Choosing the Right Cell Line for Acute Myeloid Leukemia (AML) Research
Abstract
1. Introduction
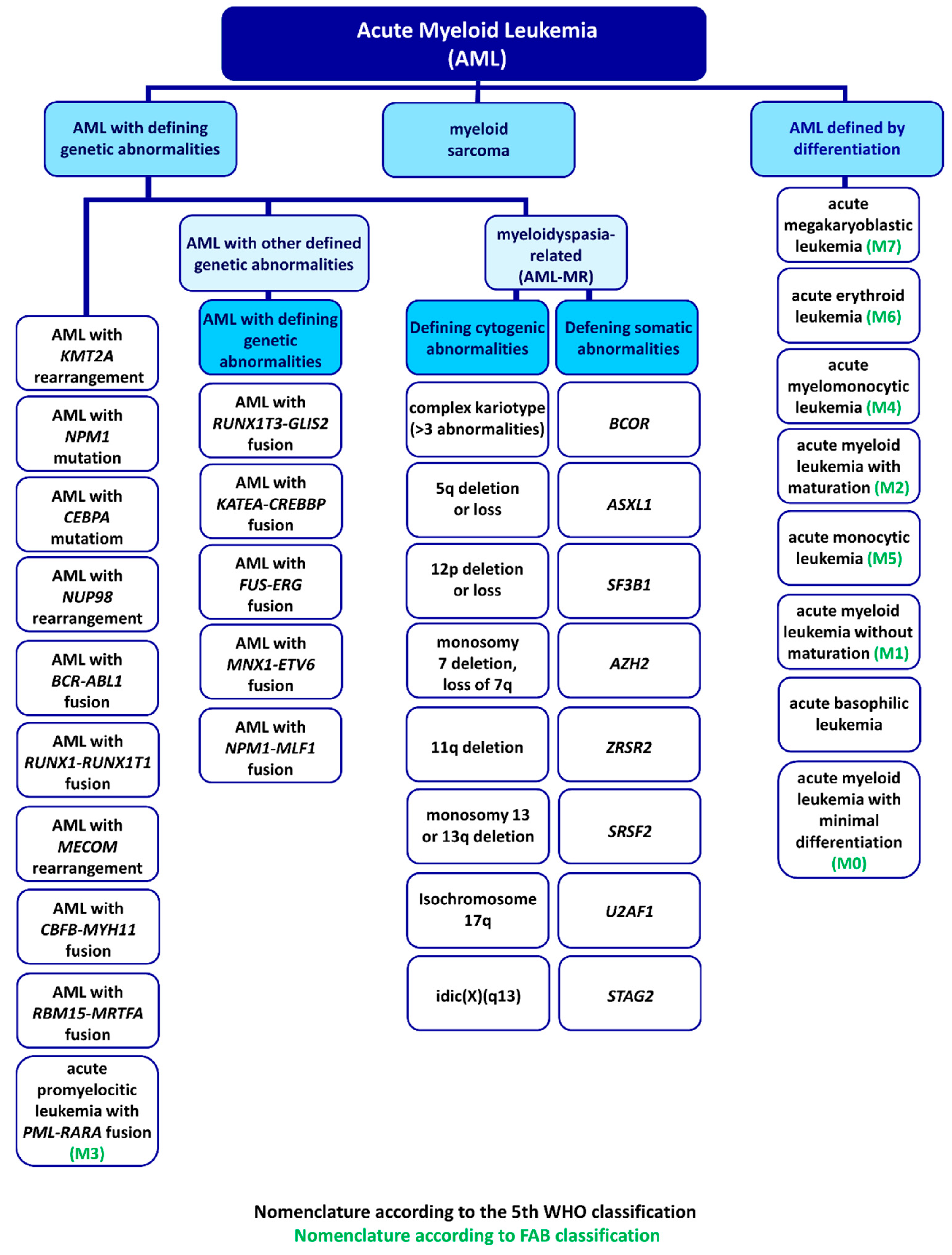
2. FAB Classification and Diagnostic Process
3. WHO Classification
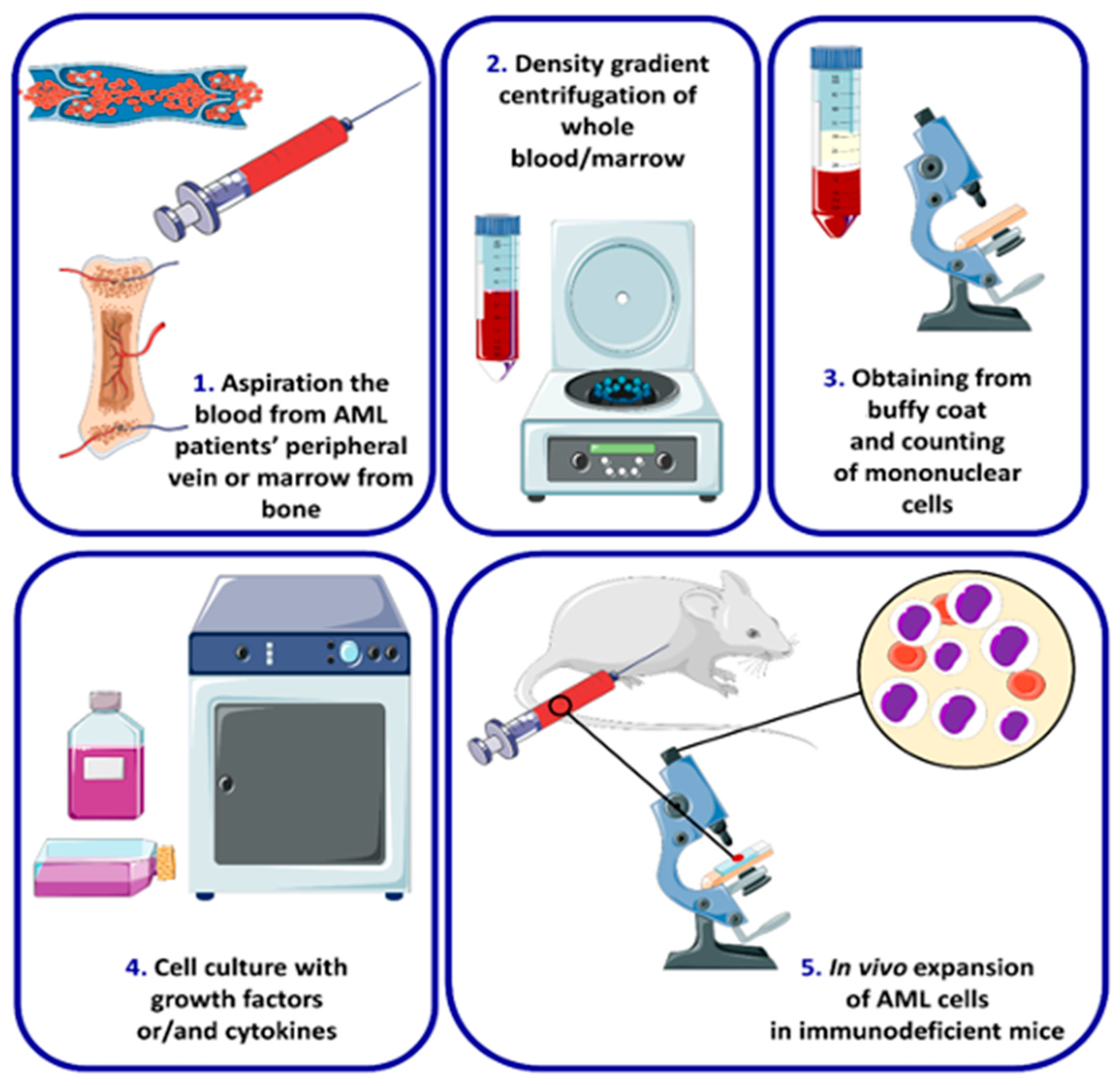
4. AML Cell Lines and Types
4.1. AML with Minimal Differentiation (M0)
4.1.1. Kasumi-3
4.1.2. MOLM-16
4.2. AML without Maturation (M1)
4.2.1. CTS
4.2.2. UoC-M1
4.2.3. KG-1
4.2.4. K-562
4.3. AML with Maturation (M2)
4.3.1. Kasumi-1
4.3.2. Kasumi-6
4.3.3. SKNO-1
4.3.4. HL-60
4.3.5. PLB-985
4.4. Acute Promyelocytic Leukemia (M3)
4.4.1. NB4
4.4.2. PL-21
4.4.3. UF-1
4.4.4. HT93
4.4.5. AP-1060
4.5. Acute Myelomonocytic Leukemia (M4)
4.5.1. OCI-AML2 and OCI-AML3
4.5.2. MUTZ-11
4.5.3. MUTZ-8
4.5.4. MUTZ-3
4.5.5. ME-1
4.6. Acute Monoblastic and Monocytic Leukemia (M5)
4.6.1. THP-1
4.6.2. U-937
4.6.3. MOLM-13 and MV4-11
4.7. Acute Erythroid Leukemia (M6)
4.7.1. HEL
4.7.2. OCI-M1
4.7.3. OCI-M2
4.7.4. F-36P
4.7.5. TF-1
4.7.6. AS-E2
4.8. Acute Megakaryoblastic Leukemia (M7)
4.8.1. CMK
4.8.2. ELF-153
4.8.3. UT-7
4.8.4. M-07
4.8.5. MEG-01
4.8.6. MEGAL
4.9. Cell-Line Markers
| Disease | Cell Line | Markers | Use |
|---|---|---|---|
| M0 | Kasumi-3 | CD2−, cy/smCD3−, CD4+, CD5−, CD7+, CD8−, CD13+, CD14+, CD15−, CD19−, CD20−, CD22+, CD25+, CD33+, CD34+,CD38+, CD56+, cyCD68+, HLA-DR+, c-Kit+ | EVI1 and BET inhibitors research; |
| Drug response in AML; | |||
| Engraftment studies | |||
| MOLM-16 | CD3−, CD9+, CD13+, CD19−, CD22+ CD31+, CD33+, CD34+, CD36+, CD38+, CD41+, CD47+, CD56+, CD61+, CD62P+, CD63+, CD71+, CD110+, CD117+, CD119+ CD151+, CD235A+, thrombospondin+, vWf+, fibrinogen+, HLA-DR- | t(6;8) (q21;q24.3) model; | |
| PIM/FLT3 signaling; | |||
| JAK2 V617F function research; | |||
| PMS2 and RSPH10B2 deletion | |||
| M1 | CTS | CD1+, CD2+, CD3+, CD4+, CD5+, CD7+, CD8+, CD10+, CD13+, CD14+, CD19+, CD20+, CD25+, CD33+, CD34+, HLA-DR+, D2-10+, P2+, HPCA-1+ | t(6;11) (q27;q23) model; |
| KMT2/AF6 research model; | |||
| GM-CSF and G-CSF differentiation; | |||
| Pluripotent stem cell research | |||
| UoC-M1 | CD7+, CD24+, CD34+, CD38+, CD45+, HLA-DR+ CD61+ | Monosomy 7 and 5q loss model; | |
| High KMT2A mRNA level | |||
| KG1 | CD3−, CD13+, CD14−, CD15+, CD19−, CD33+, CD34+, HLA-DR+ | Cell maturation studies; | |
| KMT2A and WT DNMT3A research; | |||
| Toxicology and drug testing; | |||
| Macrophage differentiation; | |||
| K-562 | CD3−, CD14−, CD15+, CD19−, CD33+, CD71+, CD235a+ | BCR-ABL1 fusion, Ph1 chromosome; | |
| Platelet-formation; | |||
| p53-deficient | |||
| M2 | Kasumi-1 | CD3−, CD4+, CD13+, CD14−, CD15+, CD19−, CD33+, CD34+, CD38+, CD71+, HLA-DR+ | t(8;21) model; |
| RUNX1-RUNX1T1 fusion research; | |||
| c-kit, TP53 mutations; | |||
| Granulocytic and macrophage differentiation; | |||
| Il-5 and TPA-induced differentiation; | |||
| Kasumi-6 | CD3−, CD4−, CD13+, CD14−, CD19−, CD33+, CD34−, cyCD68−, HLA-DR+ | FLT3, CEBPA, TP53 mutations; | |
| Model for differentiation research | |||
| TPA-induced differentiation | |||
| SKNO-1 | CD3−, CD4+, CD13+, CD14−, CD15−, CD19-, CD33+ CD1, CD2, CD3, CD4, CD5, CD6, CD7, CD8, CD11, CD13, CD14, CD15, CD19, CD20, CD33, CD34, HLA-DR | t(8;21) (q22;q22) research; | |
| RUNX1/RUNX1T1 fusion; | |||
| Myeloid leukemogenesis studies; | |||
| HL-60 | CD3−, CD4+, CD13+, CD14−, CD15+, CD19−, CD33+, CD34−, HLA-DR- | t(15;17) model; | |
| Granulocytic and mononuclear maturation; Chemotherapeutics influence; | |||
| Proliferation, apoptosis, and cell cycle study; | |||
| Chemotactic response; | |||
| miRNA studies | |||
| PLB-985 | CD3−, CD4+, CD13+, CD14−, CD15+, CD19−, CD33+, CD34− | Granulocytic, monocytic, macrophage maturation, proliferation; | |
| Neutrophil differentiation; | |||
| Maturation studies of cells in early stage | |||
| M3 | NB4 | CD3−, CD4+, CD11b−, CD13+, CD14−, CD15+, CD19−, CD33+, CD34−, CD38+, HLA-DR- | ATRA resistance mechanisms |
| PML-RARAPro900Ser mutation; | |||
| Retinoic acid, DMSO, TPA differentiation; | |||
| Drug screening | |||
| PL-21 | CD3−, CD4 (+), CD14−, CD15+, CD19−, CD33+, cyCD68+, HLA-DR- | Lack of t(15;17) | |
| KRAS, FLT3 mutations and WT P53; | |||
| Kinase inhibitors studies | |||
| UF-1 | CD3−, CD4−, CD5−, CD8−, CD11b−, CD10−, CD7+, CD13+, CD19−, CD20−, CD33+, CD34−, CD38+, CD41− [82] | WT RARA; | |
| PML-RARA research; | |||
| ATRA-resistance studies; | |||
| Multi-drug screening with ATRA | |||
| HT93 | CD3−, CD19−, CD33+, CD34+, cyCD68+, HLA-DR− | t(15;17) and t(1;12) model with PML-RARA and | |
| ETV6-ABL2 fusion; | |||
| TP53 mutation; | |||
| Differentiation, proliferation, cytokine studies | |||
| AP-1060 | CD3−, CD14−, CD15+, CD19−, CD33+, cyCD68+, HLA-DR− | t(15;17) and unique t(3;14) model; | |
| PML-RARAPro900Leu mutation; | |||
| ATRA and ATO resistance; | |||
| Cytokine-dependent growth research; | |||
| ETV6-NTRK3 fusion model; | |||
| Neutrophil maturation | |||
| M4 | OCI-AML2 | CD3−, CD4+, CD13+, CD15+, CD19−, CD33+, CD34−, cyCD68+, HLA-DR+ | Mutated DNMT3A role in leukemogenesis; |
| xenograft models | |||
| OCI-AML3 | CD3−, CD4+, CD13+, CD14−, CD15+, CD19−, cyCD68+, HLA-DR- | Mutated DNMT3A role in leukemogenesis; xenograft models; | |
| NPM1 mutation; | |||
| MUTZ-11 | CD4+, CD7+, CD13+, CD15+, CD33+, CD65+, CD68+ [102] | Response to cytokines; | |
| Dendritic cell myeloid differentiation; | |||
| KMT2A and FLT3 mutations | |||
| MUTZ-8 | CD3−, CD4−, CD13+, CD19−, CD33+, CD34+, HLA-DR+ | t(5;11) model; | |
| JAK2 V617F mutation; | |||
| Cytokine response | |||
| MUTZ-3 | CD3−, CD4+, CD5−, CD7−, CD8−, CD13+, CD14+, CD15+, CD19−, CD34+, HLA-DR+ | Role of FLT3 in leukemia pathogenesis; Cytokine response; | |
| Dendritic cell myeloid differentiation | |||
| M5 | THP-1 | CD3−, CD4+, CD13+, CD15+, CD19−, CD34−, cyCD68+, HLA-DR+ | t(9;11) (p22;q23) with KMT2A/MLLT3(AF9) fusion model; |
| Immune and inflammatory response; inflammation; | |||
| Susceptible to genetic modifications; | |||
| Skin sensitization model; | |||
| Cytokine response; | |||
| U-937 | CD3−, CD4+, CD14−, CD15+, CD19−, CD33+, CD34−, CD54+ | Monocyte and macrophage differentiation; Response to ROS; | |
| Skin sensitization model; | |||
| Model for cell apoptotic disintegration | |||
