Populus euphratica GLABRA3 Binds PLDδ Promoters to Enhance Salt Tolerance
Abstract
1. Introduction
2. Results
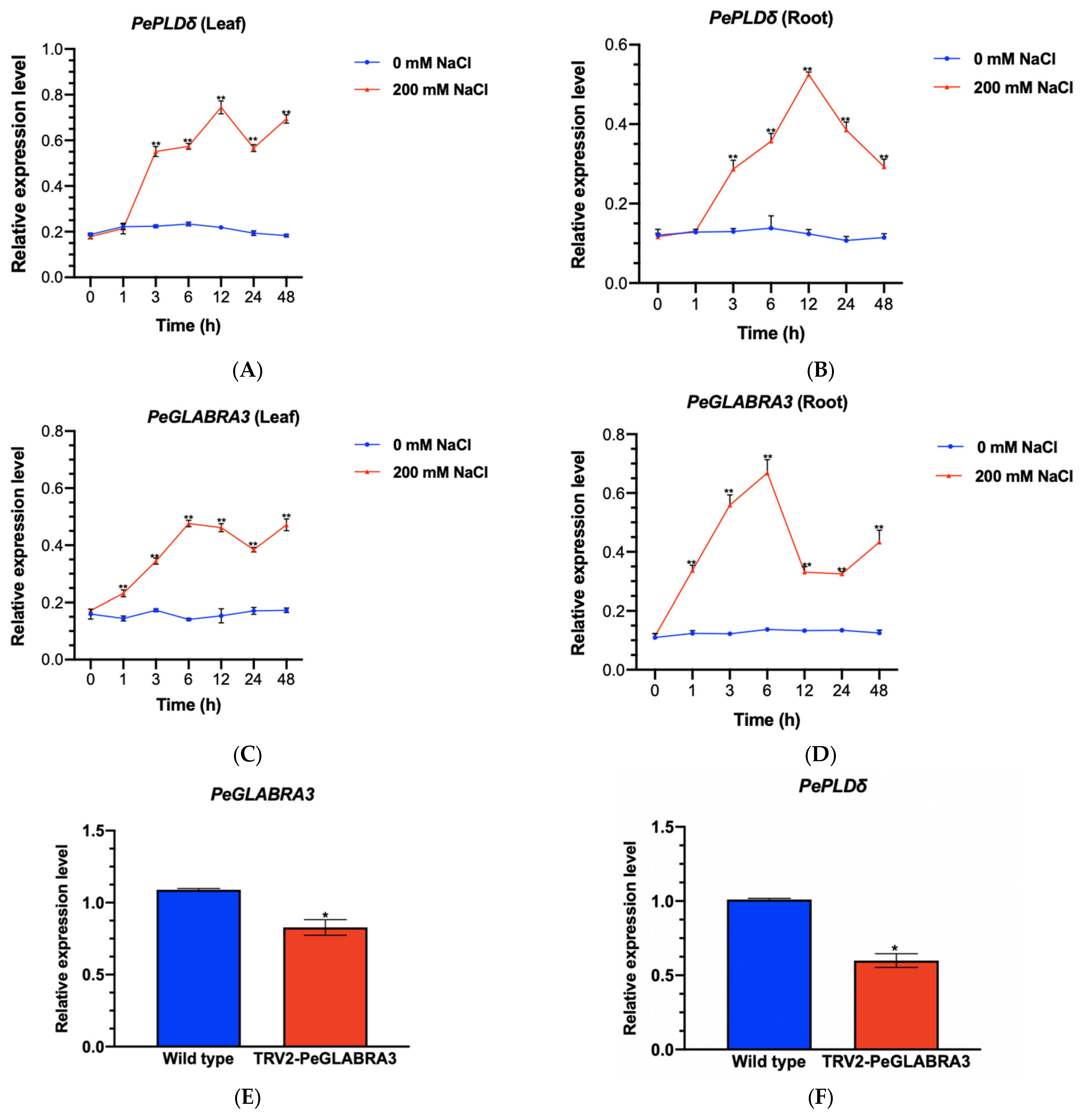
2.1. NaCl-Induced Transcription of PePLDδ in P. euphratica
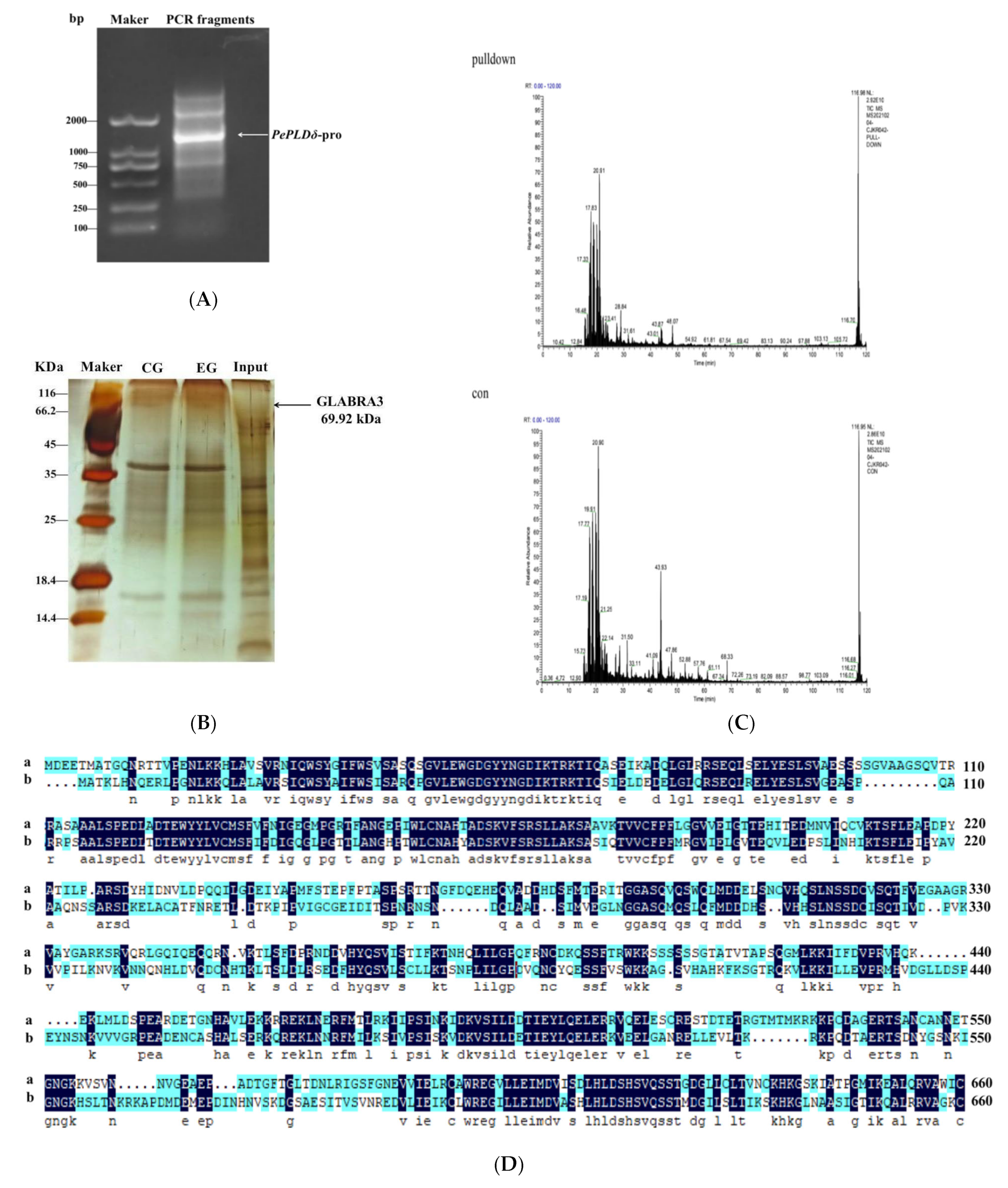
2.2. PePLDδ Promoter Cloning and Analysis
2.3. NaCl Activates the PePLDδ Promoter in Root and Leaf Tissues
2.4. Transcription Factor Identification by DNA Pull-Down
2.5. PeGLABRA3 Cloning and Sequence Analysis
2.6. NaCl-Induced PeGLABRA3 Expression
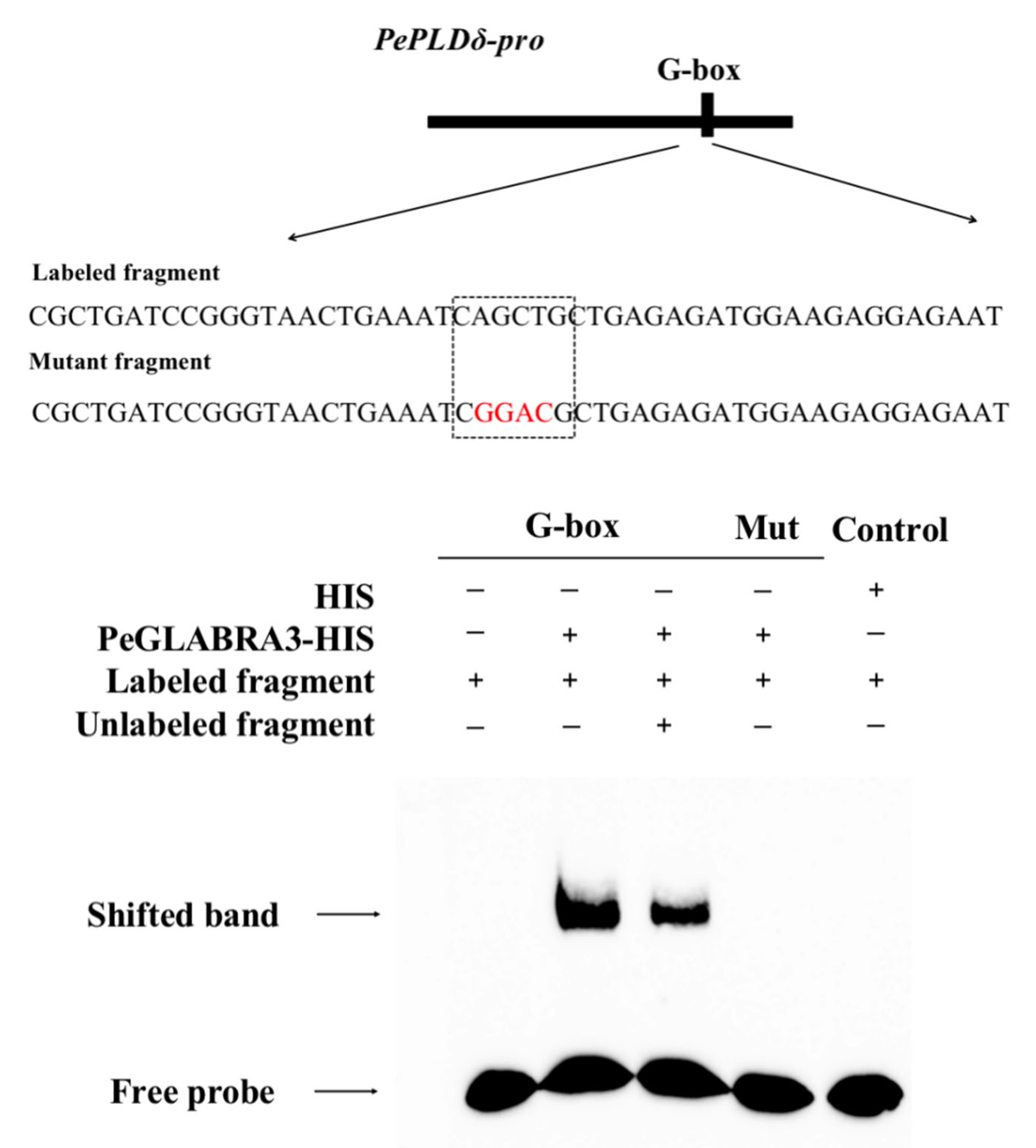
2.7. PeGLABRA3 Binds to the PePLDδ Promoter
2.7.1. Electrophoretic Mobility Shift Assay
2.7.2. Y1H Assay
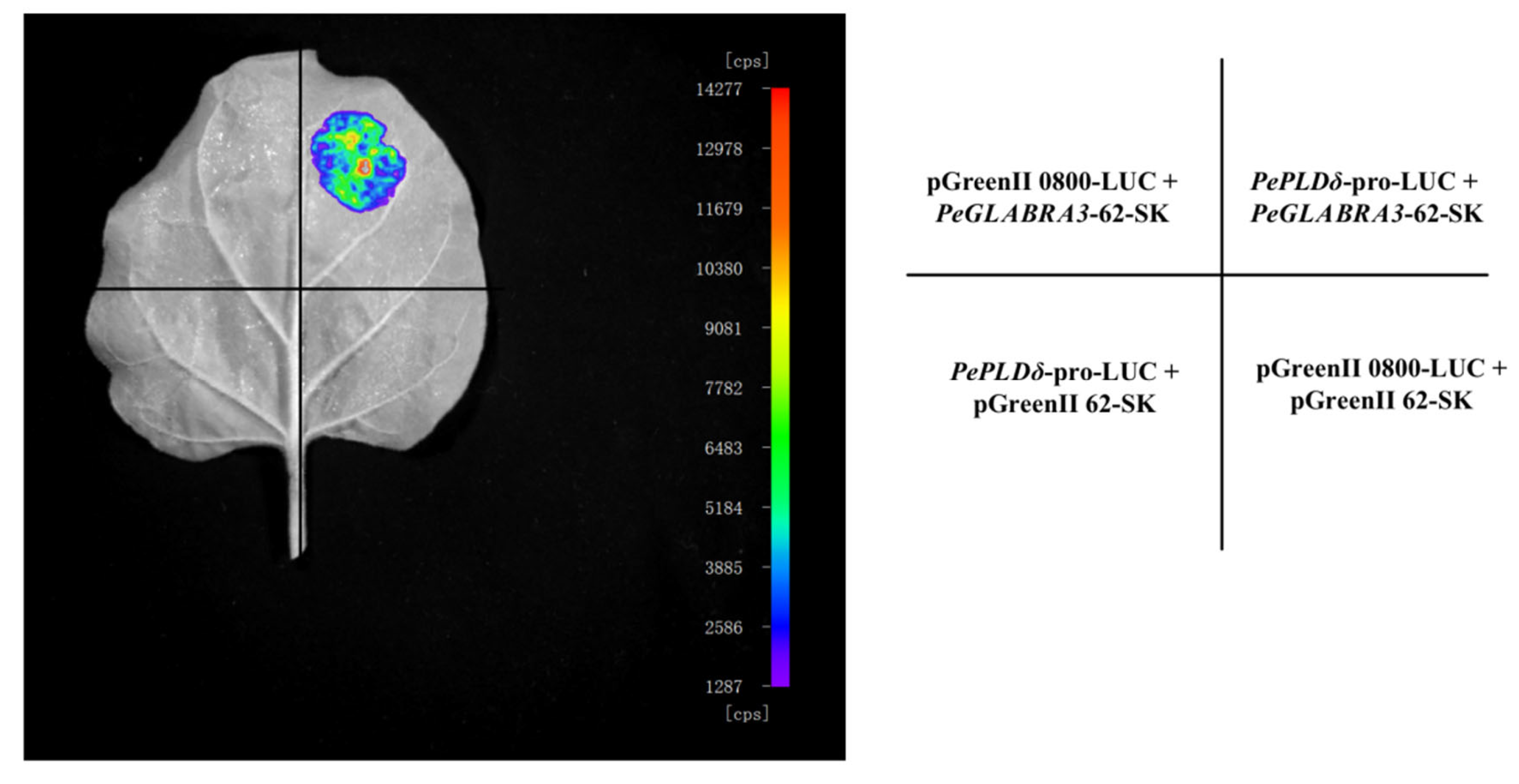
2.7.3. Luciferase Reporter Assay
2.7.4. Virus-Induced Gene Silencing
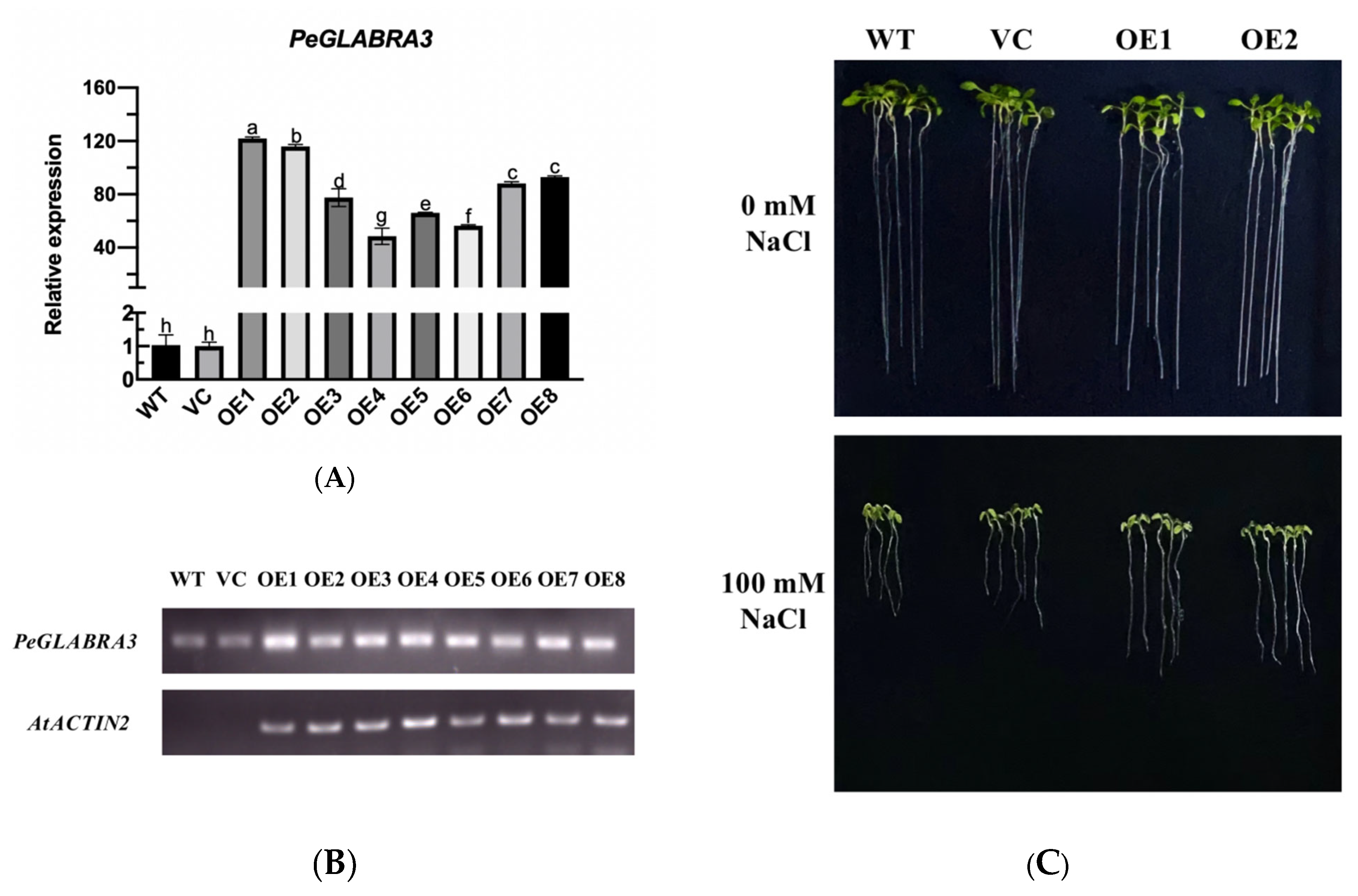
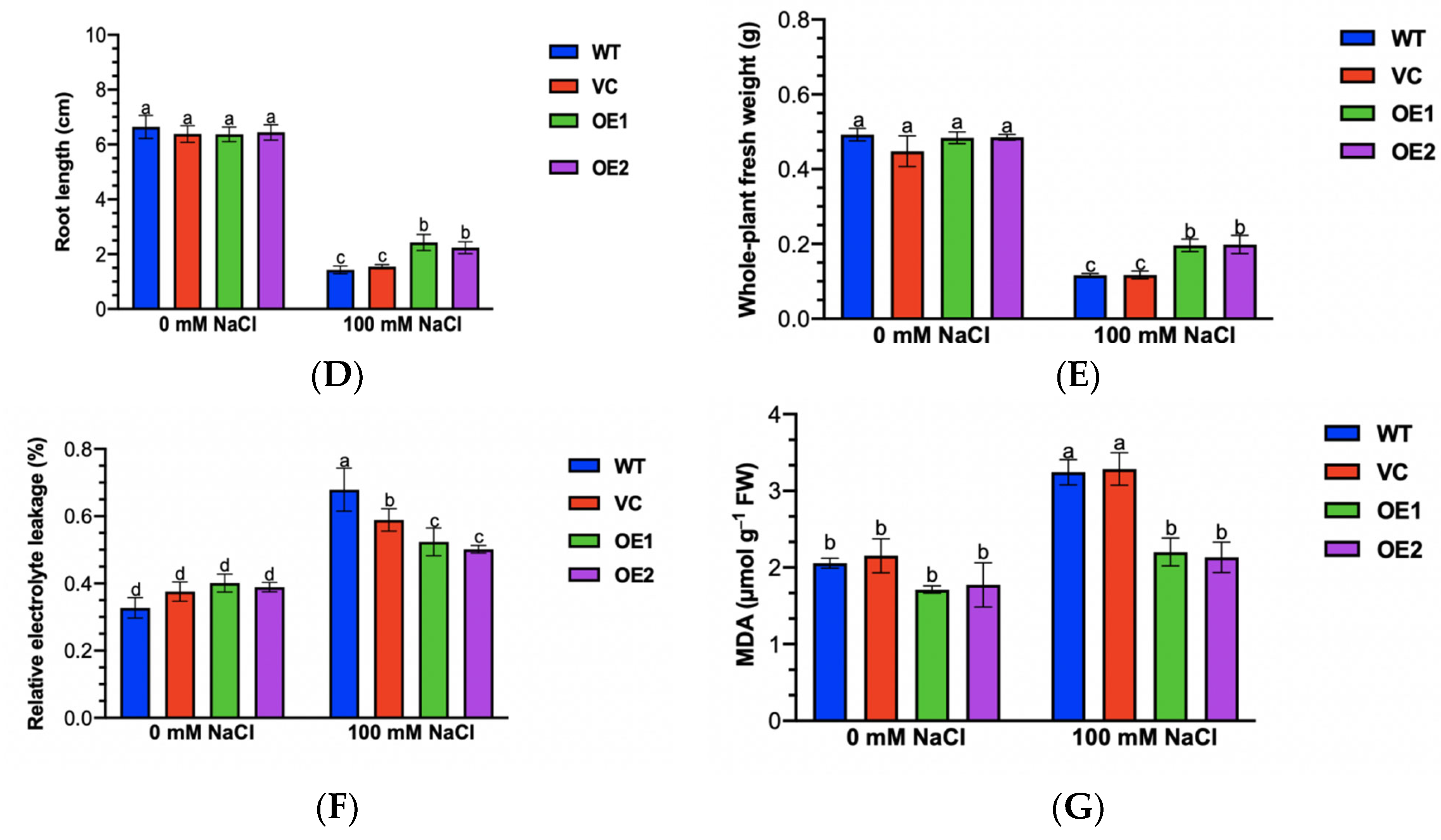
2.8. The Overexpression of PeGLABRA3 Enhances Salt Tolerance in Arabidopsis
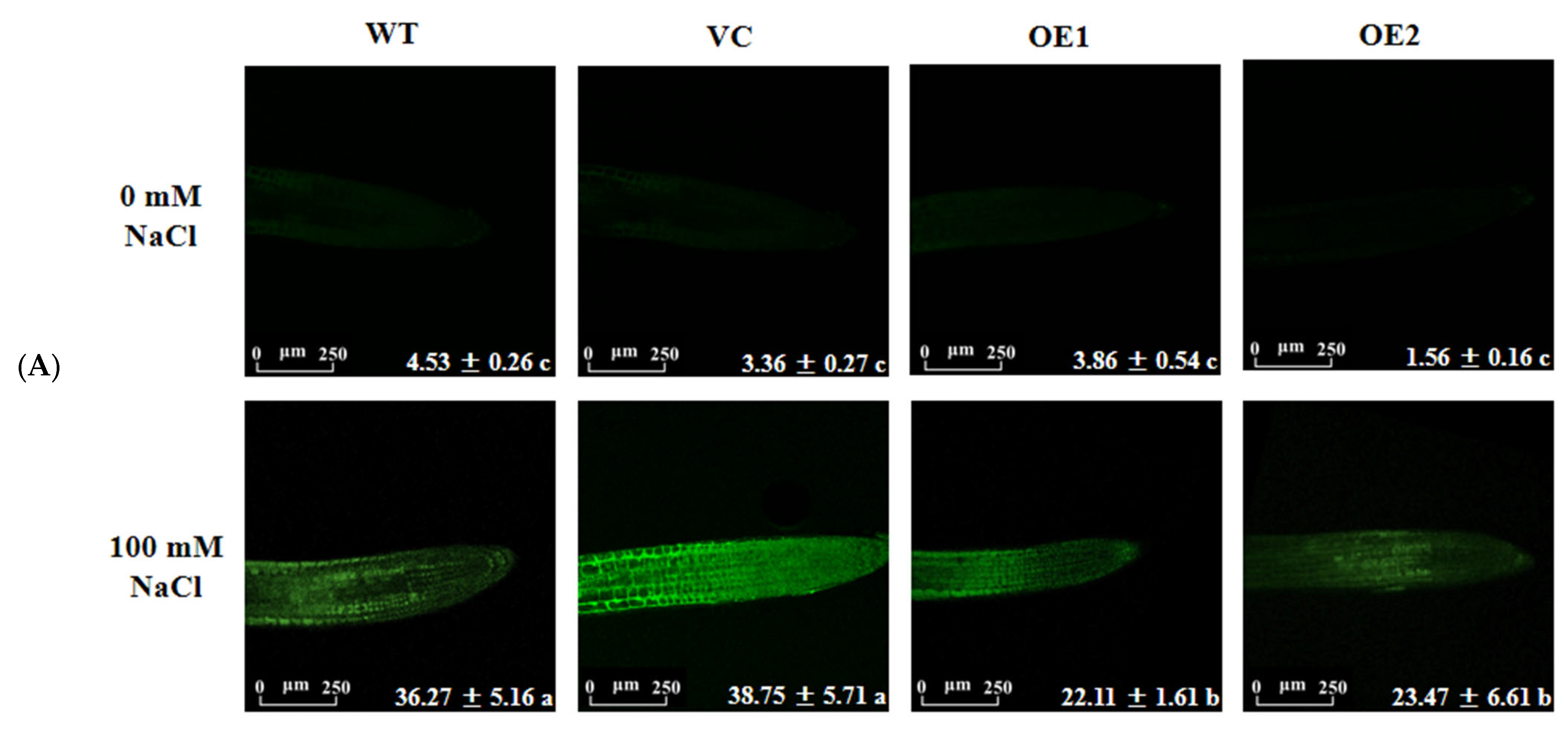
2.9. PeGLABRA3-Transgenic Plants Increased ROS Scavenging Capacity under Salt Stress
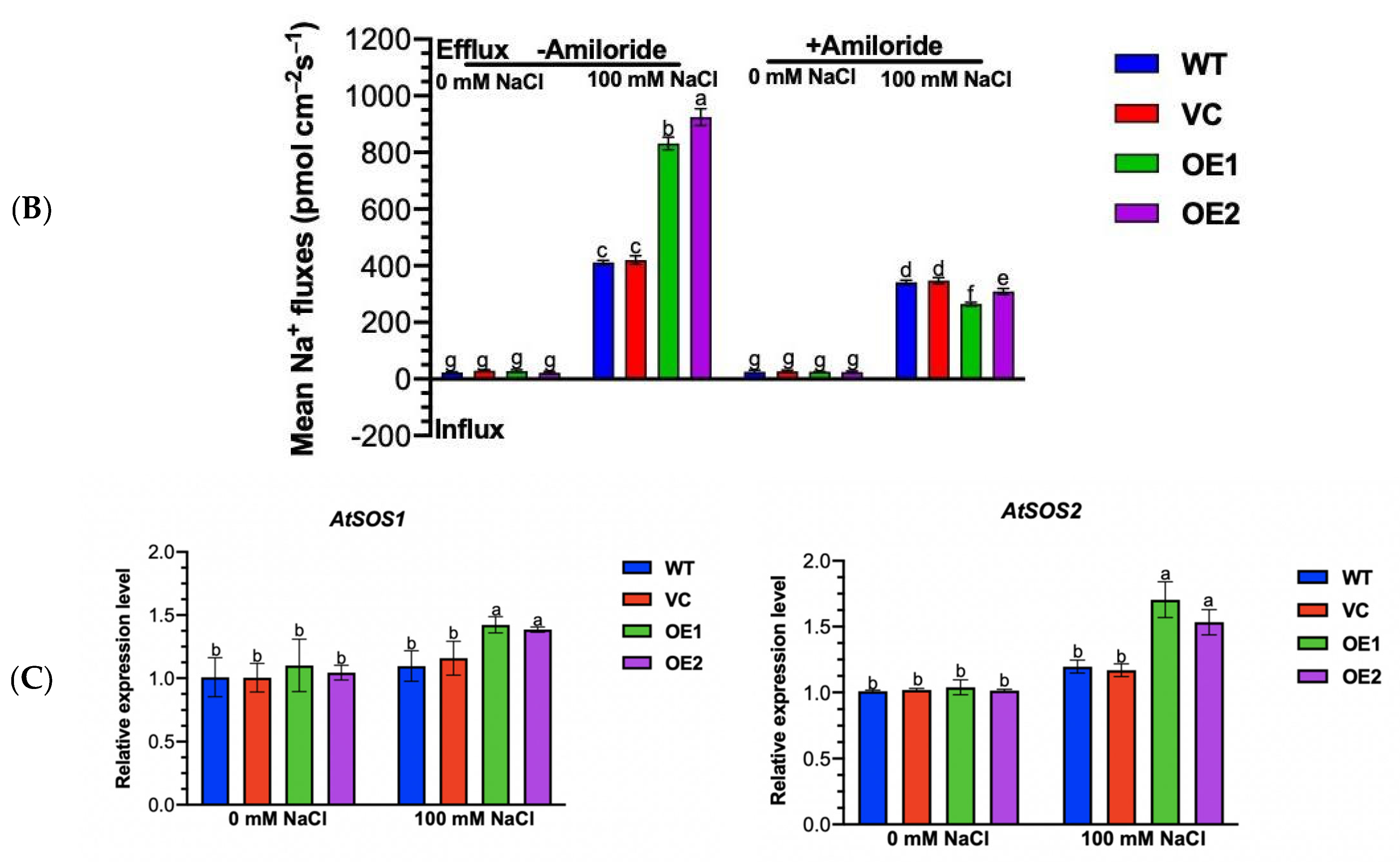
2.10. PeGLABRA3-Transgenic Plants Maintained Ionic Homeostasis under Salinity
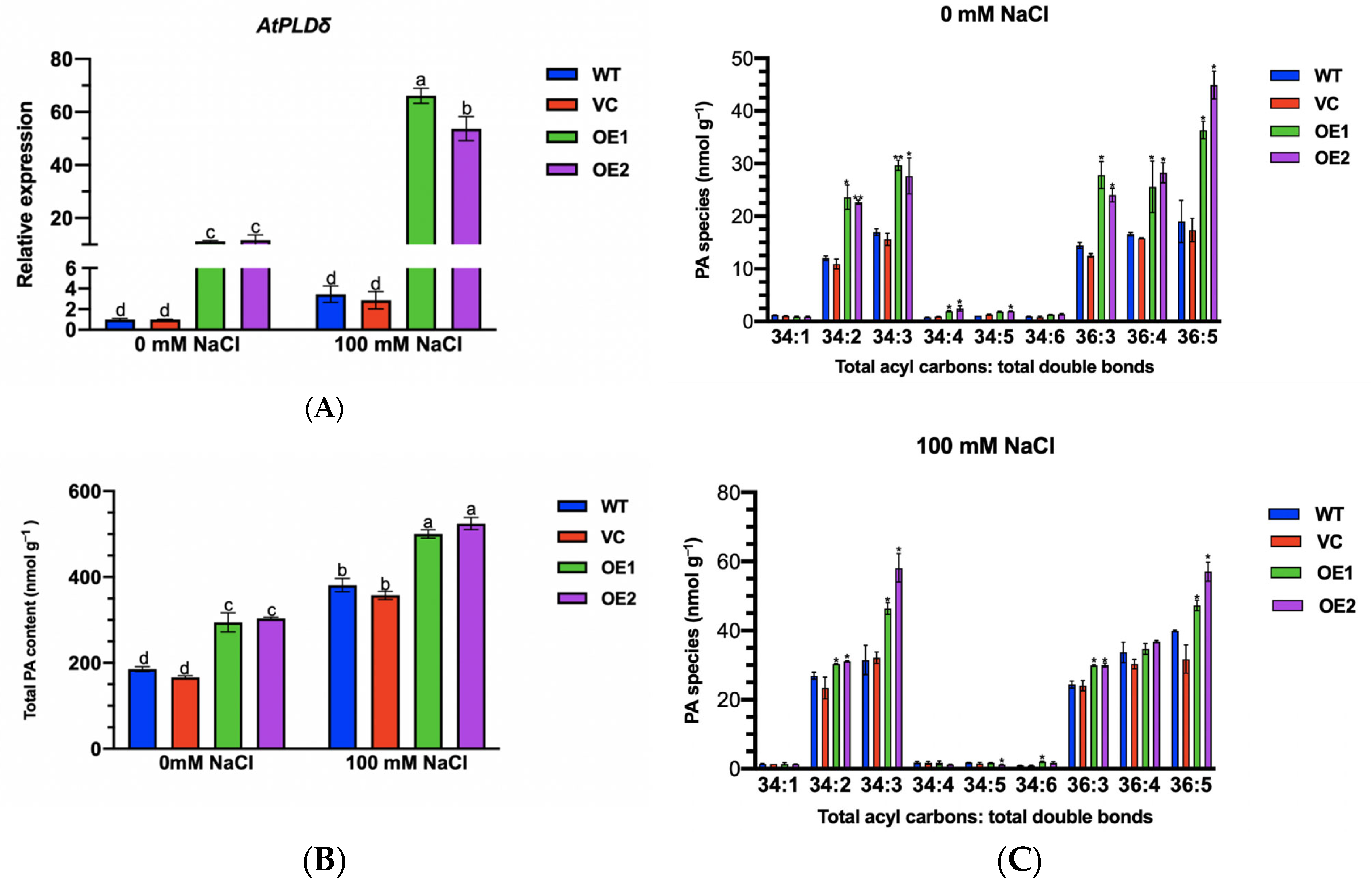
2.11. PeGLABRA3 Enhances AtPLDδ Transcription by Binding Its Promoter
2.12. Phosphatidic Acid Content of PeGLABRA3-Transgenic Lines
3. Discussion
3.1. PeGLABRA3 Interacts with PePLDδ to Increase Gene Expression under NaCl Stress
3.2. PeGLABRA3 Interacts with AtPLDδ to Mediate Ionic and ROS Homeostasis
4. Materials and Methods
4.1. Plant Material and Salt Treatment
4.2. RNA Isolation and Full-Length Cloning of PeGLABRA3
4.3. PeGLABRA3 Sequence and Phylogenetic Analysis
4.4. DNA Pull-Down
4.4.1. Probe Design and Labeling
4.4.2. DNA Pull-Down and Mass Spectrometry Analysis
4.5. Promoter Cloning and Sequence Analysis
4.6. Luciferase Reporter Assay
4.7. Yeast One-Hybrid Assay
4.8. Electrophoretic Mobility Shift Assay
