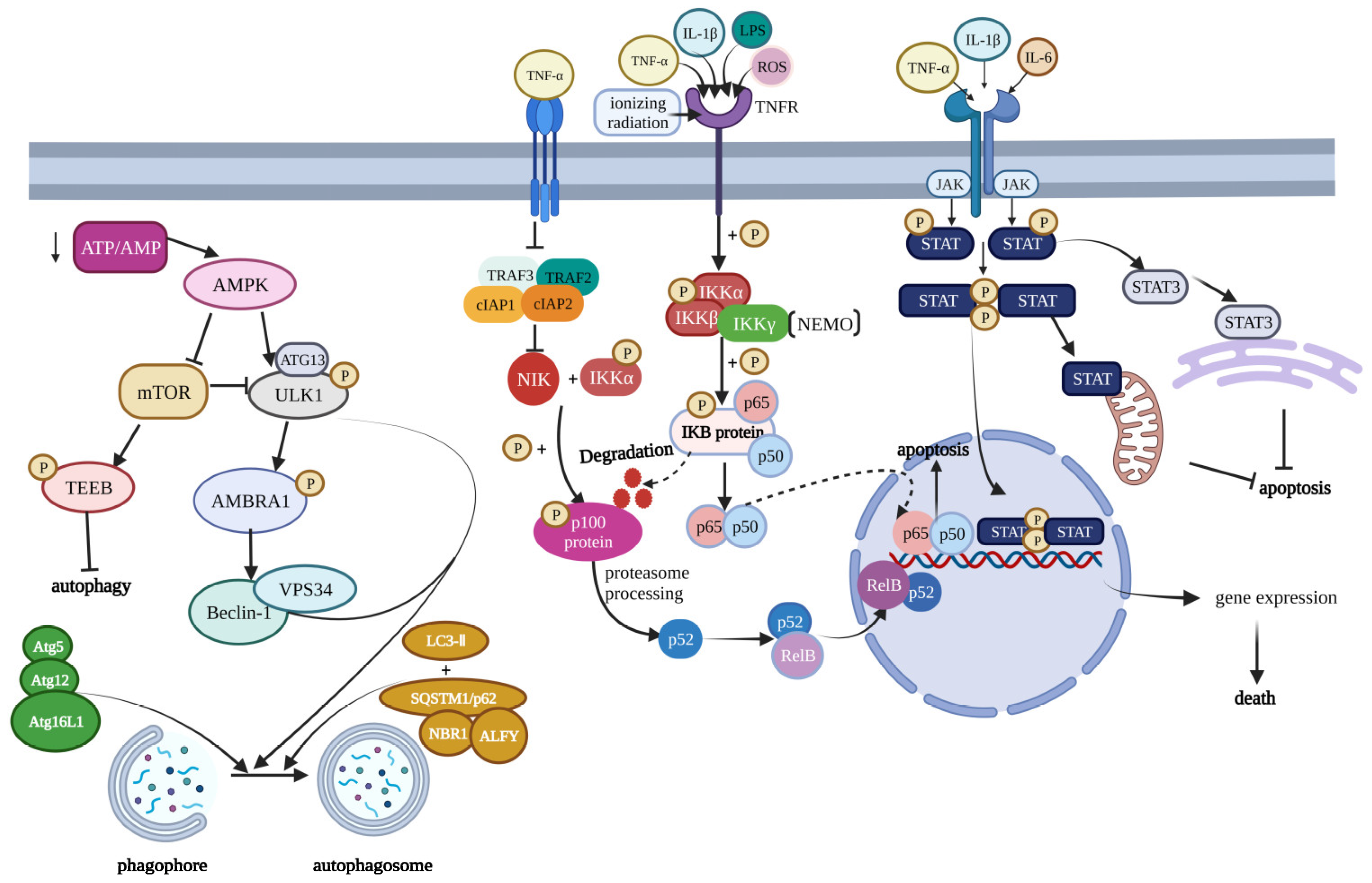
3. Modulation of Cell Death in Osteoarthritis
3.1. NF-κB Signaling Pathway
3.2. mTOR Signaling Pathway
3.3. JAK Signaling Pathway
3.4. HIF Signaling Pathway
3.5. URP Signaling Pathway
4. Therapy Options in Osteoarthritis
| Therapeutic Method | Typical Molecule or Drug | Target Site | Treatment Outcome | Mechanism | Existing Problems | Reference |
|---|---|---|---|---|---|---|
| Intra-Articular Injection | Corticosteroid, Hyaluronic acid, Platelet-rich plasma | interleukin-1, prostaglandins, leukotriene, MMP9, MMP-11 (Corticosteroid) | Reduce pain and increase joint mobility | Corticosteroid: inhibit the secretion of the molecule that can induce pain. Hyaluronic acid: artificially recover the environment of the joint. Platelet-rich plasma: promote healing of damaged joint | Long-time use damaged cartilage High-risk (HA) | [227,228,229] |
| Calcitonin treatment | Oral calcitonin, calcitonin-based nanocomplex | Subchondral bone, MMP-13, CTX-II | Reduce the degree of cartilage lesions | Reduce the MMP-13 and CTX-Ⅱurinary excretion | Clinical research is insufficient | [217] |
| NF-κB signal pathway | Small interfering RNAs, SC-514, KINK-1, PHA-408, Proteosome inhibitors, targeted IκBα ubiquination blockers, Electrophilic compounds, Morroniside | IL-1 receptor, MMP, JNK and MAPK dependent cytokine signaling, | Reduce the inflammatory response | Inhibit the transcription of NF-κB target genes or block the NF-κB signaling | In its infancy in animal models and clinical studies | [217,220] |
| Caspase-l/IL-1β inflammatory pathway | ICA, licochalcone A | NLRP1 inflammasomes, NLRP3 inflammasomes | Reduce the cartilage damage | Inhibit pyroptosis and extracelluar matrix degradation | Clinical research is insufficient | [15] |
| JAK/STAT signal pathway | AG490, Artesunate, Acteoside | STAT, SOCS | Delay deterioration | Block the JAK/STAT signaling to inhibit the development of OA | The exact effect and side effect is unknown | [163,169] |
| PI3K/AKT/ mTOR signal pathway | LY294002, Casodex, rapamycin, 17b-estradiol, FGF18, ghrelin | PI3K, AKT | Reduce the cartilage damage | 1. Inhibit the PI3K/AKT signaling to decrease sclerosis in subchondral (LY294002, Casodex, rapamycin) 2. Activate the PI3K/AKT signaling to promote the chondrocyte proliferation (17b-estradiol, FGF18, ghrelin) | Have side effects and the effect is a double-edged sword | [161] |
| OA genetics and epigenetics therapeutic option | DOMADs, HDAC inhibitor, CRISPR-Cas9 | DOMADs: highlighted gene protein in GWAS HDAC inhibitor: MMPs and IL-1 | Inhibition of OA development in a mouse model | Improving symptoms by affecting the expression of OA-related genes | Accurate delivery of drugs to the joint tissue and targeted treatment is required | [226] |
5. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Das, S.K.; Farooqi, A. Osteoarthritis. Best Pract. Res. Clin. Rheumatol. 2008, 22, 657–675. [Google Scholar] [CrossRef] [PubMed]
- Reynard, L.N.; Loughlin, J. Genetics and epigenetics of osteoarthritis. Maturitas 2012, 71, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Buckwalter, J.A.; Mankin, H.J. Articular cartilage: Degeneration and osteoarthritis, repair, regeneration, and transplantation. Instr. Course Lect. 1998, 47, 487–504. [Google Scholar] [PubMed]
- Abramoff, B.; Caldera, F.E. Osteoarthritis: Pathology, Diagnosis, and Treatment Options. Med. Clin. N. Am. 2020, 104, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Felson, D.T. Developments in the clinical understanding of osteoarthritis. Arthritis Res. Ther. 2009, 11, 203. [Google Scholar] [CrossRef]
- Musumeci, G.; Aiello, F.C.; Szychlinska, M.A.; Di Rosa, M.; Castrogiovanni, P.; Mobasheri, A. Osteoarthritis in the XXIst century: Risk factors and behaviours that influence disease onset and progression. Int. J. Mol. Sci. 2015, 16, 6093–6112. [Google Scholar] [CrossRef]
- Pereira, D.; Ramos, E.; Branco, J. Osteoarthritis. Acta Med. Port. 2015, 28, 99–106. [Google Scholar] [CrossRef]
- Sellam, J.; Berenbaum, F. Is osteoarthritis a metabolic disease? Jt. Bone Spine 2013, 80, 568–573. [Google Scholar] [CrossRef]
- Felson, D.T.; Zhang, Y.; Hannan, M.T.; Naimark, A.; Weissman, B.N.; Aliabadi, P.; Levy, D. The incidence and natural history of knee osteoarthritis in the elderly. The Framingham Osteoarthritis Study. Arthritis Rheum. 1995, 38, 1500–1505. [Google Scholar] [CrossRef]
- Moujalled, D.; Strasser, A.; Liddell, J.R. Molecular mechanisms of cell death in neurological diseases. Cell Death Differ. 2021, 28, 2029–2044. [Google Scholar] [CrossRef]
- Jorgensen, I.; Miao, E.A. Pyroptotic cell death defends against intracellular pathogens. Immunol. Rev. 2015, 265, 130–142. [Google Scholar] [CrossRef]
- D’Arcy, M.S. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef]
- Hwang, H.S.; Kim, H.A. Chondrocyte Apoptosis in the Pathogenesis of Osteoarthritis. Int. J. Mol. Sci. 2015, 16, 26035–26054. [Google Scholar] [CrossRef]
- Yu, P.; Zhang, X.; Liu, N.; Tang, L.; Peng, C.; Chen, X. Pyroptosis: Mechanisms and diseases. Signal Transduct. Target. Ther. 2021, 6, 128. [Google Scholar] [CrossRef]
- Yang, J.; Hu, S.; Bian, Y.; Yao, J.; Wang, D.; Liu, X.; Guo, Z.; Zhang, S.; Peng, L. Targeting Cell Death: Pyroptosis, Ferroptosis, Apoptosis and Necroptosis in Osteoarthritis. Front. Cell Dev. Biol. 2021, 9, 789948. [Google Scholar] [CrossRef]
- Mou, Y.; Wang, J.; Wu, J.; He, D.; Zhang, C.; Duan, C.; Li, B. Ferroptosis, a new form of cell death: Opportunities and challenges in cancer. J. Hematol. Oncol. 2019, 12, 34. [Google Scholar] [CrossRef]
- Yagoda, N.; Von Rechenberg, M.; Zaganjor, E.; Bauer, A.J.; Yang, W.S.; Fridman, D.J.; Wolpaw, A.J.; Smukste, I.; Peltier, J.M.; Boniface, J.J.; et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 2007, 447, 864–868. [Google Scholar] [CrossRef]
- Yu, H.; Guo, P.; Xie, X.; Wang, Y.; Chen, G. Ferroptosis, a new form of cell death, and its relationships with tumourous diseases. J. Cell. Mol. Med. 2017, 21, 648–657. [Google Scholar] [CrossRef]
- Latunde-Dada, G.O. Ferroptosis: Role of lipid peroxidation, iron and ferritinophagy. Biochim. Biophys. Acta. Gen. Subj. 2017, 1861, 1893–1900. [Google Scholar] [CrossRef]
- Cao, J.Y.; Dixon, S.J. Mechanisms of ferroptosis. Cell. Mol. Life Sci. 2016, 73, 2195–2209. [Google Scholar] [CrossRef]