| MOLM-13 | CD3−, CD4+, CD14−, CD15+, CD19−, CD33+, CD34−, CD68+, HLA-DR− | Drug resistance research; Leukemia xenograft models; | |
| WT TP53, FLT3-ITD+ in AML | |||
| MV4-11 | CD3−, CD4+, CD5−, CD8−, CD10−, CD14−, CD15+, CD19−, CD33+, CD34− | t(4;11) (q21;q23) model; | |
| Mechanisms of FLT3-ITD+ AML Leukemia; Xenograft models; | |||
| WT TP53, FLT3-ITD+ in AML | |||
| M6 | HEL | CD3−, CD13+, CD14−, CD19−, CD33+, CD41a+, CD71+, CD235a+ | JAK/STAT signaling pathway, |
| Differential globins expression | |||
| JAK2 mutation | |||
| OCI-M1 | CD3−, CD4−, CD15+, CD19−, CD33+, CD34−, CD41−, CD42−, CD71+, HLA-DR+ | Model for the EPO receptors studies, | |
| OCI-M2 | CD3+, CD14+, CD19−, CD33 (+), CD71+ | Studying NFkB inhibitors; | |
| NKX2-4 expression | |||
| F-36P | CD3−, CD13+, CD14−, CD15−, CD19−, CD33+, CD34+, CD41−, CD42−, CD71+, CD235a+ | Study of IL-3 and GM-CSF dependence; | |
| Primitive progenitor cell model; | |||
| Myeloid cell differentiation; | |||
| Oncogenesis | |||
| TF-1 | CD3−, CD13+, CD14−, CD15−, CD19−, CD33+, CD34+, CD71+, HLA-DR+ | TNFR2 expression; | |
| TPA macrophage differentiation; | |||
| Cytokines response; | |||
| Anti-IL-6R nanobody ALX-0061 | |||
| AS-E2 | CD2−, CD3−, CD10−, CD11b+, CD13+, CD19−, CD25−, CD33+, CD36+, CD41−, Glycophorin A+, CD71+ [199] | Erythroid progenitor cells; | |
| EPO-dependent growth; | |||
| GATA-1 expression | |||
| M7 | CMK | CD3−, CD13+, CD14−, CD15−, CD19−, CD33+, CD34+, CD71+, CD235a+ | der(17)t(11:17) model; |
| JAK2 V617F mutation; | |||
| Megakaryocytopoiesis research; | |||
| ELF-153 | CD3−, CD4+, CD13+, CD14−, CD15−, CD19−, CD33+, CD34+, HLA-DR+ | High GATA level; | |
| Megakaryocytopoiesis research | |||
| UT-7 | CD3−, CD13+, CD14−, CD15−, CD19−, CD33+, CD34−, cyCD68+ | Response to cytokines; | |
| EPO-response; | |||
| GATA expression, | |||
| M-07 | Non disclosed | Cytokine response | |
| M-07e | CD3−, CD13+, CD14−, CD19−, CD33+, HLA-DR- | Cytokine response | |
| MEG-01 | CD3−, CD13+, CD15+, CD19−, CD33+ | t(9;22) model; | |
| BCR/CBL fusion; | |||
| Megakaryocytic differentiation/maturation | |||
| MEGAL | CD3−, CD13−, CD14−, CD19−, CD33+, CD34+, CD71+, CD235a− | SET-NUP214 fusion |
4.10. Controls from Healthy Donors
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.; Gralnick, H.R.; Sultan, C. Proposals for the Classification of the Acute Leukaemias. French-American-British (FAB) Co-Operative Group. Br. J. Haematol. 1976, 33, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.; Gralnick, H.R.; Sultan, C. Proposed Revised Criteria for the Classification of Acute Myeloid Leukemia. A Report of the French-American-British Cooperative Group. Ann. Intern. Med. 1985, 103, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Naeim, F.; Rao, P.N. Chapter 11—Acute Myeloid Leukemia. In Hematopathology; Naeim, F., Rao, P.N., Grody, W.W., Eds.; Academic Press: Oxford, UK, 2008; pp. 207–255. [Google Scholar] [CrossRef]
- Ladines-Castro, W.; Barragán-Ibañez, G.; Luna-Pérez, M.A.; Santoyo-Sánchez, A.; Collazo-Jaloma, J.; Mendoza-García, E.; Ramos-Peñafiel, C.O. Morphology of Leukaemias. Rev. Médica Hosp. Gen. México 2016, 79, 107–113. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, H.; Abdullah, W.Z.; Yong, A.C.; Ahmed, S.A.; Abdullah, A.D.; Baba, A.A.; Ankathil, R.; Husin, A.; Hussein, A.R.; Mustaffa, R.; et al. A Review of AML Classification: A Single Institution Experience in a Developing Country. J. Hematopathol. 2014, 7, 3–8. [Google Scholar] [CrossRef]
- Dozzo, A.; Galvin, A.; Shin, J.-W.; Scalia, S.; O’Driscoll, C.M.; Ryan, K.B. Modelling Acute Myeloid Leukemia (AML): What’s New? A Transition from the Classical to the Modern. Drug Deliv. Transl. Res. 2022. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Pollak, A.; Leavitt, R.D.; Testa, J.R.; Schiffer, C.A. Minimally Differentiated Acute Nonlymphocytic Leukemia: A Distinct Entity. Blood 1987, 70, 1400–1406. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.L.; Hoyer, J.D.; Kurtin, P.J.; Dewald, G.W.; Hanson, C.A. Acute Myeloid Leukemia with Minimal Differentiation. A Multiple Parameter Study. Am. J. Clin. Pathol. 1998, 109, 32–38. [Google Scholar] [CrossRef]
- Shanmugam, V.; Dorfman, D.M. Acute Myeloid Leukemia with Minimal Differentiation (AML M0) Mimicking Acute Lymphoblastic Leukemia. Am. J. Hematol. 2019, 94, 955–956. [Google Scholar] [CrossRef]
- Asou, H.; Tashiro, S.; Hamamoto, K.; Otsuji, A.; Kita, K.; Kamada, N. Establishment of a Human Acute Myeloid Leukemia Cell Line (Kasumi-1) With 8;21 Chromosome Translocation. Blood 1991, 77, 2031–2036. [Google Scholar] [CrossRef]
- Kasai, F.; Asou, H.; Ozawa, M.; Kobayashi, K.; Kuramitsu, H.; Satoh, M.; Kohara, A.; Kaneko, Y.; Kawamura, M. Kasumi Leukemia Cell Lines: Characterization of Tumor Genomes with Ethnic Origin and Scales of Genomic Alterations. Hum. Cell 2020, 33, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Wang, J. EVI1 in Leukemia and Solid Tumors. Cancers 2020, 12, 2667. [Google Scholar] [CrossRef]
- Birdwell, C.; Fiskus, W.; Kadia, T.M.; DiNardo, C.D.; Mill, C.P.; Bhalla, K.N. EVI1 Dysregulation: Impact on Biology and Therapy of Myeloid Malignancies. Blood Cancer J. 2021, 11, 64. [Google Scholar] [CrossRef]
- Chin, D.; Pietsch, C.; McCabe, F.; Chippari, S.; Kaiser, E.; Hanson, R.; Lubomirski, M. Abstract 90: Development of a Systemic Kasumi-3 Acute Myeloid Leukemia Model in NSG Mice. Cancer Res. 2014, 74 (Suppl. S19), 90. [Google Scholar] [CrossRef]
- Matsuo, Y.; Drexler, H.G.; Kaneda, K.; Kojima, K.; Ohtsuki, Y.; Hara, M.; Yasukawa, M.; Tanimoto, M.; Orita, K. Megakaryoblastic Leukemia Cell Line MOLM-16 Derived from Minimally Differentiated Acute Leukemia with Myeloid/NK Precursor Phenotype. Leuk. Res. 2003, 27, 165–171. [Google Scholar] [CrossRef]
- Kaashoek, J.G.; Mout, R.; Falkenburg, J.H.; Willemze, R.; Fibbe, W.E.; Landegent, J.E. Cytokine Production by the Bladder Carcinoma Cell Line 5637: Rapid Analysis of MRNA Expression Levels Using a CDNA-PCR Procedure. Lymphokine Cytokine Res. 1991, 10, 231–235. [Google Scholar] [PubMed]
- Bruyere, H.; Al Moosawi, M. t(6;8) (p21;q24) MYC/SUPT3H. Atlasgeneticsoncology.org. Available online: https://atlasgeneticsoncology.org/haematological/2987/t(6;8)p21;q24) (accessed on 13 December 2022).
