A Cinnamate 4-HYDROXYLASE1 from Safflower Promotes Flavonoids Accumulation and Stimulates Antioxidant Defense System in Arabidopsis
Abstract
1. Introduction
2. Results
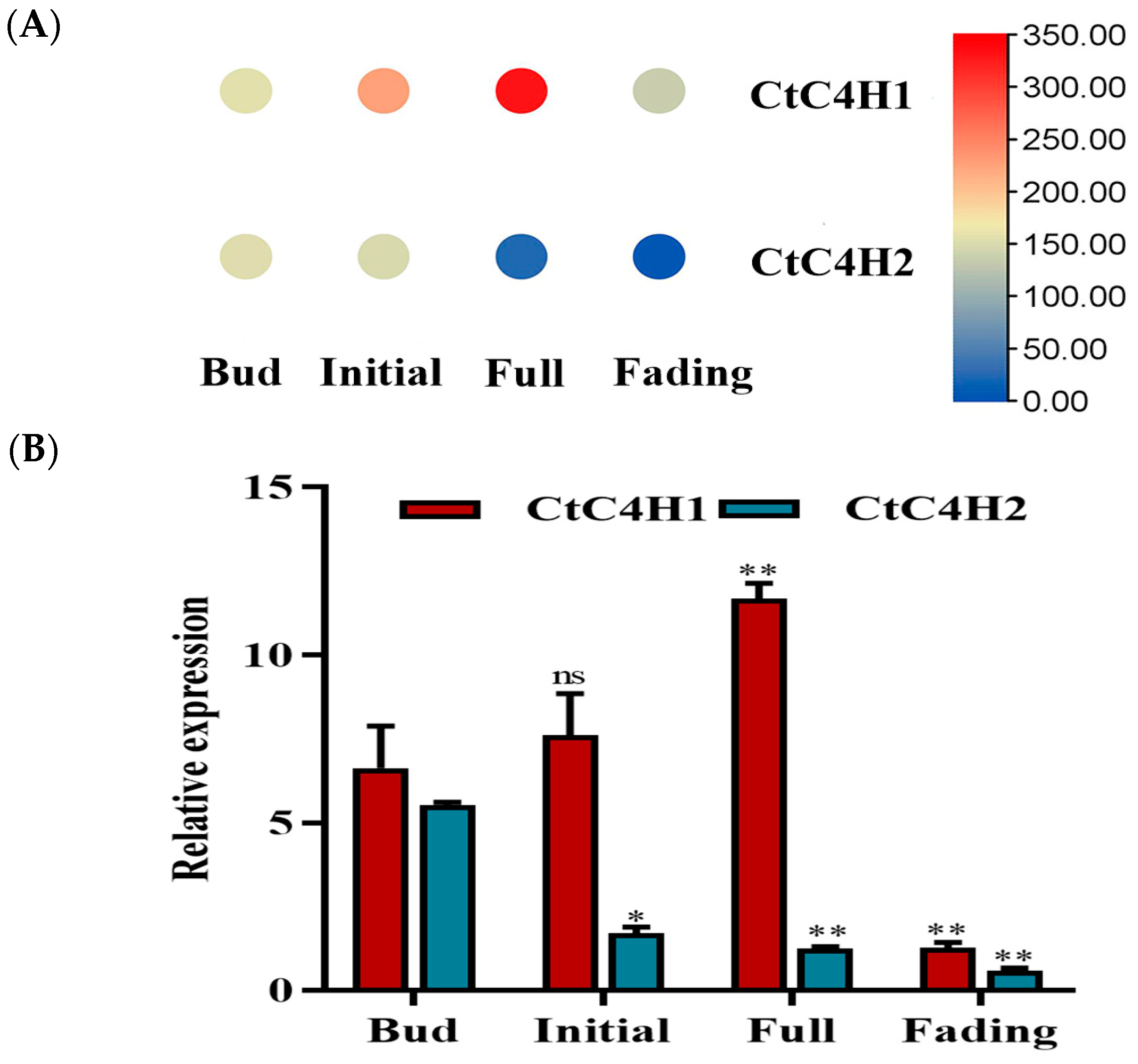
2.1. Transcriptome Analysis and Identification of CtC4H Genes in Safflower
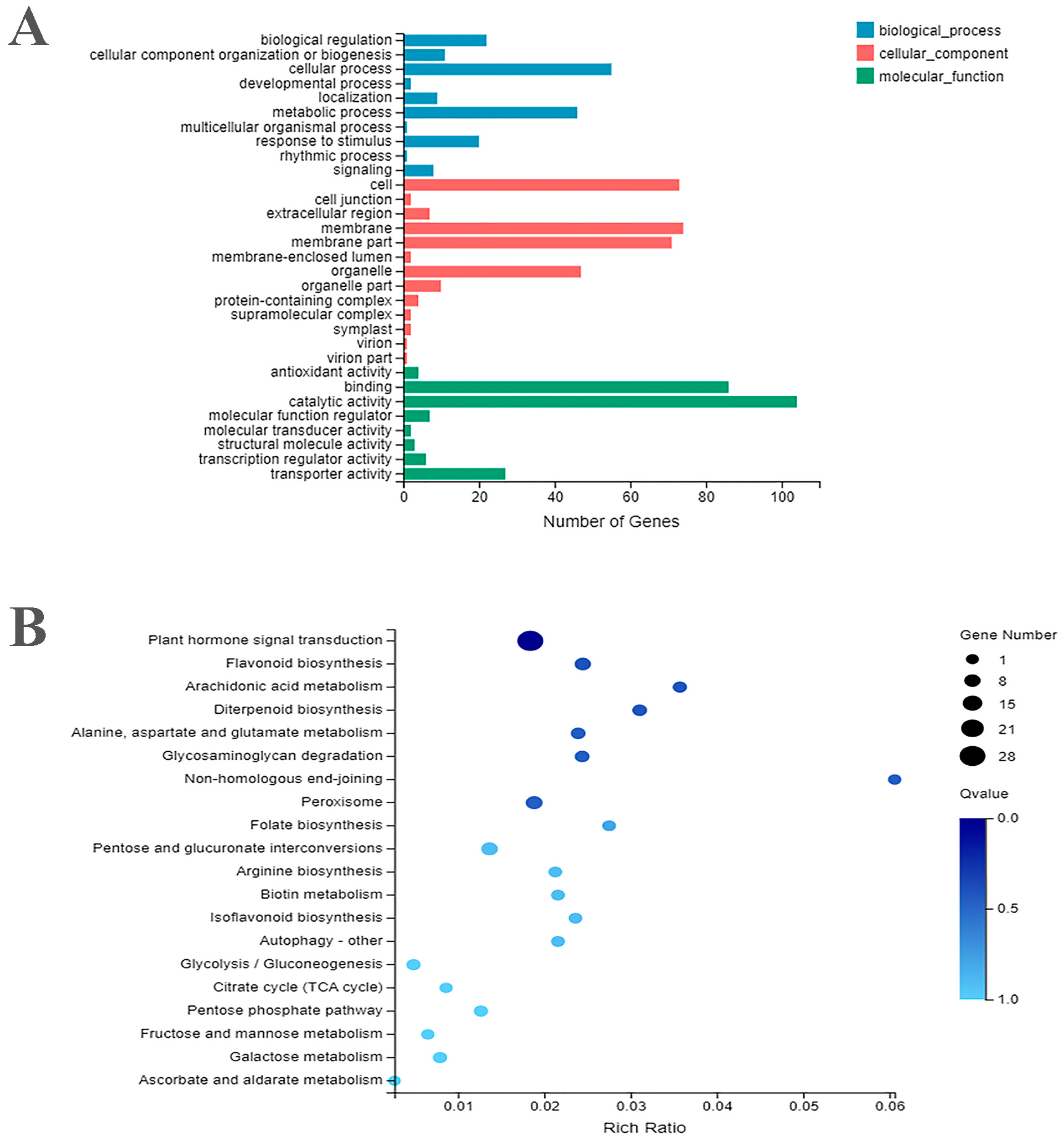
2.2. Functional Annotations of DEGS
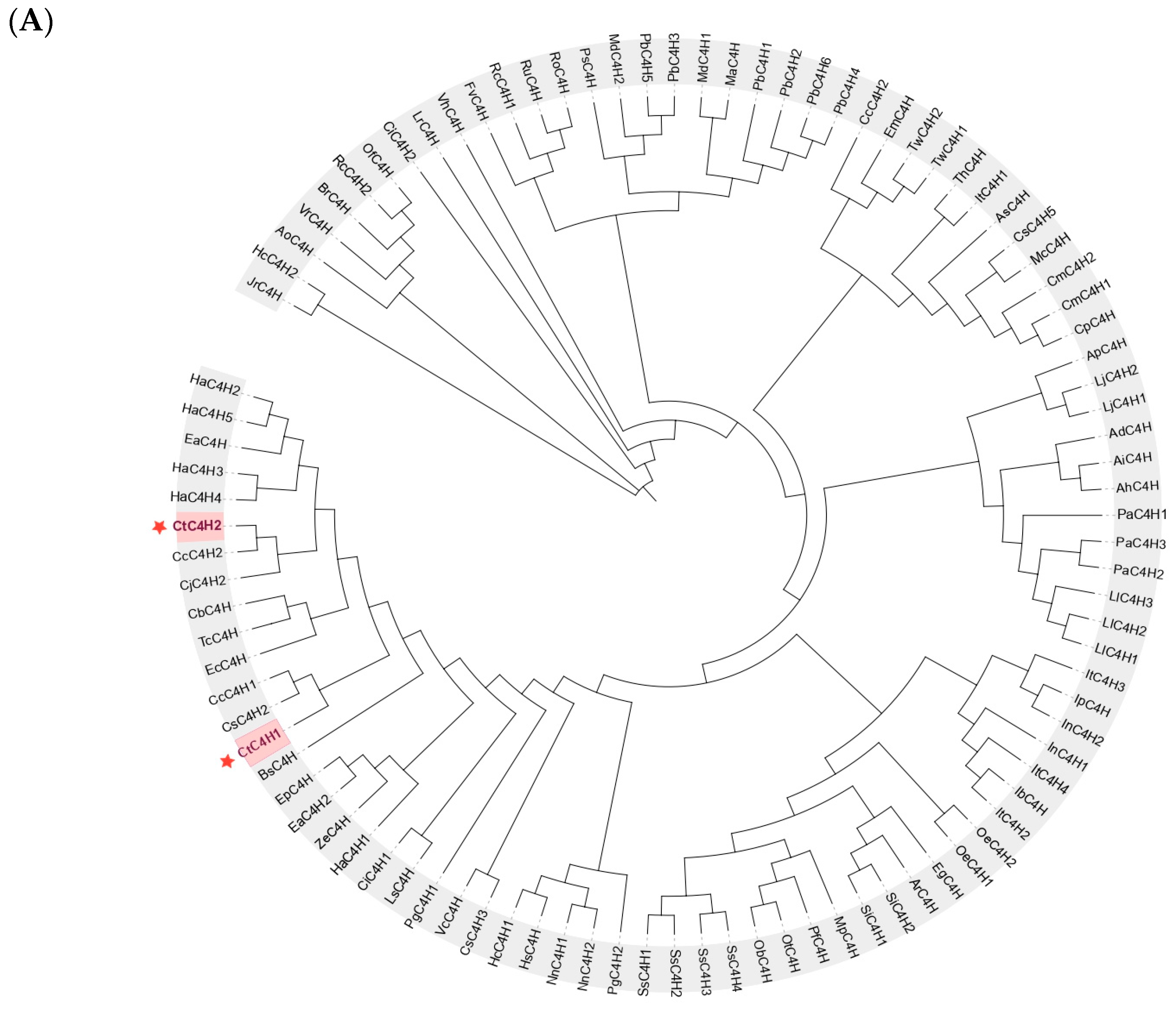
2.3. Phylogenetic Analysis and Multiple Sequence Alignments of CtC4H Proteins
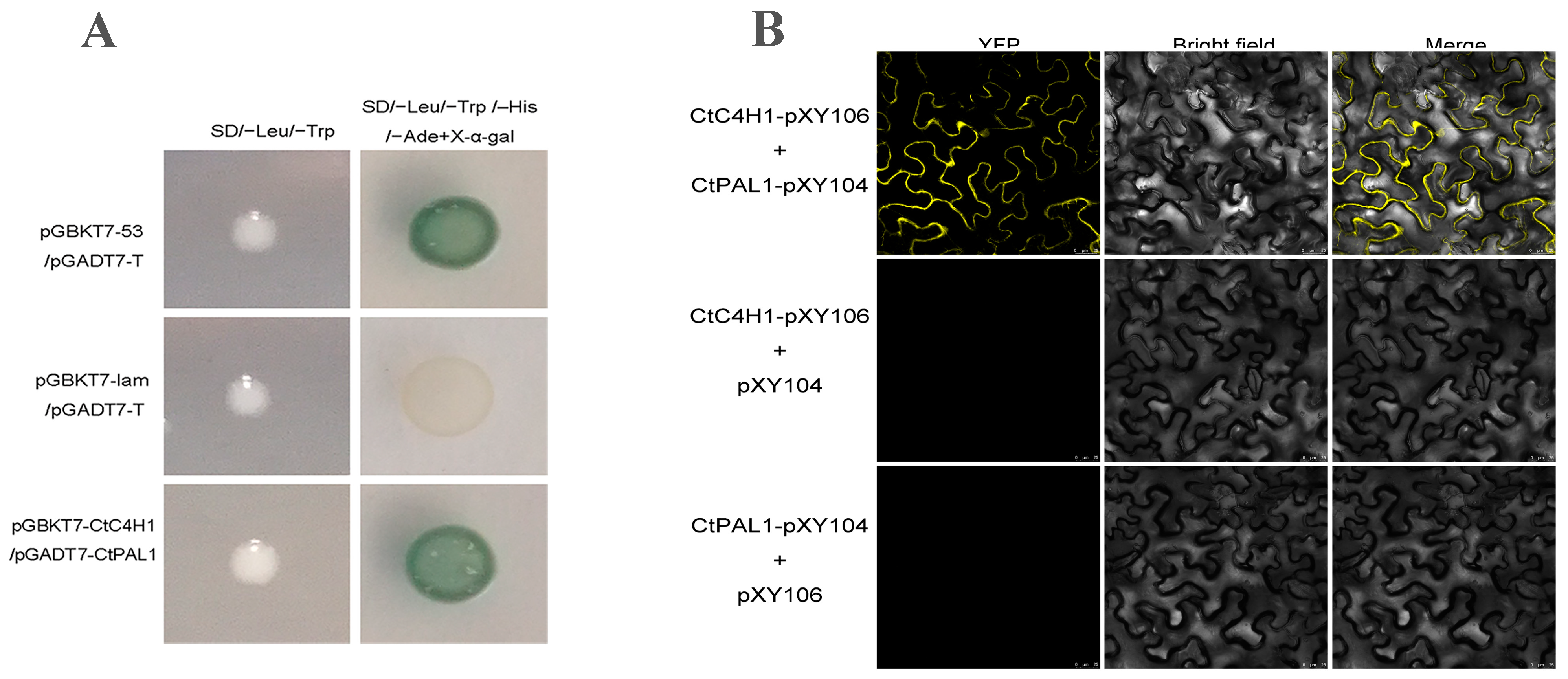
2.4. CtC4H1 Interacts with CtPAL1 In Vivo
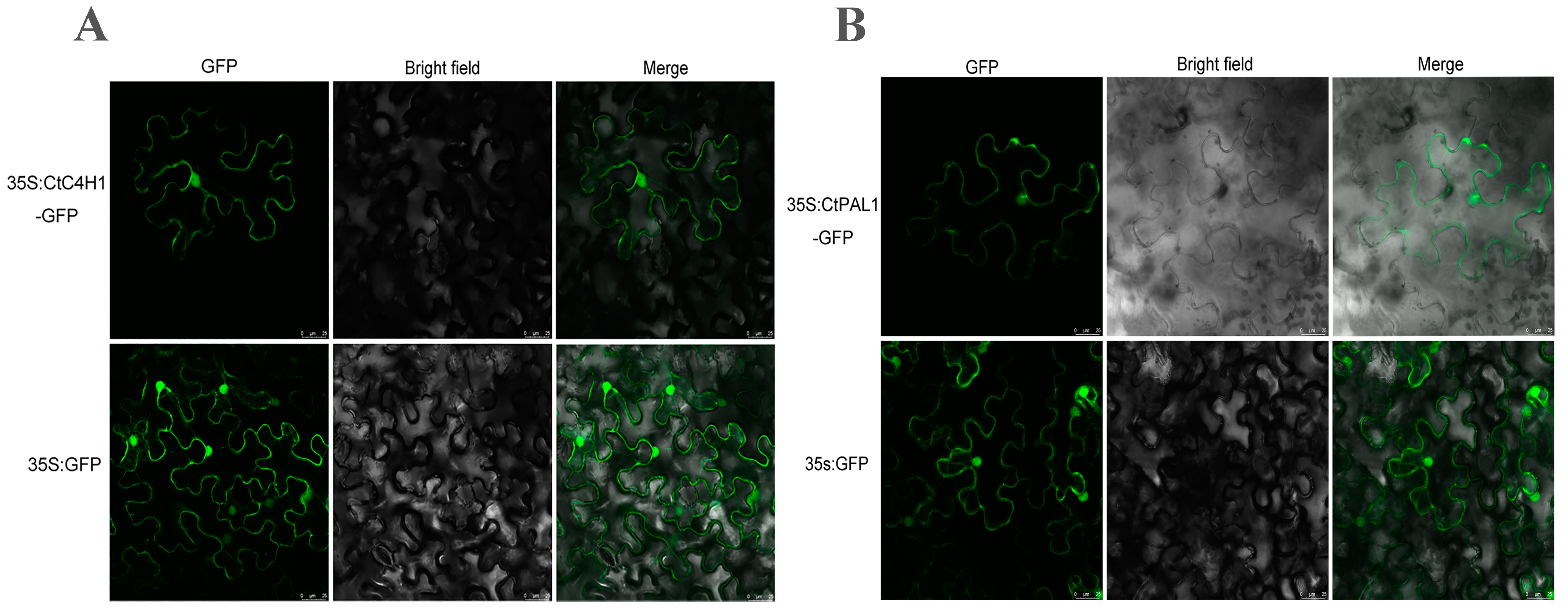
2.5. Subcellular Localization Assays
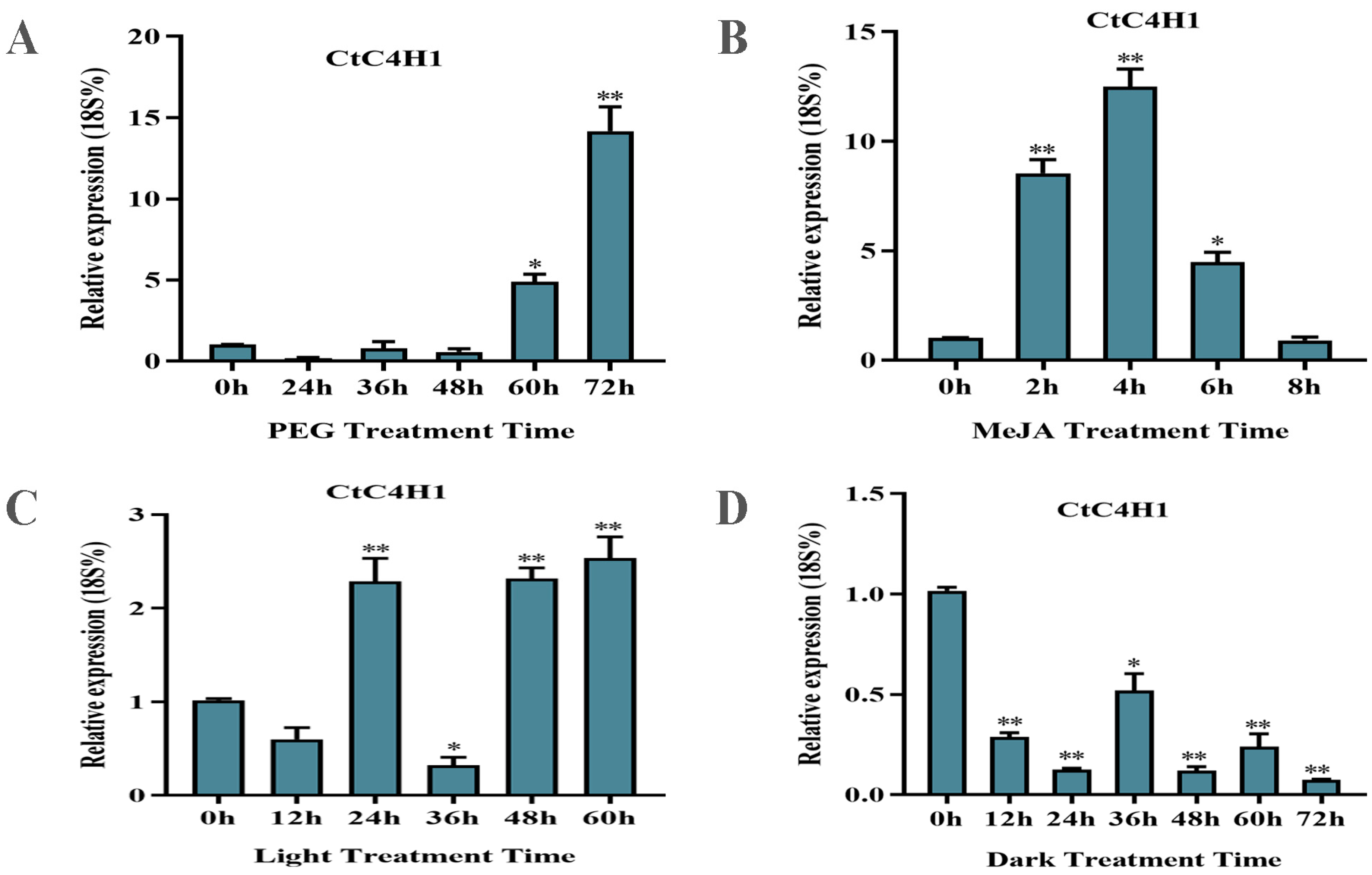
2.6. Expression Pattern of CtC4H1 Gene under Different Abiotic Stresses in Safflower
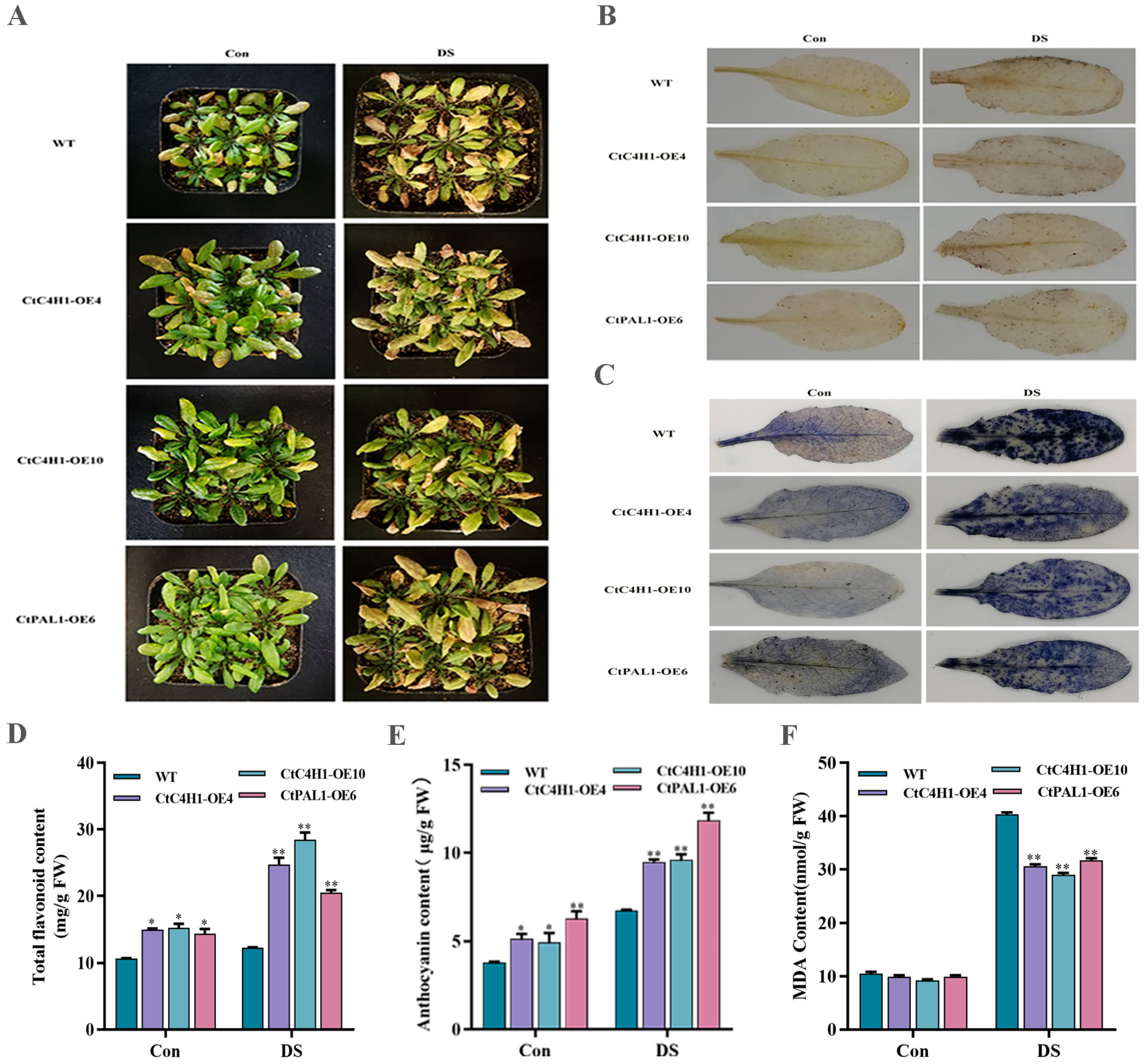
2.7. Phenotypic Analysis and Quantification of Metabolites in CtC4H1 Overexpressed Arabidopsis
2.8. Overexpression of CtC4H1 and CtPAL1 Enhanced Drought Stress Tolerance in Transgenic Arabidopsis
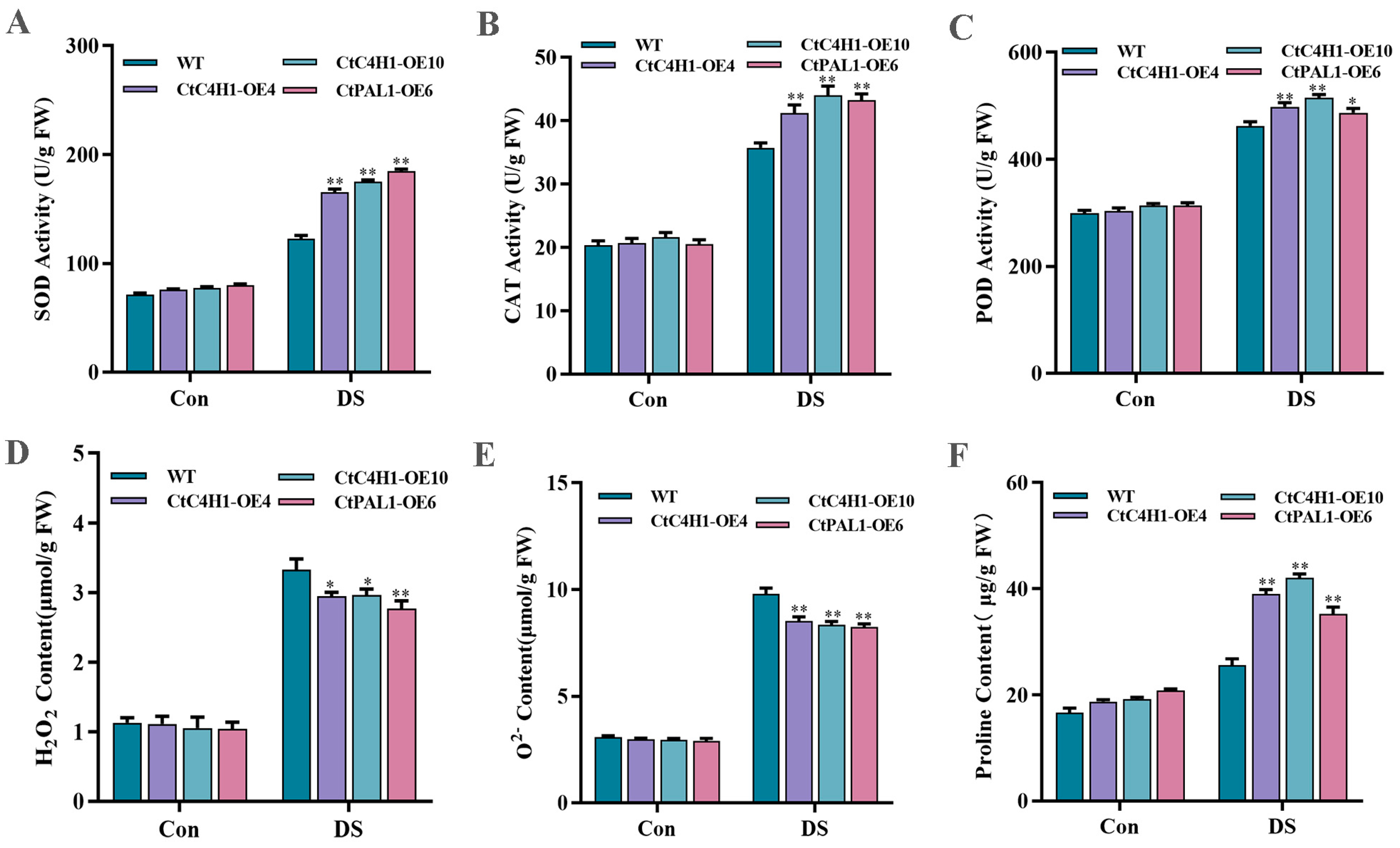
2.9. CtC4H1 Positively Regulates ROS Scavenging Ability under Drought Stress
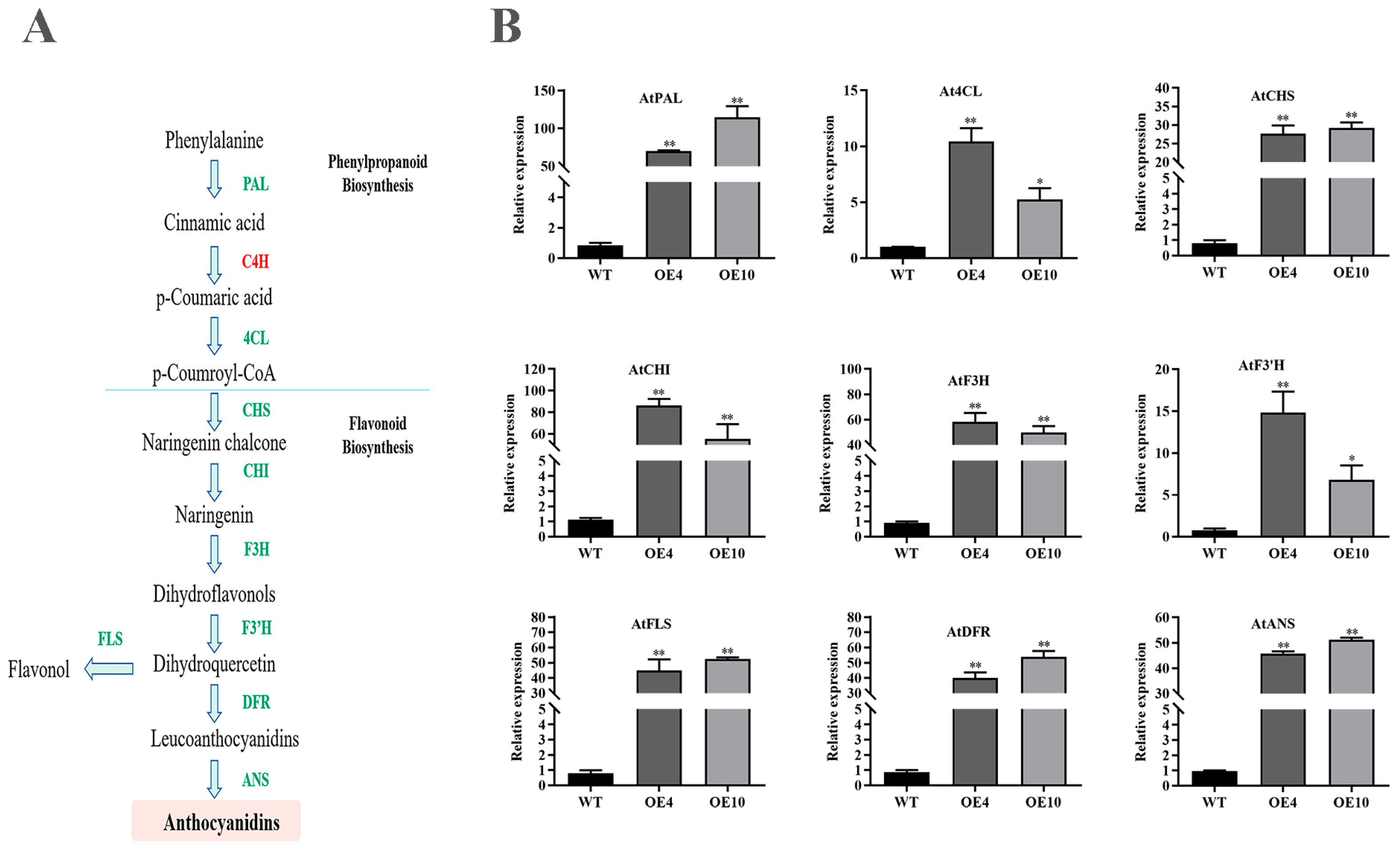
2.10. CtC4H1 Positively Regulates the Expression of Genes Involved in Flavonoid Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Treatments, and Growth Conditions
4.2. RNA-Seq, Functional Annotation and Differential Expression Analysis
4.3. Multiple Sequence Alignments and Phylogenetic Analyses
4.4. Molecular Cloning and Subcellular Localization of CtC4H1
4.5. Yeast-2-Hybrid (Y2H) Assay and Bimolecular Fluorescence Complementation (BiFC)
4.6. Construction of the Plant Expression Vector and Arabidopsis Transformation
4.7. Expression Analysis
4.8. Determination of Total Flavonoids and Anthocyanins Content