- Xie, Y.; Hou, W.; Song, X.; Yu, Y.; Huang, J.; Sun, X.; Kang, R.; Tang, D. Ferroptosis: Process and function. Cell Death Differ. 2016, 23, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Sun, K.; Yu, S.; Luo, J.; Guo, J.; Lin, J.; Wang, G.; Guo, Z.; Ye, Y.; Guo, F. Chondrocyte ferroptosis contribute to the progression of osteoarthritis. J. Orthop. Transl. 2021, 27, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Kepp, O.; Chan, F.K.; Kroemer, G. Necroptosis: Mechanisms and Relevance to Disease. Annu. Rev. Pathol. 2017, 12, 103–130. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.M.; Ding, W.X.; Gao, W. Autophagy in the liver. Hepatology 2008, 47, 1773–1785. [Google Scholar] [CrossRef]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef]
- Jing, K.; Lim, K. Why is autophagy important in human diseases? Exp. Mol. Med. 2012, 44, 69–72. [Google Scholar] [CrossRef]
- Ma, Y.; Galluzzi, L.; Zitvogel, L.; Kroemer, G. Autophagy and cellular immune responses. Immunity 2013, 39, 211–227. [Google Scholar] [CrossRef]
- Füllgrabe, J.; Klionsky, D.J.; Joseph, B. The return of the nucleus: Transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014, 15, 65–74. [Google Scholar] [CrossRef]
- Michael, J.W.; Schlüter-Brust, K.U.; Eysel, P. The epidemiology, etiology, diagnosis, and treatment of osteoarthritis of the knee. Dtsch. Arztebl. Int. 2010, 107, 152–162. [Google Scholar] [CrossRef]
- Kerr, J.F.; Wyllie, A.; Currie, A. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef]
- Sharif, M.; Whitehouse, A.; Sharman, P.; Perry, M.; Adams, M. Increased apoptosis in human osteoarthritic cartilage corresponds to reduced cell density and expression of caspase-3. Arthritis Rheum. 2004, 50, 507–515. [Google Scholar] [CrossRef]
- Hashimoto, S.; Ochs, R.L.; Komiya, S.; Lotz, M. Linkage of chondrocyte apoptosis and cartilage degradation in human osteoarthritis. Arthritis Rheum. 1998, 41, 1632–1638. [Google Scholar] [CrossRef]
- Pelletier, J.P.; Martel-Pelletier, J.; Abramson, S.B. Osteoarthritis, an inflammatory disease: Potential implication for the selection of new therapeutic targets. Arthritis Rheum. 2001, 44, 1237–1247. [Google Scholar] [CrossRef]
- Abramson, S.B.; Attur, M.; Yazici, Y. Prospects for disease modification in osteoarthritis. Nat. Clin. Pract. Rheumatol. 2006, 2, 304–312. [Google Scholar] [CrossRef]
- Mao, X.; Fu, P.; Wang, L.; Xiang, C. Mitochondria: Potential Targets for Osteoarthritis. Front. Med. 2020, 7, 581402. [Google Scholar] [CrossRef]
- Lepetsos, P.; Papavassiliou, A.G. ROS/oxidative stress signaling in osteoarthritis. Biochim. Biophys. Acta 2016, 1862, 576–591. [Google Scholar] [CrossRef]
- Kapoor, M.; Martel-Pelletier, J.; Lajeunesse, D.; Pelletier, J.P.; Fahmi, H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 33–42. [Google Scholar] [CrossRef]
- Amin, A.R.; Di Cesare, P.E.; Vyas, P.; Attur, M.; Tzeng, E.; Billiar, T.R.; Stuchin, S.A.; Abramson, S.B. The expression and regulation of nitric oxide synthase in human osteoarthritis-affected chondrocytes: Evidence for up-regulated neuronal nitric oxide synthase. J. Exp. Med. 1995, 182, 2097–2102. [Google Scholar] [CrossRef]
- Loeser, R.F.; Carlson, C.S.; Del Carlo, M.; Cole, A. Detection of nitrotyrosine in aging and osteoarthritic cartilage: Correlation of oxidative damage with the presence of interleukin-1beta and with chondrocyte resistance to insulin-like growth factor 1. Arthritis Rheum. 2002, 46, 2349–2357. [Google Scholar] [CrossRef]
- Wu, G.J.; Chen, T.G.; Chang, H.C.; Chiu, W.T.; Chang, C.C.; Chen, R.M. Nitric oxide from both exogenous and endogenous sources activates mitochondria-dependent events and induces insults to human chondrocytes. J. Cell. Biochem. 2007, 101, 1520–1531. [Google Scholar] [CrossRef] [PubMed]
- Maneiro, E.; López-Armada, M.J.; De Andres, M.C.; Caramés, B.; Martín, M.A.; Bonilla, A.; Del Hoyo, P.; Galdo, F.; Arenas, J.; Blanco, F.J. Effect of nitric oxide on mitochondrial respiratory activity of human articular chondrocytes. Ann. Rheum. Dis. 2005, 64, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Yang, H.; Xie, M.; Wen, Y.; Liu, Q.; Li, X.; Liu, J.; Xu, H.; Tang, W.; Wang, M. Chondrocyte apoptosis in rat mandibular condyles induced by dental occlusion due to mitochondrial damage caused by nitric oxide. Arch. Oral Biol. 2019, 101, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, K.; Rajpurohit, R.; Shapiro, I.M. Extracellular phosphate ions cause apoptosis of terminally differentiated epiphyseal chondrocytes. J. Cell. Physiol. 1999, 179, 276–286. [Google Scholar] [CrossRef]
- Antoniel, M.; Giorgio, V.; Fogolari, F.; Glick, G.D.; Bernardi, P.; Lippe, G. The oligomycin-sensitivity conferring protein of mitochondrial ATP synthase: Emerging new roles in mitochondrial pathophysiology. Int. J. Mol. Sci. 2014, 15, 7513–7536. [Google Scholar] [CrossRef]
- Bauer, T.M.; Murphy, E. Role of Mitochondrial Calcium and the Permeability Transition Pore in Regulating Cell Death. Circ. Res. 2020, 126, 280–293. [Google Scholar] [CrossRef]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef]
- Wenz, T. PGC-1alpha activation as a therapeutic approach in mitochondrial disease. IUBMB Life 2009, 61, 1051–1062. [Google Scholar] [CrossRef]
- Liang, S.; Sun, K.; Wang, Y.; Dong, S.; Wang, C.; Liu, L.; Wu, Y. Role of Cyt-C/caspases-9,3, Bax/Bcl-2 and the FAS death receptor pathway in apoptosis induced by zinc oxide nanoparticles in human aortic endothelial cells and the protective effect by alpha-lipoic acid. Chem. Biol. Interact. 2016, 258, 40–51. [Google Scholar] [CrossRef]
- Akhmedov, A.T.; Marín-García, J. Mitochondrial DNA maintenance: An appraisal. Mol. Cell. Biochem. 2015, 409, 283–305. [Google Scholar] [CrossRef]
- McCulloch, K.; Litherland, G.J.; Rai, T.S. Cellular senescence in osteoarthritis pathology. Aging Cell 2017, 16, 210–218. [Google Scholar] [CrossRef]
- Grishko, V.I.; Ho, R.; Wilson, G.L.; Pearsall, A.W.t. Diminished mitochondrial DNA integrity and repair capacity in OA chondrocytes. Osteoarthr. Cartil. 2009, 17, 107–113. [Google Scholar] [CrossRef]
- Blanco, F.J.; López-Armada, M.J.; Maneiro, E. Mitochondrial dysfunction in osteoarthritis. Mitochondrion 2004, 4, 715–728. [Google Scholar] [CrossRef]