- Czardybon, W.; Windak, R.; Dolata, I.; Salwińska, M.; Szydlowski, M.; Sewastianik, T.; Białopiotrowicz, E.; Mądro, E.; Lech-Marańda, E.; Budziszewska, B.K.; et al. Abstract 1749: Preclinical Characterization of SEL24-B489, a Dual PIM/FLT3 Inhibitor for the Treatment of Hematological Malignancies. Cancer Res. 2014, 74 (Suppl. S19), 1749. [Google Scholar] [CrossRef]
- Quentmeier, H.; Reinhardt, J.; Zaborski, M.; Drexler, H.G. FLT3 Mutations in Acute Myeloid Leukemia Cell Lines. Leukemia 2003, 17, 120–124. [Google Scholar] [CrossRef]
- Gonçalves, E.; Poulos, R.C.; Cai, Z.; Barthorpe, S.; Manda, S.S.; Lucas, N.; Beck, A.; Bucio-Noble, D.; Dausmann, M.; Hall, C.; et al. Pan-Cancer Proteomic Map of 949 Human Cell Lines. Cancer Cell 2022, 40, 835–849.e8. [Google Scholar] [CrossRef] [PubMed]
- Kakuda, H.; Sato, T.; Hayashi, Y.; Enomoto, Y.; Takayama, J.; Ohira, M.; Seto, M.; Ueda, R.; Fuse, A.; Niimi, H. A Novel Human Leukaemic Cell Line, CTS, Has a t(6;11) Chromosomal Translocation and Characteristics of Pluripotent Stem Cells. Br. J. Haematol. 1996, 95, 306–318. [Google Scholar] [CrossRef]
- Allen, R.J.; Smith, S.D.; Moldwin, R.L.; Lu, M.M.; Giordano, L.; Vignon, C.; Suto, Y.; Harden, A.; Tomek, R.; Veldman, T.; et al. Establishment and Characterization of a Megakaryoblast Cell Line with Amplification of MLL. Leukemia 1998, 12, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Koeffler, H.P.; Golde, D.W. Acute Myelogenous Leukemia: A Human Cell Line Responsive to Colony-Stimulating Activity. Science 1978, 200, 1153–1154. [Google Scholar] [CrossRef] [PubMed]
- Koya, J.; Kataoka, K.; Sato, T.; Bando, M.; Kato, Y.; Tsuruta-Kishino, T.; Kobayashi, H.; Narukawa, K.; Miyoshi, H.; Shirahige, K.; et al. DNMT3A R882 Mutants Interact with Polycomb Proteins to Block Haematopoietic Stem and Leukaemic Cell Differentiation. Nat. Commun. 2016, 7, 10924. [Google Scholar] [CrossRef]
- Rau, R.E.; Rodriguez, B.A.; Luo, M.; Jeong, M.; Rosen, A.; Rogers, J.H.; Campbell, C.T.; Daigle, S.R.; Deng, L.; Song, Y.; et al. DOT1L as a Therapeutic Target for the Treatment of DNMT3A-Mutant Acute Myeloid Leukemia. Blood 2016, 128, 971–981. [Google Scholar] [CrossRef]
- Mouly, E.; Planquette, C.; Rousseau, E.; Delansorne, R. Abstract 1890: Inecalcitol Respectively Induces or Increases CD38 Expression at the Surface of CD38- or CD38+ AML Cell Lines Representative of All 9 FAB Subtypes except M6. Cancer Res. 2018, 78 (Suppl. S13), 1890. [Google Scholar] [CrossRef]
- Sarde, A.; Eckard, S.; Mei, L.; Ruegg, C.; Chun, P.; Smith, V. Selectivity of T Cell Engager AMV564 Against Different Leukemic Blast Populations and Potential Application for Patient Selection. Blood 2020, 136, 25–26. [Google Scholar] [CrossRef]
- Leung, K.K.; Nguyen, A.; Shi, T.; Tang, L.; Ni, X.; Escoubet, L.; MacBeth, K.J.; DiMartino, J.; Wells, J.A. Multi-Omics Analysis of AML Cells Treated with Azacitidine Reveals Highly Variable Cell Surface Proteome Remodeling. bioRxiv 2018, 369322. [Google Scholar] [CrossRef]
- Liu, W.; Cheng, F. Circular RNA CircCRKL Inhibits the Proliferation of Acute Myeloid Leukemia Cells via the MiR-196a-5p/MiR-196b-5p/P27 Axis. Bioengineered 2021, 12, 7704–7713. [Google Scholar] [CrossRef]
- Koeffler, H.P.; Billing, R.; Lusis, A.J.; Sparkes, R.; Golde, D.W. An Undifferentiated Variant Derived from the Human Acute Myelogenous Leukemia Cell Line (KG-1). Blood 1980, 56, 265–273. [Google Scholar] [CrossRef]
- Li, Y.; Feng, Y.; Si, X.; Zhao, C.; Wang, F.; Niu, X. Genetic Expression Screening of Arsenic Trioxide-Induced Cytotoxicity in KG-1a Cells Based on Bioinformatics Technology. Front. Genet. 2021, 12, 654826. [Google Scholar] [CrossRef]
- Leung, K.K.; Nguyen, A.; Shi, T.; Tang, L.; Ni, X.; Escoubet, L.; MacBeth, K.J.; DiMartino, J.; Wells, J.A. Multiomics of Azacitidine-Treated AML Cells Reveals Variable and Convergent Targets That Remodel the Cell-Surface Proteome. Proc. Natl. Acad. Sci. USA 2019, 116, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.; Alloush, H.; Smith, M.A.; Hill, P.J.; Salisbury, V. A Rapid Assay of Cytosine Arabinoside Uptake and Metabolism by Acute Myeloid Leukaemic Cells Using a Bioluminescent Bacterial Biosensor. Blood 2007, 110, 4308. [Google Scholar] [CrossRef]
- St Louis, D.C.; Woodcock, J.B.; Franzoso, G.; Blair, P.J.; Carlson, L.M.; Murillo, M.; Wells, M.R.; Williams, A.J.; Smoot, D.S.; Kaushal, S.; et al. Evidence for Distinct Intracellular Signaling Pathways in CD34+ Progenitor to Dendritic Cell Differentiation from a Human Cell Line Model. J. Immunol. 1999, 162, 3237–3248. [Google Scholar] [CrossRef]
- Cejas, P.J.; Carlson, L.M.; Kolonias, D.; Zhang, J.; Lindner, I.; Billadeau, D.D.; Boise, L.H.; Lee, K.P. Regulation of RelB Expression during the Initiation of Dendritic Cell Differentiation. Mol. Cell. Biol. 2005, 25, 7900–7916. [Google Scholar] [CrossRef]
- Andersson, A.; Edén, P.; Lindgren, D.; Nilsson, J.; Lassen, C.; Heldrup, J.; Fontes, M.; Borg, Å.; Mitelman, F.; Johansson, B.; et al. Gene Expression Profiling of Leukemic Cell Lines Reveals Conserved Molecular Signatures among Subtypes with Specific Genetic Aberrations. Leukemia 2005, 19, 1042–1050. [Google Scholar] [CrossRef]
- Jensen, H.A.; Yourish, H.B.; Bunaciu, R.P.; Varner, J.D.; Yen, A. Induced Myelomonocytic Differentiation in Leukemia Cells Is Accompanied by Noncanonical Transcription Factor Expression. FEBS Open Bio. 2015, 5, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Lozzio, C.B.; Lozzio, B.B. Human Chronic Myelogenous Leukemia Cell-Line With Positive Philadelphia Chromosome. Blood 1975, 45, 321–334. [Google Scholar] [CrossRef]
- Raghuvar Gopal, D.V.; Narkar, A.A.; Badrinath, Y.; Mishra, K.P.; Joshi, D.S. Betulinic Acid Induces Apoptosis in Human Chronic Myelogenous Leukemia (CML) Cell Line K-562 without Altering the Levels of Bcr-Abl. Toxicol. Lett. 2005, 155, 343–351. [Google Scholar] [CrossRef]
- Drexler, H.G.; Quentmeier, H.; MacLeod, R.A.F.; Uphoff, C.C.; Hu, Z.-B. Leukemia Cell Lines: In Vitro Models for the Study of Acute Promyelocytic Leukemia. Leuk. Res. 1995, 19, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Kurkowiak, M.; Pępek, M.; Machnicki, M.M.; Solarska, I.; Borg, K.; Rydzanicz, M.; Stawiński, P.; Płoski, R.; Stokłosa, T. Genomic Landscape of Human Erythroleukemia K562 Cell Line, as Determined by next-Generation Sequencing and Cytogenetics. Acta Haematol. Pol. 2017, 48, 343–349. [Google Scholar] [CrossRef]
- Fan, Y.; Lu, H.; An, L.; Wang, C.; Zhou, Z.; Feng, F.; Ma, H.; Xu, Y.; Zhao, Q. Effect of Active Fraction of Eriocaulon Sieboldianum on Human Leukemia K562 Cells via Proliferation Inhibition, Cell Cycle Arrest and Apoptosis Induction. Environ. Toxicol. Pharmacol. 2016, 43, 13–20. [Google Scholar] [CrossRef]
- Duncan, M.T.; DeLuca, T.A.; Kuo, H.-Y.; Yi, M.; Mrksich, M.; Miller, W.M. SIRT1 Is a Critical Regulator of K562 Cell Growth, Survival, and Differentiation. Exp. Cell Res. 2016, 344, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Koeffler, H.P.; Golde, D.W. Human Myeloid Leukemia Cell Lines: A Review. Blood 1980, 56, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Kim, S.H.; Zhou, D.-C.; Ding, W.; Paietta, E.; Guidez, F.; Zelent, A.; Ramesh, K.H.; Cannizzaro, L.; Warrell, R.P.; et al. Acute Promyelocytic Leukemia Cell Line AP-1060 Established as a Cytokine-Dependent Culture from a Patient Clinically Resistant to All-Trans Retinoic Acid and Arsenic Trioxide. Leukemia 2004, 18, 1258–1269. [Google Scholar] [CrossRef] [PubMed]
- Drexler, H.G.; Matsuo, Y.; MacLeod, R.A.F. Malignant Hematopoietic Cell Lines: In Vitro Models for the Study of Erythroleukemia. Leuk. Res. 2004, 28, 1243–1251. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Wang, H.; Qin, L.; Feng, A.; Qi, D.; Wang, H.; Zhao, Y.; Kong, L.; Wang, H.; et al. JMJD1C Regulates Megakaryopoiesis in In Vitro Models through the Actin Network. Cells 2022, 11, 3660. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Miyoshi, H.; Shimizu, K.; Kozu, T.; Maseki, N.; Kaneko, Y.; Ohki, M. T(8;21) Breakpoints on Chromosome 21 in Acute Myeloid Leukemia Are Clustered within a Limited Region of a Single Gene, AML1. Proc. Natl. Acad. Sci. USA 1991, 88, 10431–10434. [Google Scholar] [CrossRef]
- Nerlov, C. C/EBPα Mutations in Acute Myeloid Leukaemias. Nat. Rev. Cancer 2004, 4, 394–400. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A.; Chávez-Valencia, V.; Gómez-Guijosa, M.Á.; Cortes-Penagos, C. Acute Myeloid Leukemia—Genetic Alterations and Their Clinical Prognosis. Int. J. Hematol. Oncol. Stem Cell Res. 2017, 11, 328–339. [Google Scholar]
- Chi, Y.; Lindgren, V.; Quigley, S.; Gaitonde, S. Acute Myelogenous Leukemia with t(6;9) (P23;Q34) and Marrow Basophilia: An Overview. Arch. Pathol. Lab. Med. 2008, 132, 1835–1837. [Google Scholar] [CrossRef]
- Thiede, C.; Steudel, C.; Mohr, B.; Schaich, M.; Schäkel, U.; Platzbecker, U.; Wermke, M.; Bornhäuser, M.; Ritter, M.; Neubauer, A.; et al. Analysis of FLT3-Activating Mutations in 979 Patients with Acute Myelogenous Leukemia: Association with FAB Subtypes and Identification of Subgroups with Poor Prognosis. Blood 2002, 99, 4326–4335. [Google Scholar] [CrossRef] [PubMed]
- Slovak, M.L.; Gundacker, H.; Bloomfield, C.D.; Dewald, G.; Appelbaum, F.R.; Larson, R.A.; Tallman, M.S.; Bennett, J.M.; Stirewalt, D.L.; Meshinchi, S.; et al. A Retrospective Study of 69 Patients with t(6;9) (P23;Q34) AML Emphasizes the Need for a Prospective, Multicenter Initiative for Rare “poor Prognosis” Myeloid Malignancies. Leukemia 2006, 20, 1295–1297. [Google Scholar] [CrossRef]