4.9. DAB and NBT Staining
4.10. Measurement of Physiological Traits
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ekin, Z. Resurgence of Safflower (Carthamus tinctorius L.) Utilization: A Global View. J. Agron. 2005, 4, 83–87. [Google Scholar] [CrossRef]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef]
- He, J.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Traka, M.H.; Mithen, R.F. Plant Science and Human Nutrition: Challenges in Assessing Health-Promoting Properties of Phytochemicals. Plant Cell 2011, 23, 2483–2497. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, J.; Xie, J.; Deng, L.; Yao, S.; Zeng, K. Transcriptomic and biochemical analysis of highlighted induction of phenylpropanoid pathway metabolism of citrus fruit in response to salicylic acid, Pichia membranaefaciens and oligochitosan. Postharvest Biol. Technol. 2018, 142, 81–92. [Google Scholar] [CrossRef]
- Torres, C.A.; Azocar, C.; Ramos, P.; Pérez-Díaz, R.; Sepulveda, G.; Moya-León, M.A. Photooxidative stress activates a complex multigenic response integrating the phenylpropanoid pathway and ethylene, leading to lignin accumulation in apple (Malus domestica Borkh.) fruit. Hortic. Res. 2020, 7, 22. [Google Scholar] [CrossRef]
- Wang, R.; Ren, C.; Dong, S.; Chen, C.; Xian, B.; Wu, Q.; Wang, J.; Pei, J.; Chen, J. Integrated Metabolomics and Transcriptome Analysis of Flavonoid Biosynthesis in Safflower (Carthamus tinctorius L.) With Different Colors. Front. Plant Sci. 2021, 12, 712038. [Google Scholar] [CrossRef] [PubMed]