4.9. Construction and Transformation of the PePLDδ-pro::GUS Gene
4.10. Virus-Induced Gene Silencing of PeGLABRA3 in P. euphratica
4.11. Generation of PeGLABRA3-Transgenic Lines
4.12. Phenotype Tests of Transgenic Lines
4.13. Na+ Flux Measurements
4.14. Cellular Na+ and H2O2 Determination Roots
4.15. Determination of Activity and Transcription of Antioxidant Enzymes
4.16. RT-qPCR Analysis
4.17. Quantitative Analysis of PA Species
4.18. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bargmann, B.; Munnik, T. The role of phospholipase D in plant stress responses. Curr. Opin. Plant Biol. 2006, 9, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Bargmann, B.O.R.; Laxalt, A.M.; Riet, B.T.; van Schooten, B.; Merquiol, E.; Testerink, C.; Haring, M.A.; Bartels, D.; Munnik, T. Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 2009, 50, 78–89. [Google Scholar] [CrossRef]
- Pleskot, R.; Li, J.; Zárský, V.; Potocký, M.; Staiger, C.J. Regulation of cytoskeletal dynamics by phospholipase D and phosphatidic acid. Trends Plant Sci. 2013, 18, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Takahashi, S.; Shinozaki, K. Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 2001, 26, 595–605. [Google Scholar] [CrossRef]
- Singh, A.; Pandey, A.; Baranwal, V.; Kapoor, S.; Pandey, G.K. Comprehensive expression analysis of rice phospholipase D gene family during abiotic stresses and development. Plant Signal. Behav. 2012, 7, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhou, D.; Zhang, Q.; Zhang, W. Genomic analysis of phospholipase D family and characterization of GmPLDαs in soybean (Glycine max). J. Plant Res. 2012, 125, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Distéfano, A.M.; Valiñas, M.A.; Scuffi, D.; Lamattina, L.; Ten Have, A.; García-Mata, C.; Laxalt, A.M. Phospholipase Dδ knock-out mutants are tolerant to severe drought stress. Plant Signal. Behav. 2015, 10, e1089371. [Google Scholar] [CrossRef] [PubMed]