- Larizza, L.; Magnani, I.; Beghini, A. The Kasumi-1 Cell Line: A t(8;21)-Kit Mutant Model for Acute Myeloid Leukemia. Leuk. Lymphoma 2005, 46, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Asou, H.; Gombart, A.F.; Takeuchi, S.; Tanaka, H.; Tanioka, M.; Matsui, H.; Kimura, A.; Inaba, T.; Koeffler, H.P. Establishment of the Acute Myeloid Leukemia Cell Line Kasumi-6 from a Patient with a Dominant-Negative Mutation in the DNA-Binding Region of the C/EBPalpha Gene. Genes Chromosomes Cancer 2003, 36, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Matozaki, S.; Nakagawa, T.; Kawaguchi, R.; Aozaki, R.; Tsutsumi, M.; Murayama, T.; Koizumi, T.; Nishimura, R.; Isobe, T.; Chihara, K. Establishment of a Myeloid Leukaemic Cell Line (SKNO-1) from a Patient with t(8;21) Who Acquired Monosomy 17 during Disease Progression. Br. J. Haematol. 1995, 89, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lv, N.; Zhou, L.; Li, Y.; Yu, L. Chidamide Inhibits t(8;21) AML Cell Proliferation and AMK1/ETO and C-KIT Expression by Inhibiting ERK1/2 Signaling Pathway. Transl. Cancer Res. 2020, 9, 827–839. [Google Scholar] [CrossRef]
- Gallagher, R.; Collins, S.; Trujillo, J.; McCredie, K.; Ahearn, M.; Tsai, S.; Metzgar, R.; Aulakh, G.; Ting, R.; Ruscetti, F.; et al. Characterization of the Continuous, Differentiating Myeloid Cell Line (HL-60) from a Patient with Acute Promyelocytic Leukemia. Blood 1979, 54, 713–733. [Google Scholar] [CrossRef]
- Sak, K.; Everaus, H. Established Human Cell Lines as Models to Study Anti-Leukemic Effects of Flavonoids. Curr. Genom. 2017, 18, 3–26. [Google Scholar] [CrossRef]
- Kwa, F.A.A.; Cole-Sinclair, M.F.; Kapuscinski, M.K. Combination Treatment of P53-Null HL-60 Cells with Histone Deacetylase Inhibitors and Chlorambucil Augments Apoptosis and Increases BCL6 and P21 Gene Expression. Curr. Mol. Pharmacol. 2019, 12, 72–81. [Google Scholar] [CrossRef]
- Al-Otaibi, N.A.S.; Cassoli, J.S.; Slater, N.K.H.; Rahmoune, H. Molecular Characterization of Human Leukemia 60 (HL-60) Cells as a Model of Acute Myelogenous Leukemia Post Cryopreservation. Methods Mol. Biol. 2019, 1916, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Newburger, P.E.; Chovaniec, M.E.; Greenberger, J.S.; Cohen, H.J. Functional Changes in Human Leukemic Cell Line HL-60. A Model for Myeloid Differentiation. J. Cell Biol. 1979, 82, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.J. The HL-60 Promyelocytic Leukemia Cell Line: Proliferation, Differentiation, and Cellular Oncogene Expression. Blood 1987, 70, 1233–1244. [Google Scholar] [CrossRef]
- Johnston, J.J.; Rintels, P.; Chung, J.; Sather, J.; Benz, E.J.; Berliner, N. Lactoferrin Gene Promoter: Structural Integrity and Nonexpression in HL60 Cells. Blood 1992, 79, 2998–3006. [Google Scholar] [CrossRef]
- Jian, P.; Li, Z.W.; Fang, T.Y.; Jian, W.; Zhuan, Z.; Mei, L.X.; Yan, W.S.; Jian, N. Retinoic Acid Induces HL-60 Cell Differentiation via the Upregulation of MiR-663. J. Hematol. Oncol. 2011, 4, 20. [Google Scholar] [CrossRef]
- Tucker, K.A.; Lilly, M.B.; Heck, L.; Rado, T.A. Characterization of a New Human Diploid Myeloid Leukemia Cell Line (PLB-985) with Granulocytic and Monocytic Differentiating Capacity. Blood 1987, 70, 372–378. [Google Scholar] [CrossRef]
- Drexler, H.G.; Dirks, W.G.; Matsuo, Y.; MacLeod, R.a.F. False Leukemia–Lymphoma Cell Lines: An Update on over 500 Cell Lines. Leukemia 2003, 17, 416–426. [Google Scholar] [CrossRef]
- Rincón, E.; Rocha-Gregg, B.L.; Collins, S.R. A Map of Gene Expression in Neutrophil-like Cell Lines. BMC Genom. 2018, 19, 573. [Google Scholar] [CrossRef] [PubMed]
- Kamath, G.R.; Tremblay, D.; Coltoff, A.; Caro, J.; Lancman, G.; Bhalla, S.; Najfeld, V.; Mascarenhas, J.; Taioli, E. Comparing the Epidemiology, Clinical Characteristics and Prognostic Factors of Acute Myeloid Leukemia with and without Acute Promyelocytic Leukemia. Carcinogenesis 2019, 40, 651–660. [Google Scholar] [CrossRef]
- Hwang, S.M. Classification of Acute Myeloid Leukemia. Blood Res. 2020, 55 (Suppl. S1), S1–S4. [Google Scholar] [CrossRef]
- de Thé, H.; Lavau, C.; Marchio, A.; Chomienne, C.; Degos, L.; Dejean, A. The PML-RAR Alpha Fusion MRNA Generated by the t(15;17) Translocation in Acute Promyelocytic Leukemia Encodes a Functionally Altered RAR. Cell 1991, 66, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Lo-Coco, F.; Ammatuna, E. The Biology of Acute Promyelocytic Leukemia and Its Impact on Diagnosis and Treatment. Hematol. Am. Soc. Hematol. Educ. Program 2006, 156–161, 514. [Google Scholar] [CrossRef] [PubMed]
- Pandolfi, P.P. In Vivo Analysis of the Molecular Genetics of Acute Promyelocytic Leukemia. Oncogene 2001, 20, 5726–5735. [Google Scholar] [CrossRef] [PubMed]
- Khanna-Gupta, A.; Kolibaba, K.; Zibello, T.A.; Berliner, N. NB4 Cells Show Bilineage Potential and an Aberrant Pattern of Neutrophil Secondary Granule Protein Gene Expression. Blood 1994, 84, 294–302. [Google Scholar] [CrossRef]
- Duprez, E.; Ruchaud, S.; Houge, G.; Martin-Thouvenin, V.; Valensi, F.; Kastner, P.; Berger, R.; Lanotte, M. A Retinoid Acid “resistant” t(15;17) Acute Promyelocytic Leukemia Cell Line: Isolation, Morphological, Immunological, and Molecular Features. Leukemia 1992, 6, 1281–1287. [Google Scholar] [PubMed]
- Dermime, S.; Grignani, F.; Rogaia, D.; Liberatore, C.; Marchesi, E.; Gambacorti-Passerini, C. Acute Promyelocytic Leukaemia Cells Resistant to Retinoic Acid Show Further Perturbation of the RARα Signal Transduction System. Leuk. Lymphoma 1995, 16, 289–295. [Google Scholar] [CrossRef]
- Kubonishi, I.; Machida, K.; Sonobe, H.; Ohtsuki, Y.; Akagi, T.; Miyoshi, I. Two New Human Myeloid Cell Lines Derived from Acute Promyelocytic Leukemia and Chronic Myelocytic Leukemia. GANN Jpn. J. Cancer Res. 1983, 74, 319–322. [Google Scholar] [CrossRef]
- Sugimoto, K.; Toyoshima, H.; Sakai, R.; Miyagawa, K.; Hagiwara, K.; Ishikawa, F.; Takaku, F.; Yazaki, Y.; Hirai, H. Frequent Mutations in the P53 Gene in Human Myeloid Leukemia Cell Lines. Blood 1992, 79, 2378–2383. [Google Scholar] [CrossRef]
- Reiter, K.; Polzer, H.; Krupka, C.; Maiser, A.; Vick, B.; Rothenberg-Thurley, M.; Metzeler, K.H.; Dörfel, D.; Salih, H.R.; Jung, G.; et al. Tyrosine Kinase Inhibition Increases the Cell Surface Localization of FLT3-ITD and Enhances FLT3-Directed Immunotherapy of Acute Myeloid Leukemia. Leukemia 2018, 32, 313–322. [Google Scholar] [CrossRef]
- Kizaki, M.; Matsushita, H.; Takayama, N.; Muto, A.; Ueno, H.; Awaya, N.; Kawai, Y.; Asou, H.; Kamada, N.; Ikeda, Y. Establishment and Characterization of a Novel Acute Promyelocytic Leukemia Cell Line (UF-1) with Retinoic Acid-Resistant Features. Blood 1996, 88, 1824–1833. [Google Scholar] [CrossRef]
- Sato, A.; Imaizumi, M.; Hoshi, Y.; Rikiishi, T.; Fujii, K.; Kizaki, M.; Kagechika, H.; Kakizuka, A.; Hayashi, Y.; Iinuma, K. Alteration in the Cellular Response to Retinoic Acid of a Human Acute Promyelocytic Leukemia Cell Line, UF-1, Carrying a Patient-Derived Mutant PML-RARα Chimeric Gene. Leuk. Res. 2004, 28, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Drexler, H.G. (Ed.) Front Matter. In The Leukemia-Lymphoma Cell Line FactsBook; Factsbook; Academic Press: London, UK, 2001; p. iii. [Google Scholar] [CrossRef]
- Takayama, N.; Kizaki, M.; Hida, T.; Kinjo, K.; Ikeda, Y. Novel Mutation in the PML/RARα Chimeric Gene Exhibits Dramatically Decreased Ligand-Binding Activity and Confers Acquired Resistance to Retinoic Acid in Acute Promyelocytic Leukemia. Exp. Hematol. 2001, 29, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, T.; Kizaki, M.; Omine, M. Induction of Differentiation of Retinoic Acid-Resistant Acute Promyelocytic Leukemia Cells by the Combination of All-Trans Retinoic Acid and Granulocyte Colony-Stimulating Factor. Leuk. Res. 2004, 28, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Witcher, M.; Shiu, H.Y.; Guo, Q.; Miller, W.H. Combination of Retinoic Acid and Tumor Necrosis Factor Overcomes the Maturation Block in a Variety of Retinoic Acid-Resistant Acute Promyelocytic Leukemia Cells. Blood 2004, 104, 3335–3342. [Google Scholar] [CrossRef]
- Nakazato, T.; Ito, K.; Miyakawa, Y.; Kinjo, K.; Yamada, T.; Hozumi, N.; Ikeda, Y.; Kizaki, M. Catechin, a Green Tea Component, Rapidly Induces Apoptosis of Myeloid Leukemic Cells via Modulation of Reactive Oxygen Species Production in Vitro and Inhibits Tumor Growth in Vivo. Haematologica 2005, 90, 317–325. [Google Scholar]
- Komura, N.; Ikeda, Y.; Masuda, N.; Umezawa, Y.; Ito, K.; Kizaki, M.; Umezawa, K. Designed ATRA Analogue Active against ATRA-Resistant Acute Promyelocytic Leukemia Cells Having a Single Nucleotide Substitution in Their Retinoic Acid Receptor. Leuk. Res. 2007, 31, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Cassinat, B.; Zassadowski, F.; Ferry, C.; Llopis, L.; Bruck, N.; Lainey, E.; Duong, V.; Cras, A.; Despouy, G.; Chourbagi, O.; et al. New Role for Granulocyte Colony-Stimulating Factor-Induced Extracellular Signal-Regulated Kinase 1/2 in Histone Modification and Retinoic Acid Receptor α Recruitment to Gene Promoters: Relevance to Acute Promyelocytic Leukemia Cell Differentiation. Mol. Cell. Biol. 2011, 31, 1409–1418. [Google Scholar] [CrossRef]