- Schilmiller, A.L.; Stout, J.; Weng, J.K.; Humphreys, J.; Ruegger, M.O.; Chapple, C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009, 60, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, N.; Wei, Y.; Zou, X.; Jiang, S.; Xu, F.; Wang, H.; Shao, X. Terpinen-4-ol Enhances Disease Resistance of Postharvest Strawberry Fruit More Effectively than Tea Tree Oil by Activating the Phenylpropanoid Metabolism Pathway. J. Agric. Food Chem. 2020, 68, 6739–6747. [Google Scholar] [CrossRef]
- Zafari, M.; Ebadi, A.; Jahanbakhsh, S.; Sedghi, M. Safflower (Carthamus tinctorius) Biochemical Properties, Yield, and Oil Content Affected by 24-Epibrassinosteroid and Genotype under Drought Stress. J. Agric. Food Chem. 2020, 68, 6040–6047. [Google Scholar] [CrossRef]
- Joshi, S.; Thoday-Kennedy, E.; Daetwyler, H.D.; Hayden, M.; Spangenberg, G.; Kant, S. High-throughput phenotyping to dissect genotypic differences in safflower for drought tolerance. PLoS ONE 2021, 16, e254908. [Google Scholar] [CrossRef] [PubMed]
- Amini, H.; Arzani, A.; Bahrami, F. Seed yield and some physiological traits of safflower as affected by water deficit stress. Int. J. Plant Prod. 2013, 7, 597–614. [Google Scholar]
- Rezaie, R.; Abdollahi Mandoulakani, B.; Fattahi, M. Cold stress changes antioxidant defense system, phenylpropanoid contents and expression of genes involved in their biosynthesis in Ocimum basilicum L. Sci. Rep. 2020, 10, 5290. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Del, R.L. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid Biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Achnine, L.; Blancaflor, E.B.; Rasmussen, S.; Dixon, R.A. Colocalization of l-Phenylalanine Ammonia-Lyase and Cinnamate 4-Hydroxylase for Metabolic Channeling in Phenylpropanoid Biosynthesis. Plant Cell 2004, 16, 3098–3109. [Google Scholar] [CrossRef] [PubMed]
- Noé, W.; Langebartels, C.; Seitz, H.U. Anthocyanin accumulation and pal activity in a suspension culture of daucus carota l. Planta 1980, 149, 283–287. [Google Scholar] [CrossRef]