- Ufer, G.; Gertzmann, A.; Gasulla, F.; Röhrig, H.; Bartels, D. Identification and characterization of the phosphatidic acid-binding A. thaliana phosphoprotein PLDrp1 that is regulated by PLDα1 in a stress-dependent manner. Plant J. 2017, 92, 276–290. [Google Scholar] [CrossRef]
- Gasulla, F.; Barreno, E.; Parages, M.L.; Cámara, J.; Jiménez, C.; Dörmann, P.; Bartels, D. The role of Phospholipase D and MAPK signaling cascades in the adaption of lichen microalgae to desiccation: Changes in membrane lipids and phosphoproteome. Plant Cell Physiol. 2016, 57, 1908–1920. [Google Scholar] [CrossRef]
- Deepika, D.; Singh, A. Plant phospholipase D: Novel structure, regulatory mechanism, and multifaceted functions with biotechnological application. Crit. Rev. Biotechnol. 2022, 42, 106–124. [Google Scholar] [CrossRef]
- Vadovič, P.; Šamajová, O.; Takáč, T.; Novák, D.; Zapletalová, V.; Colcombet, J.; Šamaj, J. Biochemical and genetic interactions of Phospholipase D alpha 1 and mitogen-activated protein kinase 3 affect Arabidopsis stress response. Front. Plant Sci. 2019, 10, 275. [Google Scholar] [CrossRef]
- Morris, P.C. Integrating lipid signalling, mitogen-activated protein kinase cascades and salt tolerance. New Phytol. 2010, 188, 640–643. [Google Scholar] [CrossRef]
- Pappan, K.L.; Wang, X. Assaying different types of plant phospholipase D activities in vitro. Methods Mol. Biol. 2013, 1009, 205–217. [Google Scholar]
- Li, S.; Huang, M.; Di, Q.; Ji, T.; Wang, X.; Wei, M.; Shi, Q.; Li, Y.; Gong, B.; Yang, F. The functions of a cucumber phospholipase D alpha gene (CsPLDα) in growth and tolerance to hyperosmotic stress. Plant Physiol. Biochem. 2015, 97, 175–186. [Google Scholar] [CrossRef]