- Iriyama, N.; Hatta, Y.; Takei, M. ETV6/ARG Oncoprotein Confers Autonomous Cell Growth by Enhancing c-Myc Expression via Signal Transducer and Activator of Transcription 5 Activation in the Acute Promyelocytic Leukemia Cell Line HT93A. Leuk. Lymphoma 2015, 56, 2416–2423. [Google Scholar] [CrossRef] [PubMed]
- Kishi, K.; Toba, K.; Azegami, T.; Tsukada, N.; Uesugi, Y.; Masuko, M.; Niwano, H.; Hashimoto, S.; Sakaue, M.; Furukawa, T.; et al. Hematopoietic Cytokine-Dependent Differentiation to Eosinophils and Neutrophils in a Newly Established Acute Promyelocytic Leukemia Cell Line with t(15;17). Exp. Hematol. 1998, 26, 135–142. [Google Scholar]
- Kitamura, K.; Kiyoi, H.; Yoshida, H.; Saito, H.; Ohno, R.; Naoe, T. Mutant AF-2 Domain of PML-RARalpha in Retinoic Acid-Resistant NB4 Cells: Differentiation Induced by RA Is Triggered Directly through PML-RARalpha and Its down-Regulation in Acute Promyelocytic Leukemia. Leukemia 1997, 11, 1950–1956. [Google Scholar] [CrossRef]
- Chen, S.; Nagel, S.; Schneider, B.; Dai, H.; Geffers, R.; Kaufman, M.; Meyer, C.; Pommerenke, C.; Thress, K.S.; Li, J.; et al. A New ETV6-NTRK3 Cell Line Model Reveals MALAT1 as a Novel Therapeutic Target—A Short Report. Cell. Oncol. 2018, 41, 93–101. [Google Scholar] [CrossRef]
- Corces-Zimmerman, M.R.; Hong, W.-J.; Weissman, I.L.; Medeiros, B.C.; Majeti, R. Preleukemic Mutations in Human Acute Myeloid Leukemia Affect Epigenetic Regulators and Persist in Remission. Proc. Natl. Acad. Sci. USA 2014, 111, 2548–2553. [Google Scholar] [CrossRef]
- Sano, H.; Shimada, A.; Taki, T.; Murata, C.; Park, M.; Sotomatsu, M.; Tabuchi, K.; Tawa, A.; Kobayashi, R.; Horibe, K.; et al. RAS Mutations Are Frequent in FAB Type M4 and M5 of Acute Myeloid Leukemia, and Related to Late Relapse: A Study of the Japanese Childhood AML Cooperative Study Group. Int. J. Hematol. 2012, 95, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Tiacci, E.; Spanhol-Rosseto, A.; Martelli, M.P.; Pasqualucci, L.; Quentmeier, H.; Grossmann, V.; Drexler, H.G.; Falini, B. The NPM1 Wild-Type OCI-AML2 and the NPM1-Mutated OCI-AML3 Cell Lines Carry DNMT3A Mutations. Leukemia 2012, 26, 554–557. [Google Scholar] [CrossRef]
- Falini, B.; Brunetti, L.; Sportoletti, P.; Martelli, M.P. NPM1-Mutated Acute Myeloid Leukemia: From Bench to Bedside. Blood 2020, 136, 1707–1721. [Google Scholar] [CrossRef] [PubMed]
- Banella, C.; Catalano, G.; Travaglini, S.; Pelosi, E.; Ottone, T.; Zaza, A.; Guerrera, G.; Angelini, D.F.; Niscola, P.; Divona, M.; et al. Ascorbate Plus Buformin in AML: A Metabolic Targeted Treatment. Cancers 2022, 14, 2565. [Google Scholar] [CrossRef] [PubMed]
- Grenier, A.; Poulain, L.; Mondesir, J.; Jacquel, A.; Bosc, C.; Stuani, L.; Mouche, S.; Larrue, C.; Sahal, A.; Birsen, R.; et al. AMPK-PERK Axis Represses Oxidative Metabolism and Enhances Apoptotic Priming of Mitochondria in Acute Myeloid Leukemia. Cell Rep. 2022, 38, 110197. [Google Scholar] [CrossRef]
- Zheng, A.; Castren, K.; Säily, M.; Savolainen, E.-R.; Koistinen, P.; Vähäkangas, K. P53 Status of Newly Established Acute Myeloid Leukaemia Cell Lines. Br. J. Cancer 1999, 79, 407–415. [Google Scholar] [CrossRef]
- Hu, Z.-B.; Quentmeier, H.; Meyer, C.; Kaufmann, M.; MacLeod, R.A.F.; Drexler, H.G. New Cytokine-Dependent Acute Myeloid Leukemia Cell Line MUTZ-11 with Disomic Chromosome Rearrangement t(16;17). Leuk. Res. 2004, 28, 509–515. [Google Scholar] [CrossRef] [PubMed]
- El Fitori, J.; Su, Y.; Büchler, P.; Ludwig, R.; Giese, N.A.; Büchler, M.W.; Quentmeier, H.; Hines, O.J.; Herr, I.; Friess, H. PKC 412 Small-Molecule Tyrosine Kinase Inhibitor: Single-Compound Therapy for Pancreatic Cancer. Cancer 2007, 110, 1457–1468. [Google Scholar] [CrossRef]
- Liu, W.; Deng, L.; Song, Y.; Redell, M. DOT1L Inhibition Sensitizes MLL-Rearranged AML to Chemotherapy. PLoS ONE 2014, 9, e98270. [Google Scholar] [CrossRef] [PubMed]
- Kühn, M.W.M.; Hadler, M.J.; Daigle, S.R.; Koche, R.P.; Krivtsov, A.V.; Olhava, E.J.; Caligiuri, M.A.; Huang, G.; Bradner, J.E.; Pollock, R.M.; et al. MLL Partial Tandem Duplication Leukemia Cells Are Sensitive to Small Molecule DOT1L Inhibition. Haematologica 2015, 100, e190–e193. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-B.; MacLeod, R.a.F.; Meyer, C.; Quentmeier, H.; Drexler, H.G. New Acute Myeloid Leukemia-Derived Cell Line: MUTZ-8 with 5q-. Leukemia 2002, 16, 1556–1561. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-B.; Quentmeier, H.; Meyer, C.; MacLeod, R.A.F.; Drexler, H.G. Establishment of the Two Cytokine-Dependent Acute Myeloid Leukemia-Derived Cell Lines MUTZ-8 and MUTZ-11. In Acute Leukemias IX; Hiddemann, W., Haferlach, T., Unterhalt, M., Büchner, T., Ritter, J., Eds.; Haematology and Blood Transfusion Hämatologie und Bluttransfusion; Springer: Berlin/Heidelberg, Germany, 2003; pp. 122–127. [Google Scholar] [CrossRef]
- Quentmeier, H.; MacLeod, R.a.F.; Zaborski, M.; Drexler, H.G. JAK2 V617F Tyrosine Kinase Mutation in Cell Lines Derived from Myeloproliferative Disorders. Leukemia 2006, 20, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Quentmeier, H.; Geffers, R.; Jost, E.; MacLeod, R.a.F.; Nagel, S.; Röhrs, S.; Romani, J.; Scherr, M.; Zaborski, M.; Drexler, H.G. SOCS2: Inhibitor of JAK2V617F-Mediated Signal Transduction. Leukemia 2008, 22, 2169–2175. [Google Scholar] [CrossRef]
- Hu, Z.B.; Ma, W.; Zaborski, M.; MacLeod, R.; Quentmeier, H.; Drexler, H.G. Establishment and Characterization of Two Novel Cytokine-Responsive Acute Myeloid and Monocytic Leukemia Cell Lines, MUTZ-2 and MUTZ-3. Leukemia 1996, 10, 1025–1040. [Google Scholar]
- Masterson, A.J.; Sombroek, C.C.; De Gruijl, T.D.; Graus, Y.M.F.; van der Vliet, H.J.J.; Lougheed, S.M.; van den Eertwegh, A.J.M.; Pinedo, H.M.; Scheper, R.J. MUTZ-3, a Human Cell Line Model for the Cytokine-Induced Differentiation of Dendritic Cells from CD34+ Precursors. Blood 2002, 100, 701–703. [Google Scholar] [CrossRef]
- Santegoets, S.J.A.M.; Schreurs, M.W.J.; Masterson, A.J.; Liu, Y.P.; Goletz, S.; Baumeister, H.; Kueter, E.W.M.; Lougheed, S.M.; van den Eertwegh, A.J.M.; Scheper, R.J.; et al. In Vitro Priming of Tumor-Specific Cytotoxic T Lymphocytes Using Allogeneic Dendritic Cells Derived from the Human MUTZ-3 Cell Line. Cancer Immunol. Immunother. 2006, 55, 1480–1490. [Google Scholar] [CrossRef]
- Santegoets, S.J.A.M.; van den Eertwegh, A.J.M.; van de Loosdrecht, A.A.; Scheper, R.J.; de Gruijl, T.D. Human Dendritic Cell Line Models for DC Differentiation and Clinical DC Vaccination Studies. J. Leukoc. Biol. 2008, 84, 1364–1373. [Google Scholar] [CrossRef]
- Quentmeier, H.; Drexler, H.G.; Fleckenstein, D.; Zaborski, M.; Armstrong, A.; Sims, J.E.; Lyman, S.D. Cloning of Human Thymic Stromal Lymphopoietin (TSLP) and Signaling Mechanisms Leading to Proliferation. Leukemia 2001, 15, 1286–1292. [Google Scholar] [CrossRef] [PubMed]
- Rasaiyaah, J.; Noursadeghi, M.; Kellam, P.; Chain, B. Transcriptional and Functional Defects of Dendritic Cells Derived from the MUTZ-3 Leukaemia Line. Immunology 2009, 127, 429–441. [Google Scholar] [CrossRef]
- Gimeno, M.; San José-Enériz, E.; Villar, S.; Agirre, X.; Prosper, F.; Rubio, A.; Carazo, F. Explainable Artificial Intelligence for Precision Medicine in Acute Myeloid Leukemia. Front. Immunol. 2022, 13, 977358. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, O.; Friedman, D.; Leshkowitz, D.; Goldenberg, D.; Orlovsky, K.; Pencovich, N.; Lotem, J.; Tanay, A.; Groner, Y. Addiction of t(8;21) and Inv(16) Acute Myeloid Leukemia to Native RUNX1. Cell Rep. 2013, 4, 1131–1143. [Google Scholar] [CrossRef]
- Wilkinson, A.C.; Ballabio, E.; Geng, H.; North, P.; Tapia, M.; Kerry, J.; Biswas, D.; Roeder, R.G.; Allis, C.D.; Melnick, A.; et al. RUNX1 Is a Key Target in t(4;11) Leukemias That Contributes to Gene Activation through an AF4-MLL Complex Interaction. Cell Rep 2013, 3, 116–127. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, Y.; Williams, J.; Hang, Y.; Richter, L.; Becker, M.; Amador, C.; Oupický, D.; Hyde, R.K. Use of Polymeric CXCR4 Inhibitors as SiRNA Delivery Vehicles for the Treatment of Acute Myeloid Leukemia. Cancer Gene Ther. 2019, 27, 45–55. [Google Scholar] [CrossRef]
- Gilby, D.C.; Sung, H.Y.; Winship, P.R.; Goodeve, A.C.; Reilly, J.T.; Kiss-Toth, E. Tribbles-1 and -2 Are Tumour Suppressors, down-Regulated in Human Acute Myeloid Leukaemia. Immunol. Lett. 2010, 130, 115–124. [Google Scholar] [CrossRef]
- Cripe, L.D.; Gelfanov, V.M.; Smith, E.A.; Spigel, D.R.; Phillips, C.A.; Gabig, T.G.; Jung, S.-H.; Fyffe, J.; Hartman, A.D.; Kneebone, P.; et al. Role for C-Jun N-Terminal Kinase in Treatment-Refractory Acute Myeloid Leukemia (AML): Signaling to Multidrug-Efflux and Hyperproliferation. Leukemia 2002, 16, 799–812. [Google Scholar] [CrossRef]
- Tallman, M.S.; Kim, H.T.; Paietta, E.; Bennett, J.M.; Dewald, G.; Cassileth, P.A.; Wiernik, P.H.; Rowe, J.M. Acute Monocytic Leukemia (French-American-British Classification M5) Does Not Have a Worse Prognosis Than Other Subtypes of Acute Myeloid Leukemia: A Report From the Eastern Cooperative Oncology Group. JCO 2004, 22, 1276–1286. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, S.; Yamabe, M.; Yamaguchi, Y.; Kobayashi, Y.; Konno, T.; Tada, K. Establishment and Characterization of a Human Acute Monocytic Leukemia Cell Line (THP-1). Int. J. Cancer 1980, 26, 171–176. [Google Scholar] [CrossRef]