- Jakubczyk, K.; Dec, K.; Kaldunska, J.; Kawczuga, D.; Kochman, J.; Janda, K. Reactive oxygen species-sources, functions, oxidative damage. Pol. Merkur. Lek. 2020, 48, 124–127. [Google Scholar]
- Xia, J.; Liu, Y.; Yao, S.; Li, M.; Zhu, M.; Huang, K.; Gao, L.; Xia, T. Characterization and Expression Profiling of Camellia sinensis Cinnamate 4-hydroxylase Genes in Phenylpropanoid Pathways. Genes 2017, 8, 193. [Google Scholar] [CrossRef]
- Zhang, B.; Lewis, K.M.; Abril, A.; Davydov, D.R.; Vermerris, W.; Sattler, S.E.; Kang, C. Structure and Function of the Cytochrome P450 Monooxygenase Cinnamate 4-hydroxylase fromSorghum bicolor. Plant Physiol. 2020, 183, 957–973. [Google Scholar] [CrossRef]
- Ding, X.; Zhu, X.; Zheng, W.; Li, F.; Xiao, S.; Duan, X. BTH Treatment Delays the Senescence of Postharvest Pitaya Fruit in Relation to Enhancing Antioxidant System and Phenylpropanoid Pathway. Foods 2021, 10, 846. [Google Scholar] [CrossRef]
- Cheniany, M.; Ganjeali, A. Developmental role of phenylalanine-ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) genes during adventitious rooting of Juglans regia L. microshoots. Acta Biol. Hung. 2016, 67, 379–392. [Google Scholar] [CrossRef]
- Silveira, E.; Nascimento, F.C.; Yujra, V.Q.; Webber, L.P.; Castilho, R.M.; Squarize, C.H. BMAL1 Modulates Epidermal Healing in a Process Involving the Antioxidative Defense Mechanism. Int. J. Mol. Sci. 2020, 21, 901. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.M.; Carey, E.E.; Rajashekar, C.B. Environmental stresses induce health-promoting phytochemicals in lettuce. Plant Physiol. Biochem. 2009, 47, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.H.; Chung, B.Y.; Kim, J.H.; Kim, J.S.; Lee, S.S.; An, B.C.; Lee, I.J.; Kim, T.H. cDNA cloning and expression pattern of cinnamate-4-hydroxylase in the Korean black raspberry. BMB Rep. 2008, 41, 529–536. [Google Scholar] [CrossRef]