- Ji, T.; Li, S.; Huang, M.; Di, Q.; Wang, X.; Wei, M.; Shi, Q.; Li, Y.; Gong, B.; Yang, F. Overexpression of cucumber phospholipase D alpha gene (CsPLDα) in tobacco enhanced salinity stress tolerance by regulating Na+–K+ balance and lipid peroxidation. Front. Plant Sci. 2017, 8, 499. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Song, Y.; Liu, Y.; Guo, X.; Zhu, C.; Wen, F. Overexpression of phospholipase Dα gene enhances drought and salt tolerance of Populus tomentosa. Chin. Sci Bull. 2008, 53, 3656–3665. [Google Scholar] [CrossRef]
- Hunter, K.; Kimura, S.; Rokka, A.; Tran, H.C.; Toyota, M.; Kukkonen, J.P.; Wrzaczek, M. CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol. 2019, 180, 2004–2021. [Google Scholar] [CrossRef] [PubMed]
- Ben Othman, A.; Ellouzi, H.; Planchais, S.; De Vos, D.; Faiyue, B.; Carol, P.; Abdelly, C.; Savouré, A. Phospholipases Dζ1 and Dζ2 have distinct roles in growth and antioxidant systems in Arabidopsis thaliana responding to salt stress. Planta 2017, 246, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hu, X.; Wang, P.; Wang, H.; Ge, X.; Li, F.; Hou, Y. The phospholipase D gene GhPLDδ confers resistance to Verticillium dahliae and improves tolerance to salt stress. Plant Sci. 2022, 321, 111322. [Google Scholar] [CrossRef]
- Zhang, Y.; Yao, J.; Yin, K.; Liu, Z.; Zhang, Y.; Deng, C.; Liu, J.; Zhang, Y.; Hou, S.; Zhang, H.; et al. Populus euphratica phospholipase Dδ increases salt tolerance by regulating K+/Na+ and ROS homeostasis in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 4911. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Jakoby, M.; Werber, M.; Weisshaar, B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 2003, 15, 2497–2502. [Google Scholar] [CrossRef]
- Heim, M.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Toda, Y.; Tanaka, M.; Ogawa, D.; Kurata, K.; Kurotani, K.; Habu, Y.; Ando, T.; Sugimoto, K.; Mitsuda, N.; Katoh, E.; et al. Rice salt sensitive3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell 2013, 25, 1709–1725. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef]