- Schuerwegh, A.J.; Dombrecht, E.J.; Stevens, W.J.; Van Offel, J.F.; Bridts, C.H.; De Clerck, L.S. Influence of pro-inflammatory (IL-1 alpha, IL-6, TNF-alpha, IFN-gamma) and anti-inflammatory (IL-4) cytokines on chondrocyte function. Osteoarthr. Cartil. 2003, 11, 681–687. [Google Scholar] [CrossRef]
- Kühn, K.; Hashimoto, S.; Lotz, M. IL-1 beta protects human chondrocytes from CD95-induced apoptosis. J. Immunol. 2000, 164, 2233–2239. [Google Scholar] [CrossRef]
- Blanco, F.J.; Ochs, R.L.; Schwarz, H.; Lotz, M. Chondrocyte apoptosis induced by nitric oxide. Am. J. Pathol. 1995, 146, 75–85. [Google Scholar]
- Guo, Q.; Chen, X.; Chen, J.; Zheng, G.; Xie, C.; Wu, H.; Miao, Z.; Lin, Y.; Wang, X.; Gao, W.; et al. STING promotes senescence, apoptosis, and extracellular matrix degradation in osteoarthritis via the NF-κB signaling pathway. Cell Death Dis. 2021, 12, 13. [Google Scholar] [CrossRef]
- Pistritto, G.; Trisciuoglio, D.; Ceci, C.; Garufi, A.; D’Orazi, G. Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging 2016, 8, 603–619. [Google Scholar] [CrossRef]
- Lee, S.W.; Lee, H.J.; Chung, W.T.; Choi, S.M.; Rhyu, S.H.; Kim, D.K.; Kim, K.T.; Kim, J.Y.; Kim, J.M.; Yoo, Y.H. TRAIL induces apoptosis of chondrocytes and influences the pathogenesis of experimentally induced rat osteoarthritis. Arthritis Rheum. 2004, 50, 534–542. [Google Scholar] [CrossRef]
- Cheng, S.; Nie, Z.; Cao, J.; Peng, H. Circ_0136474 promotes the progression of osteoarthritis by sponging mir-140-3p and upregulating MECP2. J. Mol. Histol. 2022, 54, 1–12. [Google Scholar] [CrossRef]
- Li, Z.; Yuan, B.; Pei, Z.; Zhang, K.; Ding, Z.; Zhu, S.; Wang, Y.; Guan, Z.; Cao, Y. Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis. J. Cell. Mol. Med. 2019, 23, 6554–6564. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Huang, N.; Zhang, X.; Wu, S.; Wang, L.; Ke, Q. Long non-coding RNA plasmacytoma variant translocation 1 and growth arrest specific 5 regulate each other in osteoarthritis to regulate the apoptosis of chondrocytes. Bioengineered 2022, 13, 13680–13688. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Duan, L.; Liu, S.; Yang, Y.; Qiao, Z.; Shi, L. Long intergenic non-protein coding RNA 00707 regulates chondrocyte apoptosis and proliferation in osteoarthritis by serving as a sponge for microRNA-199-3p. Bioengineered 2022, 13, 11137–11145. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Zhang, Y.; Cai, C.; Liu, C.; Xie, J.; Yi, C. Circular RNA circZNF652 is overexpressed in osteoarthritis and positively regulates LPS-induced apoptosis of chondrocytes by upregulating PTEN. Autoimmunity 2021, 54, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.L.; Deng, S.; Fang, H.S.; Du, X.J.; Peng, H.; Hu, Q.J. Circular RNA circANKRD36 regulates Casz1 by targeting miR-599 to prevent osteoarthritis chondrocyte apoptosis and inflammation. J. Cell. Mol. Med. 2021, 25, 120–131. [Google Scholar] [CrossRef]
- Wang, P.; Wang, Y.; Ma, B. Long noncoding RNA NAV2-AS5 relieves chondrocyte inflammation by targeting miR-8082/TNIP2 in osteoarthritis. Cell Cycle 2022, 2022, 1–12. [Google Scholar] [CrossRef]
- Yu, Z.; Cong, F.; Zhang, W.; Song, T.; Zhang, S.; Jiang, R. Circular RNA circ_0020014 contributes to osteoarthritis progression via miR-613/ADAMTS5 axis. Bosn. J. Basic Med. Sci. 2022, 22, 716–727. [Google Scholar] [CrossRef]
- Bach, D.H.; Lee, S.K.; Sood, A.K. Circular RNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 118–129. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, W.; Li, D.; Zheng, J. miR-146a-5p Promotes Chondrocyte Apoptosis and Inhibits Autophagy of Osteoarthritis by Targeting NUMB. Cartilage 2021, 13 (Suppl. S2), 1467s–1477s. [Google Scholar] [CrossRef]
- Yamasaki, K.; Nakasa, T.; Miyaki, S.; Ishikawa, M.; Deie, M.; Adachi, N.; Yasunaga, Y.; Asahara, H.; Ochi, M. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009, 60, 1035–1041. [Google Scholar] [CrossRef]
- Jin, L.; Zhao, J.; Jing, W.; Yan, S.; Wang, X.; Xiao, C.; Ma, B. Role of miR-146a in human chondrocyte apoptosis in response to mechanical pressure injury in vitro. Int. J. Mol. Med. 2014, 34, 451–463. [Google Scholar] [CrossRef]
- Shi, J.; Gao, W.; Shao, F. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem. Sci. 2017, 42, 245–254. [Google Scholar] [CrossRef]
- Zhaolin, Z.; Guohua, L.; Shiyuan, W.; Zuo, W. Role of pyroptosis in cardiovascular disease. Cell Prolif. 2019, 52, e12563. [Google Scholar] [CrossRef]
- Lacey, C.A.; Mitchell, W.J.; Dadelahi, A.S.; Skyberg, J.A. Caspase-1 and Caspase-11 Mediate Pyroptosis, Inflammation, and Control of Brucella Joint Infection. Infect. Immun. 2018, 86, e00361-18. [Google Scholar] [CrossRef]
- Swanson, K.V.; Deng, M.; Ting, J.P. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef]
- An, S.; Hu, H.; Li, Y.; Hu, Y. Pyroptosis Plays a Role in Osteoarthritis. Aging Dis. 2020, 11, 1146–1157. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, Z.; Chen, D.; He, Y. CY-09 attenuates the progression of osteoarthritis via inhibiting NLRP3 inflammasome-mediated pyroptosis. Biochem. Biophys. Res. Commun. 2021, 553, 119–125. [Google Scholar] [CrossRef]
- Franceschini, A.; Capece, M.; Chiozzi, P.; Falzoni, S.; Sanz, J.M.; Sarti, A.C.; Bonora, M.; Pinton, P.; Di Virgilio, F. The P2X7 receptor directly interacts with the NLRP3 inflammasome scaffold protein. FASEB J. 2015, 29, 2450–2461. [Google Scholar] [CrossRef]
- Li, Z.; Huang, Z.; Bai, L. The P2X7 Receptor in Osteoarthritis. Front. Cell Dev. Biol. 2021, 9, 628330. [Google Scholar] [CrossRef]
- Mapp, P.I.; Walsh, D.A. Mechanisms and targets of angiogenesis and nerve growth in osteoarthritis. Nat. Rev. Rheumatol. 2012, 8, 390–398. [Google Scholar] [CrossRef]
- Degterev, A.; Huang, Z.; Boyce, M.; Li, Y.; Jagtap, P.; Mizushima, N.; Cuny, G.D.; Mitchison, T.J.; Moskowitz, M.A.; Yuan, J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005, 1, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wu, R.; Wang, X.; Bao, X.; Lu, C. Roles of necroptosis in alcoholic liver disease and hepatic pathogenesis. Cell Prolif. 2022, 55, e13193. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.Z.; Crawford, N.; Longley, D.B. The role of Ubiquitination in Apoptosis and Necroptosis. Cell Death Differ. 2022, 29, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Pasparakis, M.; Vandenabeele, P. Necroptosis and its role in inflammation. Nature 2015, 517, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Noh, H.J.; Lee, H.; Park, H.H.; Ha, Y.J.; Park, S.H.; Lee, H.; Kim, S.J.; Kang, H.C.; Eyun, S.I.; et al. TRIM24-RIP3 axis perturbation accelerates osteoarthritis pathogenesis. Ann. Rheum. Dis. 2020, 79, 1635–1643. [Google Scholar] [CrossRef]