- Koopmann, E.; Logemann, E.; Hahlbrock, K. Regulation and functional expression of cinnamate 4-hydroxylase from parsley. Plant Physiol. 1999, 119, 49–56. [Google Scholar] [CrossRef]
- El Houari, I.; Van Beirs, C.; Arents, H.E.; Han, H.; Chanoca, A.; Opdenacker, D.; Pollier, J.; Storme, V.; Steenackers, W.; Quareshy, M.; et al. Seedling developmental defects upon blocking CINNAMATE-4-HYDROXYLASE are caused by perturbations in auxin transport. New Phytol. 2021, 230, 2275–2291. [Google Scholar] [CrossRef]
- Schuler, M.A.; Werck-Reichhart, D. Functional genomics of P450s. Annu. Rev. Plant Biol. 2003, 54, 629–667. [Google Scholar] [CrossRef] [PubMed]
- Ellard-Ivey, M.; Douglas, C.J. Role of Jasmonates in the Elicitor- and Wound-Inducible Expression of Defense Genes in Parsley and Transgenic Tobacco. Plant Physiol. 1996, 112, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Hidalgo-Shrestha, C.; Bonawitz, N.D.; Rochus, B.; Franke, A.C.C. Spatio-temporal control of phenylpropanoid biosynthesis by inducible complementation of a cinnamate 4-hydroxylase mutant. J. Exp. Bot. 2020, 72, 3061–3073. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.C.; Pinto, D.; Silva, A. Plant Flavonoids: Chemical Characteristics and Biological Activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, B.; Ma, L.; Zheng, X.; Gong, D.; Xue, H.; Bi, Y.; Wang, Y.; Zhang, Z.; Prusky, D. Benzo-(1,2,3)-thiadiazole-7-carbothioic acid s-methyl ester (BTH) promotes tuber wound healing of potato by elevation of phenylpropanoid metabolism. Postharvest Biol. Technol. 2019, 153, 125–132. [Google Scholar] [CrossRef]