- Ariyarathne, M.A.; Wone, B.W.M. Overexpression of the Selaginella lepidophylla bHLH transcription factor enhances water-use efficiency, growth, and development in Arabidopsis. Plant Sci. 2022, 315, 111129. [Google Scholar] [CrossRef] [PubMed]
- Onohata, T.; Gomi, K. Overexpression of jasmonate-responsive OsbHLH034 in rice results in the induction of bacterial blight resistance via an increase in lignin biosynthesis. Plant Cell Rep. 2020, 39, 1175–1184. [Google Scholar] [CrossRef]
- Moreno, J.E.; Moreno-Piovano, G.; Chan, R.L. The antagonistic basic helix-loop-helix partners BEE and IBH1 contribute to control plant tolerance to abiotic stress. Plant Sci. 2018, 271, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Kong, J.; Tao, R.; Ahmed, T.; Alamin, M.; Alotaibi, S.S.; Abdelsalam, N.R.; Xu, J.H. CRISPR/Cas9 mediated knockout of the OsbHLH024 transcription factor improves salt stress resistance in Rice (Oryza sativa L.). Plants 2022, 11, 1184. [Google Scholar] [CrossRef]
- Alam, M.S.; Yang, Z.K.; Li, C.; Yan, Y.; Liu, Z.; Nazir, M.M.; Xu, J.H. Loss-of-function mutations of OsbHLH044 transcription factor lead to salinity sensitivity and a greater chalkiness in rice (Oryza sativa L.). Plant Physiol. Biochem. 2022, 93, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Jalmi, S.K.; Bhagat, P.K.; Verma, N.; Sinha, A.K. A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 2020, 287, 2560–2576. [Google Scholar] [CrossRef]
- Waseem, M.; Rong, X.; Li, Z. Dissecting the role of a basic helix-loop-helix transcription factor, SlbHLH22, under salt and drought stresses in transgenic Solanum lycopersicum L. Front. Plant Sci. 2019, 10, 734. [Google Scholar] [CrossRef] [PubMed]
- Kavas, M.; Baloğlu, M.C.; Atabay, E.S.; Ziplar, U.T.; Daşgan, H.Y.; Ünver, T. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol. Genet. Genom. 2016, 291, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Babitha, K.C.; Vemanna, R.S.; Nataraja, K.N.; Udayakumar, M. Overexpression of EcbHLH57 transcription factor from Eleusine coracana L. in tobacco confers tolerance to salt, oxidative and drought stress. PLoS ONE 2015, 10, e0137098. [Google Scholar] [CrossRef]