- Sokolove, J.; Lepus, C.M. Role of inflammation in the pathogenesis of osteoarthritis: Latest findings and interpretations. Ther. Adv. Musculoskelet Dis. 2013, 5, 77–94. [Google Scholar] [CrossRef]
- Riegger, J.; Brenner, R.E. Evidence of necroptosis in osteoarthritic disease: Investigation of blunt mechanical impact as possible trigger in regulated necrosis. Cell Death Dis. 2019, 10, 683. [Google Scholar] [CrossRef]
- Gong, Y.; Qiu, J.; Ye, J. AZ-628 delays osteoarthritis progression via inhibiting the TNF-α-induced chondrocyte necroptosis and regulating osteoclast formation. Int. Immunopharmacol. 2022, 111, 109085. [Google Scholar] [CrossRef]
- Li, B.; Guan, G.; Mei, L.; Jiao, K.; Li, H. Pathological mechanism of chondrocytes and the surrounding environment during osteoarthritis of temporomandibular joint. J. Cell. Mol. Med. 2021, 25, 4902–4911. [Google Scholar] [CrossRef]
- Lawlor, K.E.; Khan, N.; Mildenhall, A.; Gerlic, M.; Croker, B.A.; D’Cruz, A.A.; Hall, C.; Kaur Spall, S.; Anderton, H.; Masters, S.L.; et al. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat. Commun. 2015, 6, 6282. [Google Scholar] [CrossRef]
- Dannappel, M.; Vlantis, K.; Kumari, S.; Polykratis, A.; Kim, C.; Wachsmuth, L.; Eftychi, C.; Lin, J.; Corona, T.; Hermance, N.; et al. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature 2014, 513, 90–94. [Google Scholar] [CrossRef]
- Cheng, J.; Duan, X.; Fu, X.; Jiang, Y.; Yang, P.; Cao, C.; Li, Q.; Zhang, J.; Hu, X.; Zhang, X.; et al. RIP1 Perturbation Induces Chondrocyte Necroptosis and Promotes Osteoarthritis Pathogenesis via Targeting BMP7. Front. Cell Dev. Biol. 2021, 9, 638382. [Google Scholar] [CrossRef]
- Martens, S.; Hofmans, S.; Declercq, W.; Augustyns, K.; Vandenabeele, P. Inhibitors Targeting RIPK1/RIPK3: Old and New Drugs. Trends Pharmacol. Sci. 2020, 41, 209–224. [Google Scholar] [CrossRef]
- Tong, L.; Yu, H.; Huang, X.; Shen, J.; Xiao, G.; Chen, L.; Wang, H.; Xing, L.; Chen, D. Current understanding of osteoarthritis pathogenesis and relevant new approaches. Bone Res. 2022, 10, 60. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Miao, Y.; Chen, Y.; Xue, F.; Liu, K.; Zhu, B.; Gao, J.; Yin, J.; Zhang, C.; Li, G. Contribution of ferroptosis and GPX4’s dual functions to osteoarthritis progression. EBioMedicine 2022, 76, 103847. [Google Scholar] [CrossRef]
- Yan, H.F.; Zou, T.; Tuo, Q.Z.; Xu, S.; Li, H.; Belaidi, A.A.; Lei, P. Ferroptosis: Mechanisms and links with diseases. Signal Transduct. Target. Ther. 2021, 6, 49. [Google Scholar] [CrossRef]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef]
- Mehana, E.E.; Khafaga, A.F.; El-Blehi, S.S. The role of matrix metalloproteinases in osteoarthritis pathogenesis: An updated review. Life Sci. 2019, 234, 116786. [Google Scholar] [CrossRef]
- Kim, Y.C.; Guan, K.L. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Investig. 2015, 125, 25–32. [Google Scholar] [CrossRef]
- Valenti, M.T.; Dalle Carbonare, L.; Zipeto, D.; Mottes, M. Control of the Autophagy Pathway in Osteoarthritis: Key Regulators, Therapeutic Targets and Therapeutic Strategies. Int. J. Mol. Sci. 2021, 22, 2700. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Gao, F.; Niu, D.; Sun, X.; Song, Q.; Guo, C.; Liang, Y.; Sun, W. The Role of Autophagy in Chondrocyte Metabolism and Osteoarthritis: A Comprehensive Research Review. BioMed Res. Int. 2019, 2019, 5171602. [Google Scholar] [CrossRef] [PubMed]
- Pal, B.; Endisha, H.; Zhang, Y.; Kapoor, M. mTOR: A potential therapeutic target in osteoarthritis? Drugs RD 2015, 15, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Pan, J.; Li, J.; Zeng, C.; Qi, W.; Shao, Y.; Liu, X.; Liu, L.; Xiao, G.; Zhang, H.; et al. Metformin attenuates cartilage degeneration in an experimental osteoarthritis model by regulating AMPK/mTOR. Aging 2020, 12, 1087–1103. [Google Scholar] [CrossRef]
- Collins, J.A.; Diekman, B.O.; Loeser, R.F. Targeting aging for disease modification in osteoarthritis. Curr. Opin. Rheumatol. 2018, 30, 101–107. [Google Scholar] [CrossRef]
- Wang, S.; Deng, Z.; Ma, Y.; Jin, J.; Qi, F.; Li, S.; Liu, C.; Lyu, F.J.; Zheng, Q. The Role of Autophagy and Mitophagy in Bone Metabolic Disorders. Int. J. Biol. Sci. 2020, 16, 2675–2691. [Google Scholar] [CrossRef]
- Duan, R.; Xie, H.; Liu, Z.Z. The Role of Autophagy in Osteoarthritis. Front. Cell Dev. Biol. 2020, 8, 608388. [Google Scholar] [CrossRef]
- Kahlson, M.A.; Dixon, S.J. Copper-induced cell death. Science 2022, 375, 1231–1232. [Google Scholar] [CrossRef]
- Li, S.R.; Bu, L.L.; Cai, L. Cuproptosis: Lipoylated TCA cycle proteins-mediated novel cell death pathway. Signal Transduct. Target. Ther. 2022, 7, 158. [Google Scholar] [CrossRef]
- Cobine, P.A.; Brady, D.C. Cuproptosis: Cellular and molecular mechanisms underlying copper-induced cell death. Mol. Cell 2022, 82, 1786–1787. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wu, G.; Yan, W.; Zhan, H.; Sun, P. miR-146b-5p regulates cell growth, invasion, and metabolism by targeting PDHB in colorectal cancer. Am. J. Cancer Res. 2017, 7, 1136–1150. [Google Scholar] [PubMed]
- Saunier, E.; Benelli, C.; Bortoli, S. The pyruvate dehydrogenase complex in cancer: An old metabolic gatekeeper regulated by new pathways and pharmacological agents. Int. J. Cancer 2016, 138, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Smolle, M.; Lindsay, J.G. Molecular architecture of the pyruvate dehydrogenase complex: Bridging the gap. Biochem. Soc. Trans. 2006, 34, 815–818. [Google Scholar] [CrossRef]
- Park, S.; Baek, I.J.; Ryu, J.H.; Chun, C.H.; Jin, E.J. PPARα-ACOT12 axis is responsible for maintaining cartilage homeostasis through modulating de novo lipogenesis. Nat. Commun. 2022, 13, 3. [Google Scholar] [CrossRef]
- Mayr, J.A.; Feichtinger, R.G.; Tort, F.; Ribes, A.; Sperl, W. Lipoic acid biosynthesis defects. J. Inherit. Metab. Dis. 2014, 37, 553–563. [Google Scholar] [CrossRef]
- Frommer, K.W.; Hasseli, R.; Schäffler, A.; Lange, U.; Rehart, S.; Steinmeyer, J.; Rickert, M.; Sarter, K.; Zaiss, M.M.; Culmsee, C.; et al. Free Fatty Acids in Bone Pathophysiology of Rheumatic Diseases. Front. Immunol. 2019, 10, 2757. [Google Scholar] [CrossRef]