- Medrano-Macias, J.; Leija-Martinez, P.; Gonzalez-Morales, S.; Juarez-Maldonado, A.; Benavides-Mendoza, A. Use of Iodine to Biofortify and Promote Growth and Stress Tolerance in Crops. Front. Plant Sci. 2016, 7, 1146. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Geng, P.; Zhang, S.; Liu, J.; Zhao, C.; Wu, J.; Cao, Y.; Fu, C.; Han, X.; He, H.; Zhao, Q. MYB20, MYB42, MYB43, and MYB85 Regulate Phenylalanine and Lignin Biosynthesis during Secondary Cell Wall Formation. Plant Physiol. 2020, 182, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Liu, X.; Zhang, Y.; Muhammad, A.; Han, W.; Li, D.; Cheng, X.; Cai, Y. Cloning and functional characterization of two cinnamate 4-hydroxylase genes from Pyrus bretschneideri. Plant Physiol. Biochem. 2020, 156, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Bu, J.; Yu, Y.; Aisikaer, G.; Ying, T. Postharvest UV-C irradiation inhibits the production of ethylene and the activity of cell wall-degrading enzymes during softening of tomato (Lycopersicon esculentum L.) fruit. Postharvest Biol. Technol. 2013, 86, 337–345. [Google Scholar] [CrossRef]
- Wohl, J.; Petersen, M. Functional expression and characterization of cinnamic acid 4-hydroxylase from the hornwort Anthoceros agrestis in Physcomitrella patens. Plant Cell Rep. 2020, 39, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plantarum 1999, 107, 142–149. [Google Scholar] [CrossRef]
- Ni, R.; Zhu, T.; Zhang, X.; Wang, P.; Sun, C.; Qiao, Y.; Lou, H.; Cheng, A. Identification and evolutionary analysis of chalcone isomerase-fold proteins in ferns. J. Exp. Bot. 2019, 71, 290–304. [Google Scholar] [CrossRef]
- Xu, H.; Lan, Y.; Xing, J.; Li, Y.; Liu, L.; Wang, Y. AfCHIL, a Type IV Chalcone Isomerase, Enhances the Biosynthesis of Naringenin in Metabolic Engineering. Front. Plant Sci. 2022, 13, 891066. [Google Scholar] [CrossRef]