- Humeniuk-Polaczek, R.; Marcinkowska, E. Impaired Nuclear Localization of Vitamin D Receptor in Leukemia Cells Resistant to Calcitriol-Induced Differentiation. J. Steroid Biochem. Mol. Biol. 2004, 88, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Verhoeckx, K.; Cotter, P.; López-Expósito, I.; Kleiveland, C.; Lea, T.; Mackie, A.; Requena, T.; Swiatecka, D.; Wichers, H. (Eds.) The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Schnoor, M.; Buers, I.; Sietmann, A.; Brodde, M.F.; Hofnagel, O.; Robenek, H.; Lorkowski, S. Efficient Non-Viral Transfection of THP-1 Cells. J. Immunol. Methods 2009, 344, 109–115. [Google Scholar] [CrossRef]
- Chanput, W.; Mes, J.; Vreeburg, R.A.M.; Savelkoul, H.F.J.; Wichers, H.J. Transcription Profiles of LPS-Stimulated THP-1 Monocytes and Macrophages: A Tool to Study Inflammation Modulating Effects of Food-Derived Compounds. Food Funct. 2010, 1, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Ashikaga, T.; Yoshida, Y.; Hirota, M.; Yoneyama, K.; Itagaki, H.; Sakaguchi, H.; Miyazawa, M.; Ito, Y.; Suzuki, H.; Toyoda, H. Development of an in Vitro Skin Sensitization Test Using Human Cell Lines: The Human Cell Line Activation Test (h-CLAT): I. Optimization of the h-CLAT Protocol. Toxicol. In Vitro 2006, 20, 767–773. [Google Scholar] [CrossRef]
- Sakaguchi, H.; Ashikaga, T.; Miyazawa, M.; Yoshida, Y.; Ito, Y.; Yoneyama, K.; Hirota, M.; Itagaki, H.; Toyoda, H.; Suzuki, H. Development of an in Vitro Skin Sensitization Test Using Human Cell Lines; Human Cell Line Activation Test (h-CLAT) II. An Inter-Laboratory Study of the h-CLAT. Toxicol. Vitr. 2006, 20, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Helal, R.; Melzig, M.F. New Aspects in the Synthesis and Secretion of Lysozyme by Cultured Human Monocyte Cell Lines. Vitr. Cell. Dev. Biol. Anim. 2010, 46, 492–496. [Google Scholar] [CrossRef]
- Helal, R.; Melzig, M.F. In Vitro Effects of Selected Saponins on the Production and Release of Lysozyme Activity of Human Monocytic and Epithelial Cell Lines. Sci. Pharm. 2011, 79, 337–350. [Google Scholar] [CrossRef]
- Helal, R.; Bader, G.; Melzig, M.F. Stimulation of Lysozyme Release by Selected Microbial Preparations. Die Pharm.-Int. J. Pharm. Sci. 2012, 67, 564–566. [Google Scholar] [CrossRef]
- Pandur, E.; Micalizzi, G.; Mondello, L.; Horváth, A.; Sipos, K.; Horváth, G. Antioxidant and Anti-Inflammatory Effects of Thyme (Thymus vulgaris L.) Essential Oils Prepared at Different Plant Phenophases on Pseudomonas Aeruginosa LPS-Activated THP-1 Macrophages. Antioxidants 2022, 11, 1330. [Google Scholar] [CrossRef] [PubMed]
- Bosshart, H.; Heinzelmann, M. Lipopolysaccharide-Mediated Cell Activation without Rapid Mobilization of Cytosolic Free Calcium. Mol. Immunol. 2004, 41, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Bruckmeier, M.; Kuehnl, A.; Culmes, M.; Pelisek, J.; Eckstein, H.-H. Impact of OxLDL and LPS on C-Type Natriuretic Peptide System Is Different between THP-1 Cells and Human Peripheral Blood Monocytic Cells. CPB 2012, 30, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Bosshart, H.; Heinzelmann, M. Arginine-Rich Cationic Polypeptides Amplify Lipopolysaccharide-Induced Monocyte Activation. Infect. Immun. 2002, 70, 6904–6910. [Google Scholar] [CrossRef]
- Bosshart, H.; Heinzelmann, M. THP-1 Cells as a Model for Human Monocytes. Ann. Transl. Med. 2016, 4, 438. [Google Scholar] [CrossRef]
- Sundström, C.; Nilsson, K. Establishment and Characterization of a Human Histiocytic Lymphoma Cell Line (U-937). Int. J. Cancer 1976, 17, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Aggerholm, A.; Grønbaek, K.; Guldberg, P.; Hokland, P. Mutational Analysis of the Tumour Suppressor Gene MMAC1/PTEN in Malignant Myeloid Disorders. Eur. J. Haematol. 2000, 65, 109–113. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. ClinVar, VCV000449416.13. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000449416.13 (accessed on 29 December 2022).
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 Cell Line: An in Vitro Cell Model for Immune Modulation Approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, K.; Tauber, S.; Goelz, N.; Simmet, D.M.; Engeli, S.; Birlem, M.; Dumrese, C.; Karer, A.; Hunziker, S.; Biskup, J.; et al. Severe Disruption of the Cytoskeleton and Immunologically Relevant Surface Molecules in a Human Macrophageal Cell Line in Microgravity—Results of an in Vitro Experiment on Board of the Shenzhou-8 Space Mission. Acta Astronaut. 2014, 94, 277–292. [Google Scholar] [CrossRef]
- Olsson, I.L.; Breitman, T.R. Induction of Differentiation of the Human Histiocytic Lymphoma Cell Line U-937 by Retinoic Acid and Cyclic Adenosine 3’:5’-Monophosphate-Inducing Agents. Cancer Res. 1982, 42, 3924–3927. [Google Scholar]
- Hattori, T.; Pack, M.; Bougnoux, P.; Chang, Z.L.; Hoffman, T. Interferon-Induced Differentiation of U937 Cells. Comparison with Other Agents That Promote Differentiation of Human Myeloid or Monocytelike Cell Lines. J. Clin. Investig. 1983, 72, 237–244. [Google Scholar] [CrossRef]
- Olsson, I.; Gullberg, U.; Ivhed, I.; Nilsson, K. Induction of Differentiation of the Human Histiocytic Lymphoma Cell Line U-937 by 1 Alpha,25-Dihydroxycholecalciferol. Cancer Res. 1983, 43 Pt 1, 5862–5867. [Google Scholar] [PubMed]
- Stöckbauer, P.; Malasková, V.; Soucek, J.; Chudomel, V. Differentiation of Human Myeloid Leukemia Cell Lines Induced by Tumor-Promoting Phorbol Ester (TPA). I. Changes of the Morphology, Cytochemistry and the Surface Differentiation Antigens Analyzed with Monoclonal Antibodies. Neoplasma 1983, 30, 257–272. [Google Scholar]
- Minafra, L.; Di Cara, G.; Albanese, N.N.; Cancemi, P. Proteomic Differentiation Pattern in the U937 Cell Line. Leuk. Res. 2011, 35, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.R.; Um, H.-D. Hydrogen Peroxide Suppresses U937 Cell Death by Two Different Mechanisms Depending on Its Concentration. Exp. Cell Res. 1999, 248, 430–438. [Google Scholar] [CrossRef]
- Kim, D.K.; Cho, E.S.; Lee, B.R.; Um, H.-D. NF-ΚB Mediates the Adaptation of Human U937 Cells to Hydrogen Peroxide. Free. Radic. Biol. Med. 2001, 30, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Cho, E.S.; Yang, H.S.; Kim, H.; Um, H.-D. Serum Withdrawal Kills U937 Cells by Inducing a Positive Mutual Interaction between Reactive Oxygen Species and Phosphoinositide 3-Kinase. Cell. Signal. 2005, 17, 197–204. [Google Scholar] [CrossRef]
- Song, M.; Ryoo, I.; Choi, H.; Choi, B.; Kim, S.-T.; Heo, T.-H.; Lee, J.Y.; Park, P.-H.; Kwak, M.-K. NRF2 Signaling Negatively Regulates Phorbol-12-Myristate-13-Acetate (PMA)-Induced Differentiation of Human Monocytic U937 Cells into Pro-Inflammatory Macrophages. PLoS ONE 2015, 10, e0134235. [Google Scholar] [CrossRef]
- Vu, M.; Kassouf, N.; Appiah, S. Betulinic Acid–Doxorubicin-Drug Combination Induced Apoptotic Death via ROS Stimulation in a Relapsed AML MOLM-13 Cell Model. Antioxidants 2021, 10, 1456. [Google Scholar] [CrossRef] [PubMed]
- Greco, G.; Schnekenburger, M.; Catanzaro, E.; Turrini, E.; Ferrini, F.; Sestili, P.; Diederich, M.; Fimognari, C. Discovery of Sulforaphane as an Inducer of Ferroptosis in U-937 Leukemia Cells: Expanding Its Anticancer Potential. Cancers 2022, 14, 76. [Google Scholar] [CrossRef]
- Prasad, A.; Sedlářová, M.; Balukova, A.; Ovsii, A.; Rác, M.; Křupka, M.; Kasai, S.; Pospíšil, P. Reactive Oxygen Species Imaging in U937 Cells. Front. Physiol. 2020, 11, 552569. [Google Scholar] [CrossRef]
- Nathan, I.; Dizdaroglu, M.; Bernstein, L.; Junker, U.; Lee, C.-K.; Muegge, K.; Durum, S.K. Induction of oxidative DNA damage in U937 cells by TNF or anti-Fas stimulation. Cytokine 2000, 12, 881–887. [Google Scholar] [CrossRef]
- Pina-Jiménez, E.; Calzada, F.; Bautista, E.; Ordoñez-Razo, R.M.; Velázquez, C.; Barbosa, E.; García-Hernández, N. Incomptine A Induces Apoptosis, ROS Production and a Differential Protein Expression on Non-Hodgkin’s Lymphoma Cells. Int. J. Mol. Sci. 2021, 22, 10516. [Google Scholar] [CrossRef]
- del Rosario, H.; Saavedra, E.; Brouard, I.; González-Santana, D.; García, C.; Spínola-Lasso, E.; Tabraue, C.; Quintana, J.; Estévez, F. Structure-Activity Relationships Reveal a 2-Furoyloxychalcone as a Potent Cytotoxic and Apoptosis Inducer for Human U-937 and HL-60 Leukaemia Cells. Bioorg. Chem. 2022, 127, 105926. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.-S.; Jeong, S.-I.; Hwang, B.-S.; Lee, Y.-E.; Kang, S.-H.; Lee, H.-C.; Oh, C.-H. Gallic Acid Inhibits Cell Viability and Induces Apoptosis in Human Monocytic Cell Line U937. J. Med. Food 2011, 14, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Endo, M.; Matsui, T.; Katsuda, I.; Emi, N.; Kawamoto, Y.; Koike, T.; Beppu, H. Agaritine from Agaricus Blazei Murrill Induces Apoptosis in the Leukemic Cell Line U937. Biochim. Biophys. Acta-Gen. Subj. 2011, 1810, 519–525. [Google Scholar] [CrossRef]
- Kaba, S.I.; Egorova, E.M. In Vitro Studies of the Toxic Effects of Silver Nanoparticles on HeLa and U937 Cells. Nanotechnol. Sci. Appl. 2015, 8, 19–29. [Google Scholar] [CrossRef]
- Barbasz, A.; Kreczmer, B.; Skórka, M.; Czyżowska, A. Toxicity of Pesticides toward Human Immune Cells U-937 and HL-60. J. Environ. Sci. Health B 2020, 55, 719–725. [Google Scholar] [CrossRef]
- OECD. U-SENSTM—Myeloid U937 Skin Sensitisation Test; OECD: Paris, France, 2013; Available online: https://tsar.jrc.ec.europa.eu/test-method/tm2013-02 (accessed on 29 December 2022).