- Guo, W.; Wei, B.; Sun, J.; Chen, T.; Wei, J.; Hu, Z.; Chen, S.; Xiang, M.; Shu, Y.; Peng, Z. Suppressive oligodeoxynucleotide-induced dendritic cells rein the aggravation of osteoarthritis in mice. Immunopharmacol. Immunotoxicol. 2018, 40, 430–436. [Google Scholar] [CrossRef]
- Farinelli, L.; Aquili, A.; Mattioli-Belmonte, M.; Manzotti, S.; D’Angelo, F.; Ciccullo, C.; Gigante, A. Synovial mast cells from knee and hip osteoarthritis: Histological study and clinical correlations. J. Exp. Orthop. 2022, 9, 13. [Google Scholar] [CrossRef]
- Li, Y.; Gao, H.; Brunner, T.M.; Hu, X.; Yan, Y.; Liu, Y.; Qiao, L.; Wu, P.; Li, M.; Liu, Q.; et al. Menstrual blood-derived mesenchymal stromal cells efficiently ameliorate experimental autoimmune encephalomyelitis by inhibiting T cell activation in mice. Stem Cell Res. Ther. 2022, 13, 155. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, B.; Zhang, J.; Wu, Z.; Yu, L.; Sun, Z. Chemokine (C-C Motif) Ligand 2/Chemokine Receptor 2 (CCR2) Axis Blockade to Delay Chondrocyte Hypertrophy as a Therapeutic Strategy for Osteoarthritis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2021, 27, e930053. [Google Scholar] [CrossRef]
- Choi, M.C.; Jo, J.; Park, J.; Kang, H.K.; Park, Y. NF-κB Signaling Pathways in Osteoarthritic Cartilage Destruction. Cells 2019, 8, 734. [Google Scholar] [CrossRef]
- Deng, Y.; Lu, J.; Li, W.; Wu, A.; Zhang, X.; Tong, W.; Ho, K.K.; Qin, L.; Song, H.; Mak, K.K. Reciprocal inhibition of YAP/TAZ and NF-κB regulates osteoarthritic cartilage degradation. Nat. Commun. 2018, 9, 4564. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. NF-κB in immunobiology. Cell Res. 2011, 21, 223–244. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- Fan, Y.; Mao, R.; Yang, J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Hayden, M.S.; Ghosh, S. Crosstalk in NF-κB signaling pathways. Nat. Immunol. 2011, 12, 695–708. [Google Scholar] [CrossRef]
- Morgan, M.J.; Liu, Z.G. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef]
- Sun, S.C. The noncanonical NF-κB pathway. Immunol. Rev. 2012, 246, 125–140. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Zarnegar, B.J.; Wang, Y.; Mahoney, D.J.; Dempsey, P.W.; Cheung, H.H.; He, J.; Shiba, T.; Yang, X.; Yeh, W.C.; Mak, T.W.; et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 2008, 9, 1371–1378. [Google Scholar] [CrossRef] [PubMed]
- Vallabhapurapu, S.; Matsuzawa, A.; Zhang, W.; Tseng, P.H.; Keats, J.J.; Wang, H.; Vignali, D.A.; Bergsagel, P.L.; Karin, M. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat. Immunol. 2008, 9, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Zhang, M.; Harhaj, E.W.; Sun, S.C. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 2004, 279, 26243–26250. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Harhaj, E.W.; Sun, S.C. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol. Cell 2001, 7, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Senftleben, U.; Cao, Y.; Xiao, G.; Greten, F.R.; Krähn, G.; Bonizzi, G.; Chen, Y.; Hu, Y.; Fong, A.; Sun, S.C.; et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 2001, 293, 1495–1499. [Google Scholar] [CrossRef]
- Park, D.R.; Kim, J.; Kim, G.M.; Lee, H.; Kim, M.; Hwang, D.; Lee, H.; Kim, H.S.; Kim, W.; Park, M.C.; et al. Osteoclast-associated receptor blockade prevents articular cartilage destruction via chondrocyte apoptosis regulation. Nat. Commun. 2020, 11, 4343. [Google Scholar] [CrossRef]
- Yang, H.; Wen, Y.; Zhang, M.; Liu, Q.; Zhang, H.; Zhang, J.; Lu, L.; Ye, T.; Bai, X.; Xiao, G.; et al. MTORC1 coordinates the autophagy and apoptosis signaling in articular chondrocytes in osteoarthritic temporomandibular joint. Autophagy 2020, 16, 271–288. [Google Scholar] [CrossRef]
- Kobayashi, H.; Chang, S.H.; Mori, D.; Itoh, S.; Hirata, M.; Hosaka, Y.; Taniguchi, Y.; Okada, K.; Mori, Y.; Yano, F.; et al. Biphasic regulation of chondrocytes by Rela through induction of anti-apoptotic and catabolic target genes. Nat. Commun. 2016, 7, 13336. [Google Scholar] [CrossRef]
- Wang, Y.; Li, D.L.; Zhang, X.B.; Duan, Y.H.; Wu, Z.H.; Hao, D.S.; Chen, B.S.; Qiu, G.X. Increase of TNFα-stimulated osteoarthritic chondrocytes apoptosis and decrease of matrix metalloproteinases 9 by NF-κB inhibition. Biomed. Environ. Sci. 2013, 26, 277–283. [Google Scholar] [CrossRef]
- Ijiri, K.; Zerbini, L.F.; Peng, H.; Otu, H.H.; Tsuchimochi, K.; Otero, M.; Dragomir, C.; Walsh, N.; Bierbaum, B.E.; Mattingly, D.; et al. Differential expression of GADD45beta in normal and osteoarthritic cartilage: Potential role in homeostasis of articular chondrocytes. Arthritis Rheum. 2008, 58, 2075–2087. [Google Scholar] [CrossRef]
- Murahashi, Y.; Yano, F.; Kobayashi, H.; Makii, Y.; Iba, K.; Yamashita, T.; Tanaka, S.; Saito, T. Intra-articular administration of IκBα kinase inhibitor suppresses mouse knee osteoarthritis via downregulation of the NF-κB/HIF-2α axis. Sci. Rep. 2018, 8, 16475. [Google Scholar] [CrossRef]
- Ryu, J.H.; Shin, Y.; Huh, Y.H.; Yang, S.; Chun, C.H.; Chun, J.S. Hypoxia-inducible factor-2α regulates Fas-mediated chondrocyte apoptosis during osteoarthritic cartilage destruction. Cell Death Differ. 2012, 19, 440–450. [Google Scholar] [CrossRef]
- Liu, Y.X.; Wang, G.D.; Wang, X.; Zhang, Y.L.; Zhang, T.L. Effects of TLR-2/NF-κB signaling pathway on the occurrence of degenerative knee osteoarthritis: An in vivo and in vitro study. Oncotarget 2017, 8, 38602–38617. [Google Scholar] [CrossRef]
- Abramson, S.B. Osteoarthritis and nitric oxide. Osteoarthr. Cartil. 2008, 16 (Suppl. S2), S15–S20. [Google Scholar] [CrossRef]
- Choi, Y.A.; Lee, D.J.; Lim, H.K.; Jeong, J.H.; Sonn, J.K.; Kang, S.S.; Baek, S.H. Interleukin-1beta stimulates matrix metalloproteinase-2 expression via a prostaglandin E2-dependent mechanism in human chondrocytes. Exp. Mol. Med. 2004, 36, 226–232. [Google Scholar] [CrossRef]
- Ye, W.; Zhong, Z.; Zhu, S.; Zheng, S.; Xiao, J.; Song, S.; Yu, H.; Wu, Q.; Lin, Z.; Chen, J. Advanced oxidation protein products induce catabolic effect through oxidant-dependent activation of NF-κ B pathway in human chondrocyte. Int. Immunopharmacol. 2016, 39, 149–157. [Google Scholar] [CrossRef]