- Jiang, W.; Yin, Q.; Wu, R.; Zheng, G.; Liu, J.; Dixon, R.A.; Pang, Y. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 7165–7179. [Google Scholar] [CrossRef] [PubMed]
- Waki, T.; Mameda, R.; Nakano, T.; Yamada, S.; Terashita, M.; Ito, K.; Tenma, N.; Li, Y.; Fujino, N.; Uno, K.; et al. A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat. Commun. 2020, 11, 870. [Google Scholar] [CrossRef] [PubMed]
- Srere, P.A. Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 1987, 56, 89–124. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.; Dixon, R.A. Transgene-Mediated and Elicitor-Induced Perturbation of Metabolic Channeling at the Entry Point into the Phenylpropanoid Pathway. Plant Cell 1999, 11, 1537–1551. [Google Scholar] [CrossRef]
- Gou, M.; Ran, X.; Martin, D.W.; Liu, C.J. The scaffold proteins of lignin biosynthetic cytochrome P450 enzymes. Nat. Plants 2018, 4, 299–310. [Google Scholar] [CrossRef]
- Wang, C.; Gao, B.; Chen, N.; Jiao, P.; Jiang, Z.; Zhao, C.; Ma, Y.; Guan, S.; Liu, S. A Novel Senescence-Specific Gene (ZmSAG39) Negatively Regulates Darkness and Drought Responses in Maize. Int. J. Mol. Sci. 2022, 23, 15984. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, P.; Guo, P.; Chong, L.; Yu, G.; Sun, X.; Hu, T.; Li, Y.; Hsu, C.C.; Tang, K.; et al. CDK8 is associated with RAP2.6 and SnRK2.6 and positively modulates abscisic acid signaling and drought response in Arabidopsis. New Phytol. 2020, 228, 1573–1590. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, J.; Ali, A.A.; Al-Huqail, A.A.; Qureshi, M.I. Triacontanol attenuates drought-induced oxidative stress in Brassica juncea L. by regulating lignification genes, calcium metabolism and the antioxidant system. Plant Physiol. Bioch. 