- Liu, Y.; Ji, X.; Nie, X.; Qu, M.; Zheng, L.; Tan, Z.; Zhao, H.; Huo, L.; Liu, S.; Zhang, B.; et al. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 2015, 207, 692–709. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C.; Cheng, W.H.; Hong, C.Y.; Chang, Y.S.; Chang, M.C. The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively. Rice 2018, 11, 50. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Tian, Y.; Wang, Q.; Chen, S.; Li, H.; Ma, C.; Li, H. Functional characterization of a sugar beet BvbHLH93 transcription factor in salt stress tolerance. Int. J. Mol. Sci. 2021, 22, 3669. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, B.; Deyholos, M.K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genom. 2009, 282, 503–516. [Google Scholar] [CrossRef]
- Wang, F.B.; Zhu, H.; Chen, D.H.; Li, Z.J.; Peng, R.H.; Yao, Q.H. A grape bHLH transcription factor gene, VvbHLH, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Liang, X.; Li, Y.; Yao, A.; Liu, W.; Yang, T.; Zhao, M.; Zhang, B.; Han, D. Overexpression of MxbHLH18 increased iron and high salinity stress tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 8007. [Google Scholar] [CrossRef]
- Liu, D.; Li, Y.Y.; Zhou, Z.C.; Xiang, X.; Liu, X.; Wang, J.; Hu, Z.R.; Xiang, S.P.; Li, W.; Xiao, Q.Z.; et al. Tobacco transcription factor bHLH123 improves salt tolerance by activating NADPH oxidase NtRbohE expression. Plant Physiol. 2021, 186, 1706–1720. [Google Scholar] [CrossRef]
- Rajappa, S.; Krishnamurthy, P.; Kumar, P.P. Regulation of AtKUP2 expression by bHLH and WRKY transcription factors helps to confer increased salt tolerance to Arabidopsis thaliana plants. Front. Plant Sci. 2020, 11, 1311. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, P.; Vishal, B.; Khoo, K.; Rajappa, S.; Loh, C.S.; Kumar, P.P. Expression of AoNHX1 increases salt tolerance of rice and Arabidopsis, and bHLH transcription factors regulate AtNHX1 and AtNHX6 in Arabidopsis. Plant Cell Rep. 2019, 38, 1299–1315. [Google Scholar] [CrossRef]