- Thedieck, K.; Polak, P.; Kim, M.L.; Molle, K.D.; Cohen, A.; Jenö, P.; Arrieumerlou, C.; Hall, M.N. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS ONE 2007, 2, e1217. [Google Scholar] [CrossRef]
- Shimobayashi, M.; Hall, M.N. Making new contacts: The mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 2014, 15, 155–162. [Google Scholar] [CrossRef]
- Rosa, S.C.; Rufino, A.T.; Judas, F.; Tenreiro, C.; Lopes, M.C.; Mendes, A.F. Expression and function of the insulin receptor in normal and osteoarthritic human chondrocytes: Modulation of anabolic gene expression, glucose transport and GLUT-1 content by insulin. Osteoarthr. Cartil. 2011, 19, 719–727. [Google Scholar] [CrossRef]
- De Santis, M.C.; Gulluni, F.; Campa, C.C.; Martini, M.; Hirsch, E. Targeting PI3K signaling in cancer: Challenges and advances. Biochim. Biophys. Acta. Rev. Cancer 2019, 1871, 361–366. [Google Scholar] [CrossRef]
- Guo, H.; German, P.; Bai, S.; Barnes, S.; Guo, W.; Qi, X.; Lou, H.; Liang, J.; Jonasch, E.; Mills, G.B.; et al. The PI3K/AKT Pathway and Renal Cell Carcinoma. J. Genet. Genom. 2015, 42, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y.; Wang, D.; Xu, Y.; Dong, R.; Yang, Y.; Lv, Q.; Chen, X.; Zhang, Z. The Upstream Pathway of mTOR-Mediated Autophagy in Liver Diseases. Cells 2019, 8, 1597. [Google Scholar] [CrossRef] [PubMed]
- Downward, J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 2003, 3, 11–22. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Kempf, C.R.; Chappell, W.H.; Abrams, S.L.; Stivala, F.; Malaponte, G.; Nicoletti, F.; Libra, M.; Bäsecke, J.; et al. Therapeutic resistance resulting from mutations in Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR signaling pathways. J. Cell. Physiol. 2011, 226, 2762–2781. [Google Scholar] [CrossRef]
- Samatar, A.A.; Poulikakos, P.I. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat. Rev. Drug Discov. 2014, 13, 928–942. [Google Scholar] [CrossRef]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef]
- Xiang, H.; Zhang, J.; Lin, C.; Zhang, L.; Liu, B.; Ouyang, L. Targeting autophagy-related protein kinases for potential therapeutic purpose. Acta Pharm. Sin. B 2020, 10, 569–581. [Google Scholar] [CrossRef]
- Roczniak-Ferguson, A.; Petit, C.S.; Froehlich, F.; Qian, S.; Ky, J.; Angarola, B.; Walther, T.C.; Ferguson, S.M. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 2012, 5, ra42. [Google Scholar] [CrossRef]
- Kim, Y.M.; Jung, C.H.; Seo, M.; Kim, E.K.; Park, J.M.; Bae, S.S.; Kim, D.H. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol. Cell 2015, 57, 207–218. [Google Scholar] [CrossRef]
- Sun, K.; Luo, J.; Guo, J.; Yao, X.; Jing, X.; Guo, F. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: A narrative review. Osteoarthr. Cartil. 2020, 28, 400–409. [Google Scholar] [CrossRef]
- Park, C.; Jeong, J.W.; Lee, D.S.; Yim, M.J.; Lee, J.M.; Han, M.H.; Kim, S.; Kim, H.S.; Kim, G.Y.; Park, E.K.; et al. Sargassum serratifolium Extract Attenuates Interleukin-1β-Induced Oxidative Stress and Inflammatory Response in Chondrocytes by Suppressing the Activation of NF-κB, p38 MAPK, and PI3K/Akt. Int. J. Mol. Sci. 2018, 19, 2308. [Google Scholar] [CrossRef]
- Malemud, C.J. Negative Regulators of JAK/STAT Signaling in Rheumatoid Arthritis and Osteoarthritis. Int. J. Mol. Sci. 2017, 18, 484. [Google Scholar] [CrossRef]
- Pérez-Baos, S.; Prieto-Potin, I.; Román-Blas, J.A.; Sánchez-Pernaute, O.; Largo, R.; Herrero-Beaumont, G. Mediators and Patterns of Muscle Loss in Chronic Systemic Inflammation. Front. Physiol. 2018, 9, 409. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Wang, L.; He, C. Nrf2-mediated anti-inflammatory polarization of macrophages as therapeutic targets for osteoarthritis. Front. Immunol. 2022, 13, 967193. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Schwartz, D.M.; Villarino, A.V.; Gadina, M.; McInnes, I.B.; Laurence, A. The JAK-STAT pathway: Impact on human disease and therapeutic intervention. Annu. Rev. Med. 2015, 66, 311–328. [Google Scholar] [CrossRef]
- Durham, G.A.; Williams, J.J.L.; Nasim, M.T.; Palmer, T.M. Targeting SOCS Proteins to Control JAK-STAT Signalling in Disease. Trends Pharmacol. Sci. 2019, 40, 298–308. [Google Scholar] [CrossRef]
- Zhou, Q.; Ren, Q.; Jiao, L.; Huang, J.; Yi, J.; Chen, J.; Lai, J.; Ji, G.; Zheng, T. The potential roles of JAK/STAT signaling in the progression of osteoarthritis. Front. Endocrinol. 2022, 13, 1069057. [Google Scholar] [CrossRef]
- Bharadwaj, U.; Kasembeli, M.M.; Robinson, P.; Tweardy, D.J. Targeting Janus Kinases and Signal Transducer and Activator of Transcription 3 to Treat Inflammation, Fibrosis, and Cancer: Rationale, Progress, and Caution. Pharmacol. Rev. 2020, 72, 486–526. [Google Scholar] [CrossRef]
- Limnander, A.; Rothman, P.B. Abl oncogene bypasses normal regulation of Jak/STAT activation. Cell Cycle 2004, 3, 1486–1488. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Xue, Y. Silence of TANK-binding kinase 1 (TBK1) regulates extracellular matrix degradation of chondrocyte in osteoarthritis by janus kinase (JAK)-signal transducer of activators of transcription (STAT) signaling. Bioengineered 2022, 13, 1872–1879. [Google Scholar] [CrossRef] [PubMed]
- Grimmer, C.; Pfander, D.; Swoboda, B.; Aigner, T.; Mueller, L.; Hennig, F.F.; Gelse, K. Hypoxia-inducible factor 1alpha is involved in the prostaglandin metabolism of osteoarthritic cartilage through up-regulation of microsomal prostaglandin E synthase 1 in articular chondrocytes. Arthritis Rheum. 2007, 56, 4084–4094. [Google Scholar] [CrossRef] [PubMed]
- Bohensky, J.; Terkhorn, S.P.; Freeman, T.A.; Adams, C.S.; Garcia, J.A.; Shapiro, I.M.; Srinivas, V. Regulation of autophagy in human and murine cartilage: Hypoxia-inducible factor 2 suppresses chondrocyte autophagy. Arthritis Rheum. 2009, 60, 1406–1415. [Google Scholar] [CrossRef]
- Schipani, E.; Ryan, H.E.; Didrickson, S.; Kobayashi, T.; Knight, M.; Johnson, R.S. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 2001, 15, 2865–2876. [Google Scholar] [CrossRef]
- Bohensky, J.; Shapiro, I.M.; Leshinsky, S.; Terkhorn, S.P.; Adams, C.S.; Srinivas, V. HIF-1 regulation of chondrocyte apoptosis: Induction of the autophagic pathway. Autophagy 2007, 3, 207–214. [Google Scholar] [CrossRef]
- Bohensky, J.; Leshinsky, S.; Srinivas, V.; Shapiro, I.M. Chondrocyte autophagy is stimulated by HIF-1 dependent AMPK activation and mTOR suppression. Pediatr. Nephrol. 2010, 25, 633–642. [Google Scholar] [CrossRef]