2021, 166, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Shah Jahan, M.; Wang, Y.; Shu, S.; Zhong, M.; Chen, Z.; Wu, J.; Sun, J.; Guo, S. Exogenous salicylic acid increases the heat tolerance in Tomato (Solanum lycopersicum L) by enhancing photosynthesis efficiency and improving antioxidant defense system through scavenging of reactive oxygen species. Sci. Hortic. 2019, 247, 421–429. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Y.; Wang, Y.; Liu, X.; Ahmad, N.; Wang, N.; Jin, L.; Yao, N.; Liu, X. A Cinnamate 4-HYDROXYLASE1 from Safflower Promotes Flavonoids Accumulation and Stimulates Antioxidant Defense System in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 5393. https://doi.org/10.3390/ijms24065393
Hou Y, Wang Y, Liu X, Ahmad N, Wang N, Jin L, Yao N, Liu X. A Cinnamate 4-HYDROXYLASE1 from Safflower Promotes Flavonoids Accumulation and Stimulates Antioxidant Defense System in Arabidopsis. International Journal of Molecular Sciences. 2023; 24(6):5393. https://doi.org/10.3390/ijms24065393
Chicago/Turabian StyleHou, Yuying, Yufei Wang, Xiaoyu Liu, Naveed Ahmad, Nan Wang, Libo Jin, Na Yao, and Xiuming Liu. 2023. "A Cinnamate 4-HYDROXYLASE1 from Safflower Promotes Flavonoids Accumulation and Stimulates Antioxidant Defense System in Arabidopsis" International Journal of Molecular Sciences 24, no. 6: 5393. https://doi.org/10.3390/ijms24065393
APA StyleHou, Y., Wang, Y., Liu, X., Ahmad, N., Wang, N., Jin, L., Yao, N., & Liu, X. (2023). A Cinnamate 4-HYDROXYLASE1 from Safflower Promotes Flavonoids Accumulation and Stimulates Antioxidant Defense System in Arabidopsis. International Journal of Molecular Sciences, 24(6), 5393. https://doi.org/10.3390/ijms24065393