- Ahmad, A.; Niwa, Y.; Goto, S.; Ogawa, T.; Shimizu, M.; Suzuki, A.; Kobayashi, K.; Kobayashi, H. bHLH106 integrates functions of multiple genes through their G-Box to confer salt tolerance on Arabidopsis. PLoS ONE 2015, 10, e0126872. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, J.K.; Shin, J.S.; Suh, M.C. The SebHLH transcription factor mediates trans-activation of the SeFAD2 gene promoter through binding to E- and G-box elements. Plant Mol. Biol. 2007, 64, 453–466. [Google Scholar] [CrossRef]
- Yao, J.; Shen, Z.; Zhang, Y.; Wu, X.; Wang, J.; Sang, G.; Zhang, Y.; Zhang, H.; Deng, C.; Liu, J.; et al. Populus euphratica WRKY1 binds the promoter of H+-ATPase gene to enhance gene expression and salt tolerance. J. Exp. Bot. 2020, 71, 1527–1539. [Google Scholar] [CrossRef]
- Zhou, J.; Li, F.; Wang, J.L.; Ma, Y.; Chong, K.; Xu, Y.Y. Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt and osmotic stress in Arabidopsis. J. Plant Physiol. 2009, 166, 1296–1306. [Google Scholar] [CrossRef]
- Zhao, M.; Liu, Q.; Zhang, Y.; Yang, N.; Wu, G.; Li, Q.; Wang, W. Alleviation of osmotic stress by H2S is related to regulated PLD alpha 1 and suppressed ROS in Arabidopsis thaliana. J. Plant Res. 2020, 133, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Jia, Q.; Chen, L.; Jin, X.; Xiao, X.; Li, L.; Chen, H.; Qu, Y.; Su, Y.; Zhang, W.; et al. Involvement of Arabidopsis phospholipase D δ in regulation of ROS-mediated microtubule organization and stomatal movement upon heat shock. J. Exp. Bot. 2020, 71, 6555–6570. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Nie, J.; Cao, C.; Jin, Y.; Yan, M.; Wang, F. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010, 188, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]