- Hu, S.; Zhang, C.; Ni, L.; Huang, C.; Chen, D.; Shi, K.; Jin, H.; Zhang, K.; Li, Y.; Xie, L.; et al. Stabilization of HIF-1α alleviates osteoarthritis via enhancing mitophagy. Cell Death Dis. 2020, 11, 481. [Google Scholar] [CrossRef]
- Chen, G.; Gao, X.; Wang, J.; Yang, C.; Wang, Y.; Liu, Y.; Zou, W.; Liu, T. Hypoxia-induced microRNA-146a represses Bcl-2 through Traf6/IRAK1 but not Smad4 to promote chondrocyte autophagy. Biol. Chem. 2017, 398, 499–507. [Google Scholar] [CrossRef]
- Yudoh, K.; Nakamura, H.; Masuko-Hongo, K.; Kato, T.; Nishioka, K. Catabolic stress induces expression of hypoxia-inducible factor (HIF)-1 alpha in articular chondrocytes: Involvement of HIF-1 alpha in the pathogenesis of osteoarthritis. Arthritis Res. Ther. 2005, 7, R904–R914. [Google Scholar] [CrossRef]
- Chen, G.; Liu, T.; Yu, B.; Wang, B.; Peng, Q. CircRNA-UBE2G1 regulates LPS-induced osteoarthritis through miR-373/HIF-1a axis. Cell Cycle 2020, 19, 1696–1705. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Huang, Z.; Xing, R.; Li, X.; Yin, S.; Mao, J.; Zhang, N.; Mei, W.; Ding, L.; et al. Increased HIF-1α in Knee Osteoarthritis Aggravate Synovial Fibrosis via Fibroblast-Like Synoviocyte Pyroptosis. Oxidative Med. Cell. Longev. 2019, 2019, 6326517. [Google Scholar] [CrossRef]
- Liang, F.; Zhang, F.; Zhang, L.; Wei, W. The advances in pyroptosis initiated by inflammasome in inflammatory and immune diseases. Inflamm. Res. 2020, 69, 159–166. [Google Scholar] [CrossRef]
- Zhang, F.J.; Luo, W.; Lei, G.H. Role of HIF-1α and HIF-2α in osteoarthritis. Jt. Bone Spine 2015, 82, 144–147. [Google Scholar] [CrossRef]
- Hirata, M.; Kugimiya, F.; Fukai, A.; Saito, T.; Yano, F.; Ikeda, T.; Mabuchi, A.; Sapkota, B.R.; Akune, T.; Nishida, N.; et al. C/EBPβ and RUNX2 cooperate to degrade cartilage with MMP-13 as the target and HIF-2α as the inducer in chondrocytes. Hum. Mol. Genet. 2012, 21, 1111–1123. [Google Scholar] [CrossRef]
- Yang, S.; Kim, J.; Ryu, J.H.; Oh, H.; Chun, C.H.; Kim, B.J.; Min, B.H.; Chun, J.S. Hypoxia-inducible factor-2alpha is a catabolic regulator of osteoarthritic cartilage destruction. Nat. Med. 2010, 16, 687–693. [Google Scholar] [CrossRef]
- Saito, T.; Fukai, A.; Mabuchi, A.; Ikeda, T.; Yano, F.; Ohba, S.; Nishida, N.; Akune, T.; Yoshimura, N.; Nakagawa, T.; et al. Transcriptional regulation of endochondral ossification by HIF-2alpha during skeletal growth and osteoarthritis development. Nat. Med. 2010, 16, 678–686. [Google Scholar] [CrossRef]
- Zhou, Y.; Ming, J.; Deng, M.; Li, Y.; Li, B.; Li, J.; Ma, Y.; Chen, Z.; Wang, G.; Liu, S. Chemically modified curcumin (CMC2.24) alleviates osteoarthritis progression by restoring cartilage homeostasis and inhibiting chondrocyte apoptosis via the NF-κB/HIF-2α axis. J. Mol. Med. 2020, 98, 1479–1491. [Google Scholar] [CrossRef]
- Weng, T.; Xie, Y.; Yi, L.; Huang, J.; Luo, F.; Du, X.; Chen, L.; Liu, C.; Chen, D.; Chen, L. Loss of Vhl in cartilage accelerated the progression of age-associated and surgically induced murine osteoarthritis. Osteoarthr. Cartil. 2014, 22, 1197–1205. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, Y.; Sun, W.; Zhang, Z.; Liu, J.; Yang, W.; Yuan, W.; Yi, Y.; Wang, J.; Liu, J. D-mannose alleviates osteoarthritis progression by inhibiting chondrocyte ferroptosis in a HIF-2α-dependent manner. Cell Prolif. 2021, 54, e13134. [Google Scholar] [CrossRef]
- Huang, S.; Xing, Y.; Liu, Y. Emerging roles for the ER stress sensor IRE1α in metabolic regulation and disease. J. Biol. Chem. 2019, 294, 18726–18741. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, N.; Chiang, W.C.; Kurt, T.D.; Sigurdson, C.J.; Lin, J.H. Multiple Mechanisms of Unfolded Protein Response-Induced Cell Death. Am. J. Pathol. 2015, 185, 1800–1808. [Google Scholar] [CrossRef] [PubMed]
- Husa, M.; Petursson, F.; Lotz, M.; Terkeltaub, R.; Liu-Bryan, R. C/EBP homologous protein drives pro-catabolic responses in chondrocytes. Arthritis Res. Ther. 2013, 15, R218. [Google Scholar] [CrossRef] [PubMed]
- Oliver, B.L.; Cronin, C.G.; Zhang-Benoit, Y.; Goldring, M.B.; Tanzer, M.L. Divergent stress responses to IL-1beta, nitric oxide, and tunicamycin by chondrocytes. J. Cell. Physiol. 2005, 204, 45–50. [Google Scholar] [CrossRef]
- Briggs, M.D.; Dennis, E.P.; Dietmar, H.F.; Pirog, K.A. New developments in chondrocyte ER stress and related diseases. F1000Research 2020, 9, 22275. [Google Scholar] [CrossRef]
- Guo, F.J.; Xiong, Z.; Lu, X.; Ye, M.; Han, X.; Jiang, R. ATF6 upregulates XBP1S and inhibits ER stress-mediated apoptosis in osteoarthritis cartilage. Cell. Signal. 2014, 26, 332–342. [Google Scholar] [CrossRef]
- Huang, R.; Hui, Z.; Wei, S.; Li, D.; Li, W.; Daping, W.; Alahdal, M. IRE1 signaling regulates chondrocyte apoptosis and death fate in the osteoarthritis. J. Cell. Physiol. 2022, 237, 118–127. [Google Scholar] [CrossRef]
- Takada, K.; Hirose, J.; Senba, K.; Yamabe, S.; Oike, Y.; Gotoh, T.; Mizuta, H. Enhanced apoptotic and reduced protective response in chondrocytes following endoplasmic reticulum stress in osteoarthritic cartilage. Int. J. Exp. Pathol. 2011, 92, 232–242. [Google Scholar] [CrossRef]
- Tang, Y.H.; Yue, Z.S.; Zheng, W.J.; Shen, H.F.; Zeng, L.R.; Hu, Z.Q.; Xiong, Z.F. 4-Phenylbutyric acid presents therapeutic effect on osteoarthritis via inhibiting cell apoptosis and inflammatory response induced by endoplasmic reticulum stress. Biotechnol. Appl. Biochem. 2018, 65, 540–546. [Google Scholar] [CrossRef]
- Feng, K.; Ge, Y.; Chen, Z.; Li, X.; Liu, Z.; Li, X.; Li, H.; Tang, T.; Yang, F.; Wang, X. Curcumin Inhibits the PERK-eIF2α-CHOP Pathway through Promoting SIRT1 Expression in Oxidative Stress-induced Rat Chondrocytes and Ameliorates Osteoarthritis Progression in a Rat Model. Oxidative Med. Cell. Longev. 2019, 2019, 8574386. [Google Scholar] [CrossRef]
- Uehara, Y.; Hirose, J.; Yamabe, S.; Okamoto, N.; Okada, T.; Oyadomari, S.; Mizuta, H. Endoplasmic reticulum stress-induced apoptosis contributes to articular cartilage degeneration via C/EBP homologous protein. Osteoarthr. Cartil. 2014, 22, 1007–1017. [Google Scholar] [CrossRef]
- Wu, L.; Liu, H.; Li, L.; Xu, D.; Gao, Y.; Guan, Y.; Chen, Q. 5,7,3′,4′-Tetramethoxyflavone protects chondrocytes from ER stress-induced apoptosis through regulation of the IRE1α pathway. Connect. Tissue Res. 2018, 59, 157–166. [Google Scholar] [CrossRef]