- Passmore, J.S.; Lukey, P.T.; Ress, S.R. The Human Macrophage Cell Line U937 as an in Vitro Model for Selective Evaluation of Mycobacterial Antigen-Specific Cytotoxic T-Cell Function. Immunology 2001, 102, 146–156. [Google Scholar] [CrossRef]
- Kaszubowska, L.; Engelmann, H.; Gotartowska, M.; Iliszko, M.; Bigda, J. Identification of two U937 cell sublines exhibiting different patterns of response to tumour necrosis factor. Cytokine 2001, 13, 365–370. [Google Scholar] [CrossRef]
- Stasiłojć, G.; Pinto, S.; Wyszkowska, R.; Wejda, M.; Słomińska, E.M.; Filipska, M.; Koszałka, P.; Swierczyński, J.; O’Connor, J.E.; Bigda, J.J. U937 Variant Cells as a Model of Apoptosis without Cell Disintegration. Cell. Mol. Biol. Lett. 2013, 18, 249–262. [Google Scholar] [CrossRef]
- Reid, Y.A.; McGuire, L.; O’Neill, K.; Macy, M.; Chen, T.R.; McClintock, P.; Dorotinsky, C.; Hay, R. Cell Line Cross-Contamination of U-937 [Correction of U-397]. J. Leukoc. Biol. 1995, 57, 804. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; MacLeod, R.A.; Uphoff, C.C.; Drexler, H.G.; Nishizaki, C.; Katayama, Y.; Kimura, G.; Fujii, N.; Omoto, E.; Harada, M.; et al. Two Acute Monocytic Leukemia (AML-M5a) Cell Lines (MOLM-13 and MOLM-14) with Interclonal Phenotypic Heterogeneity Showing MLL-AF9 Fusion Resulting from an Occult Chromosome Insertion, Ins(11;9) (Q23;P22p23). Leukemia 1997, 11, 1469–1477. [Google Scholar] [CrossRef]
- Lange, B.; Valtieri, M.; Santoli, D.; Caracciolo, D.; Mavilio, F.; Gemperlein, I.; Griffin, C.; Emanuel, B.; Finan, J.; Nowell, P. Growth Factor Requirements of Childhood Acute Leukemia: Establishment of GM-CSF-Dependent Cell Lines. Blood 1987, 70, 192–199. [Google Scholar] [CrossRef]
- Stong, R.C.; Kersey, J.H. In Vitro Culture of Leukemic Cells in t(4;11) Acute Leukemia. Blood 1985, 66, 439–443. [Google Scholar] [CrossRef]
- Parkin, J.L.; Arthur, D.C.; Abramson, C.S.; McKenna, R.W.; Kersey, J.H.; Heideman, R.L.; Brunning, R.D. Acute Leukemia Associated with the t(4;11) Chromosome Rearrangement: Ultrastructural and Immunologic Characteristics. Blood 1982, 60, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Chen, Q.; Xu, J.; Li, W.; Xu, B.; Qiu, Y. Low-Frequency TP53 Hotspot Mutation Contributes to Chemoresistance through Clonal Expansion in Acute Myeloid Leukemia. Leukemia 2020, 34, 1816–1827. [Google Scholar] [CrossRef]
- Long, J.; Jia, M.-Y.; Fang, W.-Y.; Chen, X.-J.; Mu, L.-L.; Wang, Z.-Y.; Shen, Y.; Xiang, R.-F.; Wang, L.-N.; Wang, L.; et al. FLT3 Inhibition Upregulates HDAC8 via FOXO to Inactivate P53 and Promote Maintenance of FLT3-ITD+ Acute Myeloid Leukemia. Blood 2020, 135, 1472–1483. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.; McQueen, T.; Chen, Y.; Jacamo, R.; Konopleva, M.; Shinojima, N.; Shpall, E.; Huang, X.; Andreeff, M. P53 Activation of Mesenchymal Stromal Cells Partially Abrogates Microenvironment-Mediated Resistance to FLT3 Inhibition in AML through HIF-1α–Mediated down-Regulation of CXCL12. Blood 2011, 118, 4431–4439. [Google Scholar] [CrossRef] [PubMed]
- Medina-Medina, I.; Martínez-Sánchez, M.; Hernández-Monge, J.; Fahraeus, R.; Muller, P.; Olivares-Illana, V. P53 Promotes Its Own Polyubiquitination by Enhancing the HDM2 and HDMX Interaction. Protein Sci. 2018, 27, 976–986. [Google Scholar] [CrossRef]
- Zimmerman, E.I.; Turner, D.C.; Buaboonnam, J.; Hu, S.; Orwick, S.; Roberts, M.S.; Janke, L.J.; Ramachandran, A.; Stewart, C.F.; Inaba, H.; et al. Crenolanib Is Active against Models of Drug-Resistant FLT3-ITD−positive Acute Myeloid Leukemia. Blood 2013, 122, 3607–3615. [Google Scholar] [CrossRef]
- Lu, J.-W.; Wang, A.-N.; Liao, H.-A.; Chen, C.-Y.; Hou, H.-A.; Hu, C.-Y.; Tien, H.-F.; Ou, D.-L.; Lin, L.-I. Cabozantinib Is Selectively Cytotoxic in Acute Myeloid Leukemia Cells with FLT3-Internal Tandem Duplication (FLT3-ITD). Cancer Lett. 2016, 376, 218–225. [Google Scholar] [CrossRef]
- Capelli, D.; Menotti, D.; Fiorentini, A.; Saraceni, F.; Olivieri, A. Overcoming Resistance: FLT3 Inhibitors Past, Present, Future and the Challenge of Cure. Cancers 2022, 14, 4315. [Google Scholar] [CrossRef]
- Yu, Z.; Du, J.; Hui, H.; Kan, S.; Huo, T.; Zhao, K.; Wu, T.; Guo, Q.; Lu, N. LT-171-861, a Novel FLT3 Inhibitor, Shows Excellent Preclinical Efficacy for the Treatment of FLT3 Mutant Acute Myeloid Leukemia. Theranostics 2021, 11, 93–106. [Google Scholar] [CrossRef]
- Hart, S.; Goh, K.C.; Novotny-Diermayr, V.; Tan, Y.C.; Madan, B.; Amalini, C.; Ong, L.C.; Kheng, B.; Cheong, A.; Zhou, J.; et al. Pacritinib (SB1518), a JAK2/FLT3 Inhibitor for the Treatment of Acute Myeloid Leukemia. Blood Cancer J. 2011, 1, e44. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Kaneko, N.; Ueno, Y.; Yamada, M.; Tanaka, R.; Saito, R.; Shimada, I.; Mori, K.; Kuromitsu, S. Gilteritinib, a FLT3/AXL Inhibitor, Shows Antileukemic Activity in Mouse Models of FLT3 Mutated Acute Myeloid Leukemia. Investig. New Drugs 2017, 35, 556–565. [Google Scholar] [CrossRef]
- Cervera, N.; Carbuccia, N.; Garnier, S.; Guille, A.; Adélaïde, J.; Murati, A.; Vey, N.; Mozziconacci, M.-J.; Chaffanet, M.; Birnbaum, D.; et al. Molecular Characterization of Acute Erythroid Leukemia (M6-AML) Using Targeted next-Generation Sequencing. Leukemia 2016, 30, 966–970. [Google Scholar] [CrossRef] [PubMed]
- Naeim, F.; Nagesh Rao, P.; Song, S.X.; Grody, W.W. 21—Acute Myeloid Leukemia, Not Otherwise Specified. In Atlas of Hematopathology; Naeim, F., Nagesh Rao, P., Song, S.X., Grody, W.W., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 259–281. [Google Scholar] [CrossRef]
- Martin, P.; Papayannopoulou, T. HEL Cells: A New Human Erythroleukemia Cell Line with Spontaneous and Induced Globin Expression. Science 1982, 216, 1233–1235. [Google Scholar] [CrossRef] [PubMed]
- Petiti, J.; Rosso, V.; Lo Iacono, M.; Panuzzo, C.; Calabrese, C.; Signorino, E.; Pironi, L.; Cartellà, A.; Bracco, E.; Pergolizzi, B.; et al. Curcumin Induces Apoptosis in JAK2-mutated Cells by the Inhibition of JAK2/STAT and MTORC1 Pathways. J. Cell. Mol. Med. 2019, 23, 4349–4357. [Google Scholar] [CrossRef] [PubMed]
- Boddu, P.; Benton, C.B.; Wang, W.; Borthakur, G.; Khoury, J.D.; Pemmaraju, N. Erythroleukemia-Historical Perspectives and Recent Advances in Diagnosis and Management. Blood Reviews 2018, 32, 96–105. [Google Scholar] [CrossRef]
- Santos, F.P.S.; Verstovsek, S. JAK2 Inhibitors: Are They the Solution? Clin. Lymphoma Myeloma Leuk. 2011, 11, S28–S36. [Google Scholar] [CrossRef]
- Venugopal, S.; Bar-Natan, M.; Mascarenhas, J.O. JAKs to STATs: A Tantalizing Therapeutic Target in Acute Myeloid Leukemia. Blood Rev. 2020, 40, 100634. [Google Scholar] [CrossRef]
- Broudy, V.C.; Lin, N.; Egrie, J.; de Haën, C.; Weiss, T.; Papayannopoulou, T.; Adamson, J.W. Identification of the Receptor for Erythropoietin on Human and Murine Erythroleukemia Cells and Modulation by Phorbol Ester and Dimethyl Sulfoxide. Proc. Natl. Acad. Sci. USA 1988, 85, 6513–6517. [Google Scholar] [CrossRef] [PubMed]
- Nagel, S.; Pommerenke, C.; Meyer, C.; MacLeod, R.A.F. NKL Homeobox Genes NKX2-3 and NKX2-4 Deregulate Megakaryocytic-Erythroid Cell Differentiation in AML. Int. J. Mol. Sci. 2021, 22, 11434. [Google Scholar] [CrossRef] [PubMed]
- Estrov, Z.; Shishodia, S.; Faderl, S.; Harris, D.; Van, Q.; Kantarjian, H.M.; Talpaz, M.; Aggarwal, B.B. Resveratrol Blocks Interleukin-1beta-Induced Activation of the Nuclear Transcription Factor NF-KappaB, Inhibits Proliferation, Causes S-Phase Arrest, and Induces Apoptosis of Acute Myeloid Leukemia Cells. Blood 2003, 102, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Papayannopoulou, T.; Nakamoto, B.; Kurachi, S.; Tweeddale, M.; Messner, H. Surface Antigenic Profile and Globin Phenotype of Two New Human Erythroleukemia Lines: Characterization and Interpretations. Blood 1988, 72, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Takaku, F.; Tange, T.; Shibuya, K.; Misawa, C.; Sasaki, K.; Miyagawa, K.; Yazaki, Y.; Hirai, H. Establishment and Erythroid Differentiation of a Cytokine-Dependent Human Leukemic Cell Line F-36: A Parental Line Requiring Granulocyte- Macrophage Colony-Stimulating Factor or Interleukin-3, and a Subline Requiring Erythropoietin. Blood 1991, 78, 2261–2268. [Google Scholar] [CrossRef]
- Drexler, H.G.; Zaborski, M.; Quentmeier, H. Cytokine Response Profiles of Human Myeloid Factor-Dependent Leukemia Cell Lines. Leukemia 1997, 11, 701–708. [Google Scholar] [CrossRef]
- Kitamura, T.; Tojo, A.; Kuwaki, T.; Chiba, S.; Miyazono, K.; Urabe, A.; Takaku, F. Identification and Analysis of Human Erythropoietin Receptors on a Factor-Dependent Cell Line, TF-1. Blood 1989, 73, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Tange, T.; Terasawa, T.; Chiba, S.; Kuwaki, T.; Miyagawa, K.; Piao, Y.-F.; Miyazono, K.; Urabe, A.; Takaku, F. Establishment and Characterization of a Unique Human Cell Line That Proliferates Dependently on GM-CSF, IL-3, or Erythropoietin. J. Cell. Physiol. 1989, 140, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Chrétien, S.; Varlet, P.; Verdier, F.; Gobert, S.; Cartron, J.P.; Gisselbrecht, S.; Mayeux, P.; Lacombe, C. Erythropoietin-Induced Erythroid Differentiation of the Human Erythroleukemia Cell Line TF-1 Correlates with Impaired STAT5 Activation. EMBO J. 1996, 15, 4174–4181. [Google Scholar] [CrossRef] [PubMed]