- Ramegowda, V.; Mysore, K.S.; Senthil-Kumar, M. Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front. Plant Sci. 2014, 5, 323. [Google Scholar] [CrossRef]
- Voinnet, O. Non-cell autonomous RNA silencing. FEBS Lett. 2005, 579, 5858–5871. [Google Scholar] [CrossRef]
- Sun, J.; Chen, S.; Dai, S.; Wang, R.; Li, N.; Shen, X.; Zhou, X.; Lu, C.; Zheng, X.; Hu, Z.; et al. NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiol. 2009, 149, 1141–1153. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Devaiah, S.P.; Roth, M.R.; Baughman, E.; Li, M.; Tamura, P.; Jeannotte, R.; Welti, R.; Wang, X. Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dα1 knockout mutant. Phytochemistry 2006, 67, 1907–1924. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Yin, K.; Yao, J.; Zhao, Z.; Liu, Z.; Yan, C.; Zhang, Y.; Liu, J.; Li, J.; Zhao, N.; et al. Populus euphratica GLABRA3 Binds PLDδ Promoters to Enhance Salt Tolerance. Int. J. Mol. Sci. 2023, 24, 8208. https://doi.org/10.3390/ijms24098208
Zhang Y, Yin K, Yao J, Zhao Z, Liu Z, Yan C, Zhang Y, Liu J, Li J, Zhao N, et al. Populus euphratica GLABRA3 Binds PLDδ Promoters to Enhance Salt Tolerance. International Journal of Molecular Sciences. 2023; 24(9):8208. https://doi.org/10.3390/ijms24098208
Chicago/Turabian StyleZhang, Ying, Kexin Yin, Jun Yao, Ziyan Zhao, Zhe Liu, Caixia Yan, Yanli Zhang, Jian Liu, Jing Li, Nan Zhao, and et al. 2023. "Populus euphratica GLABRA3 Binds PLDδ Promoters to Enhance Salt Tolerance" International Journal of Molecular Sciences 24, no. 9: 8208. https://doi.org/10.3390/ijms24098208
APA StyleZhang, Y., Yin, K., Yao, J., Zhao, Z., Liu, Z., Yan, C., Zhang, Y., Liu, J., Li, J., Zhao, N., Zhao, R., Zhou, X., & Chen, S. (2023). Populus euphratica GLABRA3 Binds PLDδ Promoters to Enhance Salt Tolerance. International Journal of Molecular Sciences, 24(9), 8208. https://doi.org/10.3390/ijms24098208