- Siwecka, N.; Rozpędek-Kamińska, W.; Wawrzynkiewicz, A.; Pytel, D.; Diehl, J.A.; Majsterek, I. The Structure, Activation and Signaling of IRE1 and Its Role in Determining Cell Fate. Biomedicines 2021, 9, 156. [Google Scholar] [CrossRef]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Hollien, J.; Lin, J.H.; Li, H.; Stevens, N.; Walter, P.; Weissman, J.S. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009, 186, 323–331. [Google Scholar] [CrossRef]
- Hollien, J.; Weissman, J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006, 313, 104–107. [Google Scholar] [CrossRef]
- Maurel, M.; Chevet, E.; Tavernier, J.; Gerlo, S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014, 39, 245–254. [Google Scholar] [CrossRef]
- Nishitoh, H.; Matsuzawa, A.; Tobiume, K.; Saegusa, K.; Takeda, K.; Inoue, K.; Hori, S.; Kakizuka, A.; Ichijo, H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002, 16, 1345–1355. [Google Scholar] [CrossRef]
- Urano, F.; Wang, X.; Bertolotti, A.; Zhang, Y.; Chung, P.; Harding, H.P.; Ron, D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 2000, 287, 664–666. [Google Scholar] [CrossRef]
- Wang, Q.; Luo, S.; Yang, J.; Li, J.; Huan, S.; She, G.; Zha, Z. Circ_0114876 promoted IL-1β-induced chondrocyte injury by targeting miR-671/TRAF2 axis. Biotechnol. Lett. 2021, 43, 791–802. [Google Scholar] [CrossRef]
- Hochberg, M.C. Osteoarthritis year 2012 in review: Clinical. Osteoarthr. Cartil. 2012, 20, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Messier, S.P.; Legault, C.; Loeser, R.F.; Van Arsdale, S.J.; Davis, C.; Ettinger, W.H.; DeVita, P. Does high weight loss in older adults with knee osteoarthritis affect bone-on-bone joint loads and muscle forces during walking? Osteoarthr. Cartil. 2011, 19, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yue, J.; Golianu, B.; Sun, Z.; Lu, Y. Updated systematic review and meta-analysis of acupuncture for chronic knee pain. Acupunct. Med. 2017, 35, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Rutjes, A.W.; Nüesch, E.; Sterchi, R.; Jüni, P. Therapeutic ultrasound for osteoarthritis of the knee or hip. Cochrane Database Syst. Rev. 2010, 1, Cd003132. [Google Scholar] [CrossRef] [PubMed]
- Welch, V.; Brosseau, L.; Peterson, J.; Shea, B.; Tugwell, P.; Wells, G. Therapeutic ultrasound for osteoarthritis of the knee. Cochrane Database Syst. Rev. 2001, 4, Cd003132. [Google Scholar] [CrossRef]
- Yu, S.P.; Hunter, D.J. Managing osteoarthritis. Aust. Prescr. 2015, 38, 115–119. [Google Scholar] [CrossRef]
- Mobasheri, A. The future of osteoarthritis therapeutics: Emerging biological therapy. Curr. Rheumatol. Rep. 2013, 15, 385. [Google Scholar] [CrossRef]
- Liu, G.; Liu, Q.; Yan, B.; Zhu, Z.; Xu, Y. USP7 Inhibition Alleviates H2O2-Induced Injury in Chondrocytes via Inhibiting NOX4/NLRP3 Pathway. Front. Pharmacol. 2020, 11, 617270. [Google Scholar] [CrossRef]
- Li, C.; Li, L.; Lan, T. Co-treatment with disulfiram and glycyrrhizic acid suppresses the inflammatory response of chondrocytes. J. Orthop. Surg. Res. 2021, 16, 132. [Google Scholar] [CrossRef]
- Marcu, K.B.; Otero, M.; Olivotto, E.; Borzi, R.M.; Goldring, M.B. NF-kappaB signaling: Multiple angles to target OA. Curr. Drug Targets 2010, 11, 599–613. [Google Scholar] [CrossRef]
- Huang, C.Y.; Lai, K.Y.; Hung, L.F.; Wu, W.L.; Liu, F.C.; Ho, L.J. Advanced glycation end products cause collagen II reduction by activating Janus kinase/signal transducer and activator of transcription 3 pathway in porcine chondrocytes. Rheumatology 2011, 50, 1379–1389. [Google Scholar] [CrossRef]
- Lim, H.; Kim, H.P. Matrix metalloproteinase-13 expression in IL-1β-treated chondrocytes by activation of the p38 MAPK/c-Fos/AP-1 and JAK/STAT pathways. Arch. Pharmacal Res. 2011, 34, 109–117. [Google Scholar] [CrossRef]
- Malemud, C.J. Matrix Metalloproteinases and Synovial Joint Pathology. Prog. Mol. Biol. Transl. Sci. 2017, 148, 305–325. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, Q.; Wang, K. Artesunate attenuates ACLT-induced osteoarthritis by suppressing osteoclastogenesis and aberrant angiogenesis. Biomed. Pharmacother. 2017, 96, 410–416. [Google Scholar] [CrossRef]
- Kong, Y.; Zhang, Y.; Zhao, X.; Wang, G.; Liu, Q. Carboxymethyl-chitosan attenuates inducible nitric oxide synthase and promotes interleukin-10 production in rat chondrocytes. Exp. Ther. Med. 2017, 14, 5641–5646. [Google Scholar] [CrossRef]
- Aubourg, G.; Rice, S.J.; Bruce-Wootton, P.; Loughlin, J. Genetics of osteoarthritis. Osteoarthr. Cartil. 2022, 30, 636–649. [Google Scholar] [CrossRef]
- Jang, S.; Lee, K.; Ju, J.H. Recent Updates of Diagnosis, Pathophysiology, and Treatment on Osteoarthritis of the Knee. Int. J. Mol. Sci. 2021, 22, 2619. [Google Scholar] [CrossRef]
- Pourcho, A.M.; Smith, J.; Wisniewski, S.J.; Sellon, J.L. Intraarticular platelet-rich plasma injection in the treatment of knee osteoarthritis: Review and recommendations. Am. J. Phys. Med. Rehabil. 2014, 93 (Suppl. S3), S108–S121. [Google Scholar] [CrossRef]
- Katz, J.N.; Arant, K.R.; Loeser, R.F. Diagnosis and Treatment of Hip and Knee Osteoarthritis: A Review. JAMA 2021, 325, 568–578. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Pan, Y.; Li, T.; Zou, M.; Liu, W.; Li, Q.; Wan, H.; Peng, J.; Hao, L. The Role of Regulated Programmed Cell Death in Osteoarthritis: From Pathogenesis to Therapy. Int. J. Mol. Sci. 2023, 24, 5364. https://doi.org/10.3390/ijms24065364
Liu S, Pan Y, Li T, Zou M, Liu W, Li Q, Wan H, Peng J, Hao L. The Role of Regulated Programmed Cell Death in Osteoarthritis: From Pathogenesis to Therapy. International Journal of Molecular Sciences. 2023; 24(6):5364. https://doi.org/10.3390/ijms24065364
Chicago/Turabian StyleLiu, Suqing, Yurong Pan, Ting Li, Mi Zou, Wenji Liu, Qingqing Li, Huan Wan, Jie Peng, and Liang Hao. 2023. "The Role of Regulated Programmed Cell Death in Osteoarthritis: From Pathogenesis to Therapy" International Journal of Molecular Sciences 24, no. 6: 5364. https://doi.org/10.3390/ijms24065364
APA StyleLiu, S., Pan, Y., Li, T., Zou, M., Liu, W., Li, Q., Wan, H., Peng, J., & Hao, L. (2023). The Role of Regulated Programmed Cell Death in Osteoarthritis: From Pathogenesis to Therapy. International Journal of Molecular Sciences, 24(6), 5364. https://doi.org/10.3390/ijms24065364