- Steube, K.G.; Meyer, C.; Tachibana, M.; Murai, M.; Drexler, H.G. Bladder Carcinoma Cell Line KU-19-19-Derived Cytokines Support Proliferation of Growth Factor-Dependent Hematopoietic Cell Lines: Modulation by Phorbol Ester, Interferon-γ and Interleukin-1β. Biochem. Biophys. Res. Commun. 1998, 242, 497–501. [Google Scholar] [CrossRef]
- Tucker, S.J.; Rae, C.; Littlejohn, A.F.; Paul, A.; MacEwan, D.J. Switching Leukemia Cell Phenotype between Life and Death. Proc. Natl. Acad. Sci. USA 2004, 101, 12940–12945. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Kuriyama, K.; Higuchi, M.; Tsushima, H.; Sohda, H.; Imai, N.; Saito, M.; Kondo, T.; Tomonaga, M. Establishment and Characterization of a New Erythropoietin-Dependent Acute Myeloid Leukemia Cell Line, AS-E2. Leukemia 1997, 11, 1941–1949. [Google Scholar] [CrossRef]
- Vicente, C.; Conchillo, A.; García-Sánchez, M.A.; Odero, M.D. The Role of the GATA2 Transcription Factor in Normal and Malignant Hematopoiesis. Crit. Rev. Oncol./Hematol. 2012, 82, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Wu, W.; Wang, X.; Gu, J. Clinical Diagnosis of Adult Patients with Acute Megakaryocytic Leukemia. Oncol. Lett. 2018, 16, 6988–6997. [Google Scholar] [CrossRef]
- De Marchi, F.; Araki, M.; Komatsu, N. Molecular Features, Prognosis, and Novel Treatment Options for Pediatric Acute Megakaryoblastic Leukemia. Expert Rev. Hematol. 2019, 12, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.W.; Li, B.; Prouet, P.; Giri, S.; Pathak, R.; Martin, M.G. Acute Megakaryocytic Leukemia: What Have We Learned. Blood Rev. 2016, 30, 49–53. [Google Scholar] [CrossRef]
- Yoshida, K.; Toki, T.; Okuno, Y.; Kanezaki, R.; Shiraishi, Y.; Sato-Otsubo, A.; Sanada, M.; Park, M.; Terui, K.; Suzuki, H.; et al. The Landscape of Somatic Mutations in Down Syndrome–Related Myeloid Disorders. Nat. Genet. 2013, 45, 1293–1299. [Google Scholar] [CrossRef] [PubMed]
- Hitzler, J.K.; Cheung, J.; Li, Y.; Scherer, S.W.; Zipursky, A. GATA1 Mutations in Transient Leukemia and Acute Megakaryoblastic Leukemia of Down Syndrome. Blood 2003, 101, 4301–4304. [Google Scholar] [CrossRef]
- Hitzler, J.K. Acute Megakaryoblastic Leukemia in Down Syndrome. Pediatr. Blood Cancer 2007, 49 (Suppl. S7), 1066–1069. [Google Scholar] [CrossRef] [PubMed]
- De Rooij, J.D.E.; Branstetter, C.; Ma, J.; Li, Y.; Walsh, M.P.; Cheng, J.; Obulkasim, A.; Dang, J.; Easton, J.; Verboon, L.J.; et al. Pediatric Non-Down Syndrome Acute Megakaryoblastic Leukemia Is Characterized by Distinct Genomic Subsets with Varying Outcomes. Nat. Genet. 2017, 49, 451–456. [Google Scholar] [CrossRef]
- Komatsu, N.; Suda, T.; Moroi, M.; Tokuyama, N.; Sakata, Y.; Okada, M.; Nishida, T.; Hirai, Y.; Sato, T.; Fuse, A. Growth and Differentiation of a Human Megakaryoblastic Cell Line, CMK. Blood 1989, 74, 42–48. [Google Scholar] [CrossRef]
- Sato, T.; Fuse, A.; Eguchi, M.; Hayashi, Y.; Ryo, R.; Adachi, M.; Kishimoto, Y.; Teramura, M.; Mizoguchi, H.; Shima, Y.; et al. Establishment of a Human Leukaemic Cell Line (CMK) with Megakaryocytic Characteristics from a Down’s Syndrome Patient with Acute Megakaryoblastic Leukaemia. Br. J. Haematol. 1989, 72, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Mouthon, M.A.; Freund, M.; Titeux, M.; Katz, A.; Guichard, J.; Breton-Gorius, J.; Vainchenker, W. Growth and Differentiation of the Human Megakaryoblastic Cell Line (ELF-153): A Model for Early Stages of Megakaryocytopoiesis. Blood 1994, 84, 1085–1097. [Google Scholar] [CrossRef]
- Saito, H.; Hayakawa, M.; Kamoshita, N.; Yasumoto, A.; Suzuki-Inoue, K.; Yatomi, Y.; Ohmori, T. Establishment of a Megakaryoblastic Cell Line for Conventional Assessment of Platelet Calcium Signaling. Int. J. Hematol. 2020, 111, 786–794. [Google Scholar] [CrossRef]
- Avanzi, G.C.; Lista, P.; Giovinazzo, B.; Miniero, R.; Saglio, G.; Benetton, G.; Coda, R.; Cattoretti, G.; Pegoraro, L. Selective Growth Response to IL-3 of a Human Leukaemic Cell Line with Megakaryoblastic Features. Br. J. Haematol. 1988, 69, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Brizzi, M.F.; Avanzi, G.C.; Veglia, F.; Clark, S.C.; Pegoraro, L. Expression and Modulation of IL-3 and GM-CSF Receptors in Human Growth Factor Dependent Leukaemic Cells. Br. J. Haematol. 1990, 76, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Avanzi, G.C.; Brizzi, M.F.; Giannotti, J.; Ciarletta, A.; Yang, Y.-C.; Pegoraro, L.; Clark, S.C. M-07e Human Leukemic Factor-Dependent Cell Line Provides a Rapid and Sensitive Bioassay for the Human Cytokines GM-CSF and IL-3. J. Cell. Physiol. 1990, 145, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Ogura, M.; Morishima, Y.; Ohno, R.; Kato, Y.; Hirabayashi, N.; Nagura, H.; Saito, H. Establishment of a Novel Human Megakaryoblastic Leukemia Cell Line, MEG-01, with Positive Philadelphia Chromosome. Blood 1985, 66, 1384–1392. [Google Scholar] [CrossRef]
- Drexler, H.G.; MacLeod, R.A.; Uphoff, C.C. Leukemia Cell Lines: In Vitro Models for the Study of Philadelphia Chromosome-Positive Leukemia. Leuk. Res. 1999, 23, 207–215. [Google Scholar] [CrossRef]
- Wertheim, J.A.; Forsythe, K.; Druker, B.J.; Hammer, D.; Boettiger, D.; Pear, W.S. BCR-ABL-Induced Adhesion Defects Are Tyrosine Kinase-Independent. Blood 2002, 99, 4122–4130. [Google Scholar] [CrossRef]
- Takeuchi, K.; Ogura, M.; Saito, H.; Satoh, M.; Takeuchi, M. Production of Platelet-like Particles by a Human Megakaryoblastic Leukemia Cell Line (MEG-01). Exp. Cell Res. 1991, 193, 223–226. [Google Scholar] [CrossRef]
- Murate, T.; Hotta, T.; Tsushita, K.; Suzuki, M.; Yoshida, T.; Saga, S.; Saito, H.; Yoshida, S. Aphidicolin, an Inhibitor of DNA Replication, Blocks the TPA-Induced Differentiation of a Human Megakaryoblastic Cell Line, MEG-O1. Blood 1991, 78, 3168–3177. [Google Scholar] [CrossRef] [PubMed]
- Quentmeier, H.; Pommerenke, C.; Dirks, W.G.; Eberth, S.; Koeppel, M.; MacLeod, R.A.F.; Nagel, S.; Steube, K.; Uphoff, C.C.; Drexler, H.G. The LL-100 Panel: 100 Cell Lines for Blood Cancer Studies. Sci. Rep. 2019, 9, 8218. [Google Scholar] [CrossRef]
- Quentmeier, H.; Schneider, B.; Röhrs, S.; Romani, J.; Zaborski, M.; Macleod, R.A.F.; Drexler, H.G. SET-NUP214 Fusion in Acute Myeloid Leukemia- and T-Cell Acute Lymphoblastic Leukemia-Derived Cell Lines. J. Hematol. Oncol. 2009, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A. The Cellosaurus, a Cell-Line Knowledge Resource. J. Biomol. Tech. 2018, 29, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Koblitz, J.; Dirks, W.G.; Eberth, S.; Nagel, S.; Steenpass, L.; Pommerenke, C. DSMZCellDive: Diving into High-Throughput Cell Line Data. F1000Research 2022, 11, 420. [Google Scholar] [CrossRef] [PubMed]
- Mihara, K.; Imai, C.; Coustan-Smith, E.; Dome, J.S.; Dominici, M.; Vanin, E.; Campana, D. Development and Functional Characterization of Human Bone Marrow Mesenchymal Cells Immortalized by Enforced Expression of Telomerase. Br. J. Haematol. 2003, 120, 846–849. [Google Scholar] [CrossRef]
- Böcker, W.; Yin, Z.; Drosse, I.; Haasters, F.; Rossmann, O.; Wierer, M.; Popov, C.; Locher, M.; Mutschler, W.; Docheva, D.; et al. Introducing a Single-Cell-Derived Human Mesenchymal Stem Cell Line Expressing HTERT after Lentiviral Gene Transfer. J. Cell. Mol. Med. 2008, 12, 1347–1359. [Google Scholar] [CrossRef]
- Galarza Torre, A.; Shaw, J.E.; Wood, A.; Gilbert, H.T.J.; Dobre, O.; Genever, P.; Brennan, K.; Richardson, S.M.; Swift, J. An Immortalised Mesenchymal Stem Cell Line Maintains Mechano-Responsive Behaviour and Can Be Used as a Reporter of Substrate Stiffness. Sci. Rep. 2018, 8, 8981. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skopek, R.; Palusińska, M.; Kaczor-Keller, K.; Pingwara, R.; Papierniak-Wyglądała, A.; Schenk, T.; Lewicki, S.; Zelent, A.; Szymański, Ł. Choosing the Right Cell Line for Acute Myeloid Leukemia (AML) Research. Int. J. Mol. Sci. 2023, 24, 5377. https://doi.org/10.3390/ijms24065377
Skopek R, Palusińska M, Kaczor-Keller K, Pingwara R, Papierniak-Wyglądała A, Schenk T, Lewicki S, Zelent A, Szymański Ł. Choosing the Right Cell Line for Acute Myeloid Leukemia (AML) Research. International Journal of Molecular Sciences. 2023; 24(6):5377. https://doi.org/10.3390/ijms24065377
Chicago/Turabian StyleSkopek, Rafał, Małgorzata Palusińska, Katarzyna Kaczor-Keller, Rafał Pingwara, Anna Papierniak-Wyglądała, Tino Schenk, Sławomir Lewicki, Artur Zelent, and Łukasz Szymański. 2023. "Choosing the Right Cell Line for Acute Myeloid Leukemia (AML) Research" International Journal of Molecular Sciences 24, no. 6: 5377. https://doi.org/10.3390/ijms24065377
APA StyleSkopek, R., Palusińska, M., Kaczor-Keller, K., Pingwara, R., Papierniak-Wyglądała, A., Schenk, T., Lewicki, S., Zelent, A., & Szymański, Ł. (2023). Choosing the Right Cell Line for Acute Myeloid Leukemia (AML) Research. International Journal of Molecular Sciences, 24(6), 5377. https://doi.org/10.3390/ijms24065377





