Comprehensive Analysis of Universal Stress Protein Family Genes and Their Expression in Fusarium oxysporum Response of Populus davidiana × P. alba var. pyramidalis Louche Based on the Transcriptome
Abstract
:1. Introduction
2. Results
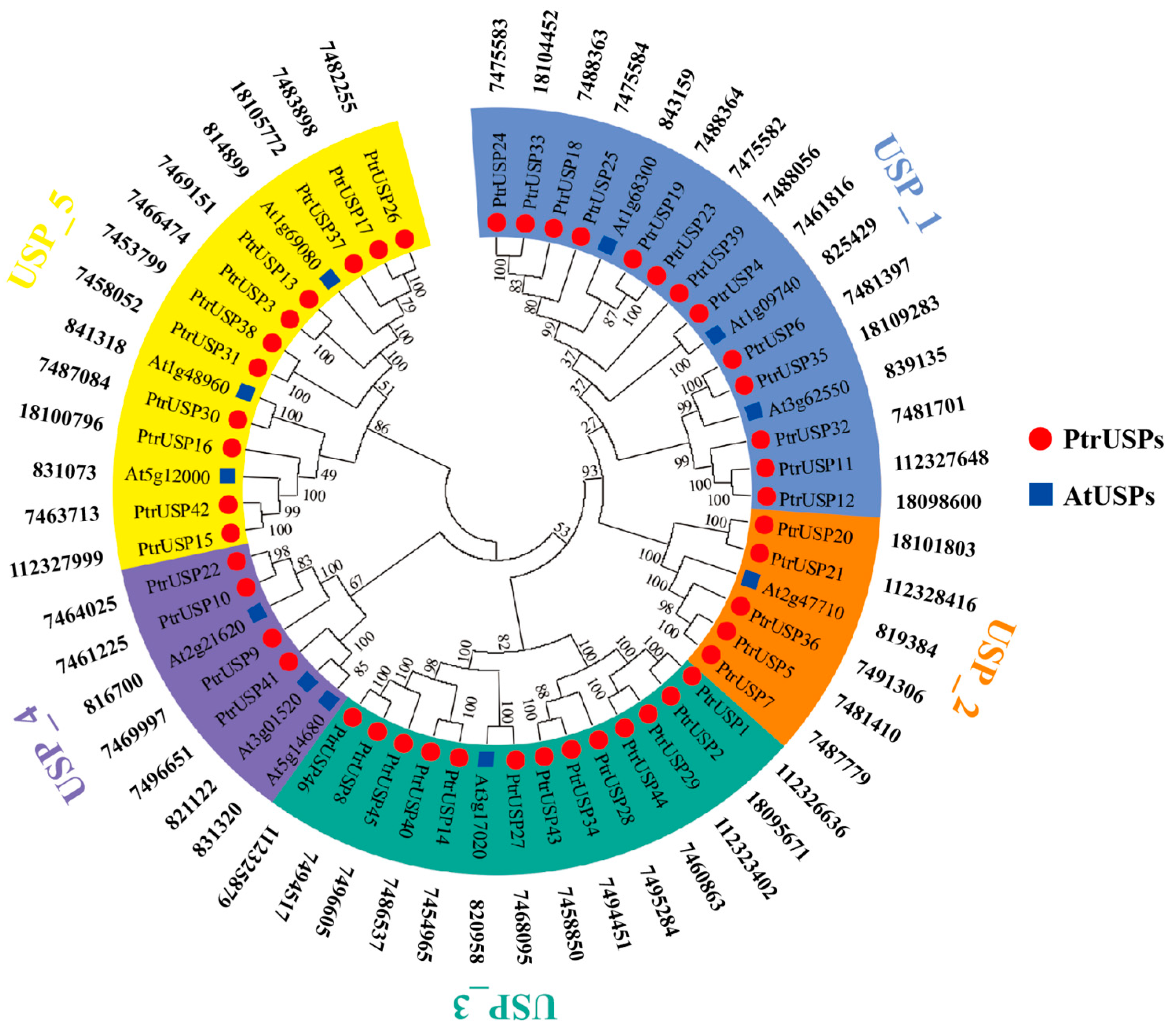
2.1. Phylogenetic Relationships and Physicochemical Properties of PtrUSPs
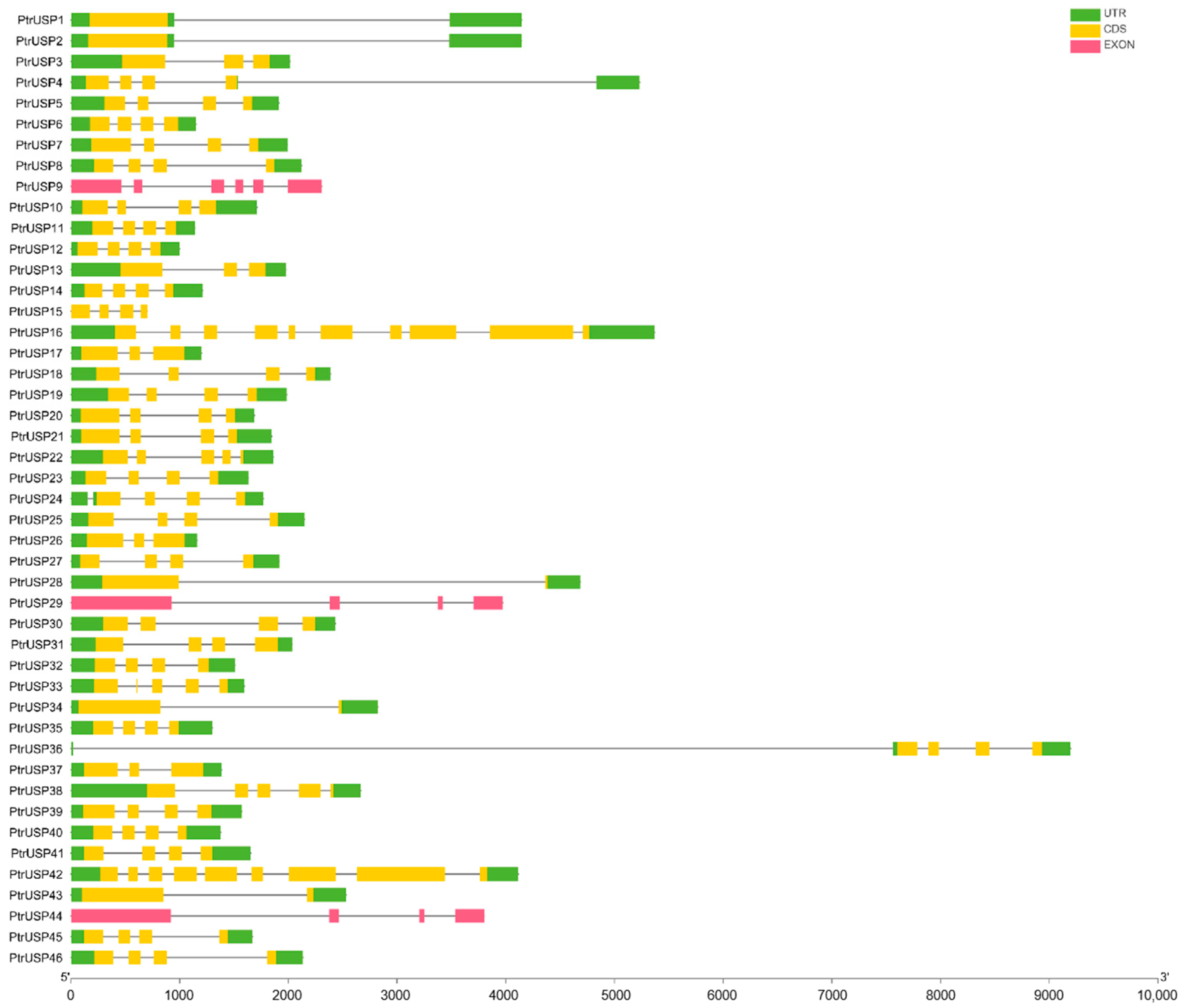
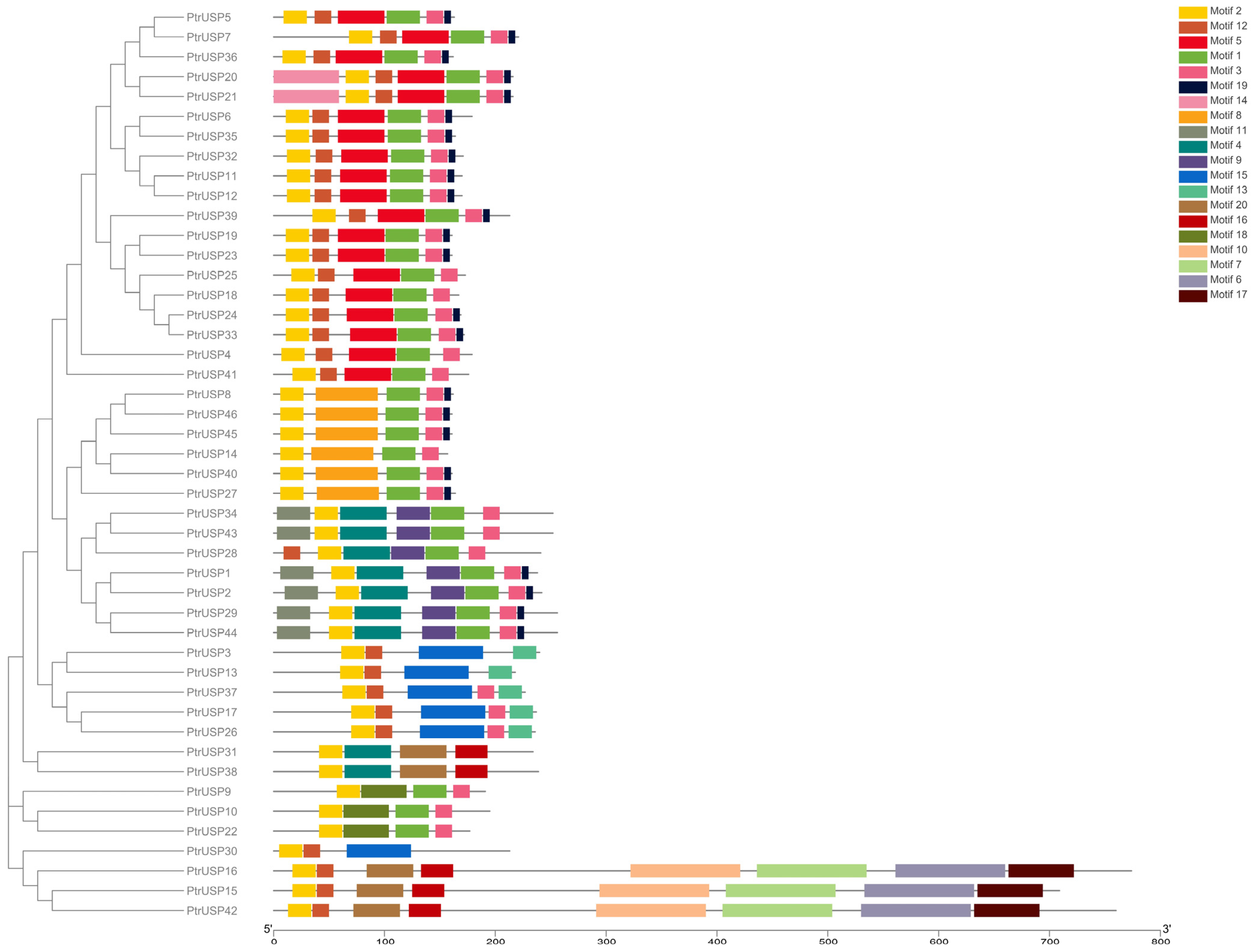
2.2. Gene Structure Analysis of PtrUSPs
2.3. Secondary and Tertiary Structure Analysis of PtrUSPs
2.4. Topological Heterogeneity Model and Subcellular Localization Analysis of PtrUSPs
2.5. Cis-Acting Elements Analysis in Promoters of PtrUSPs
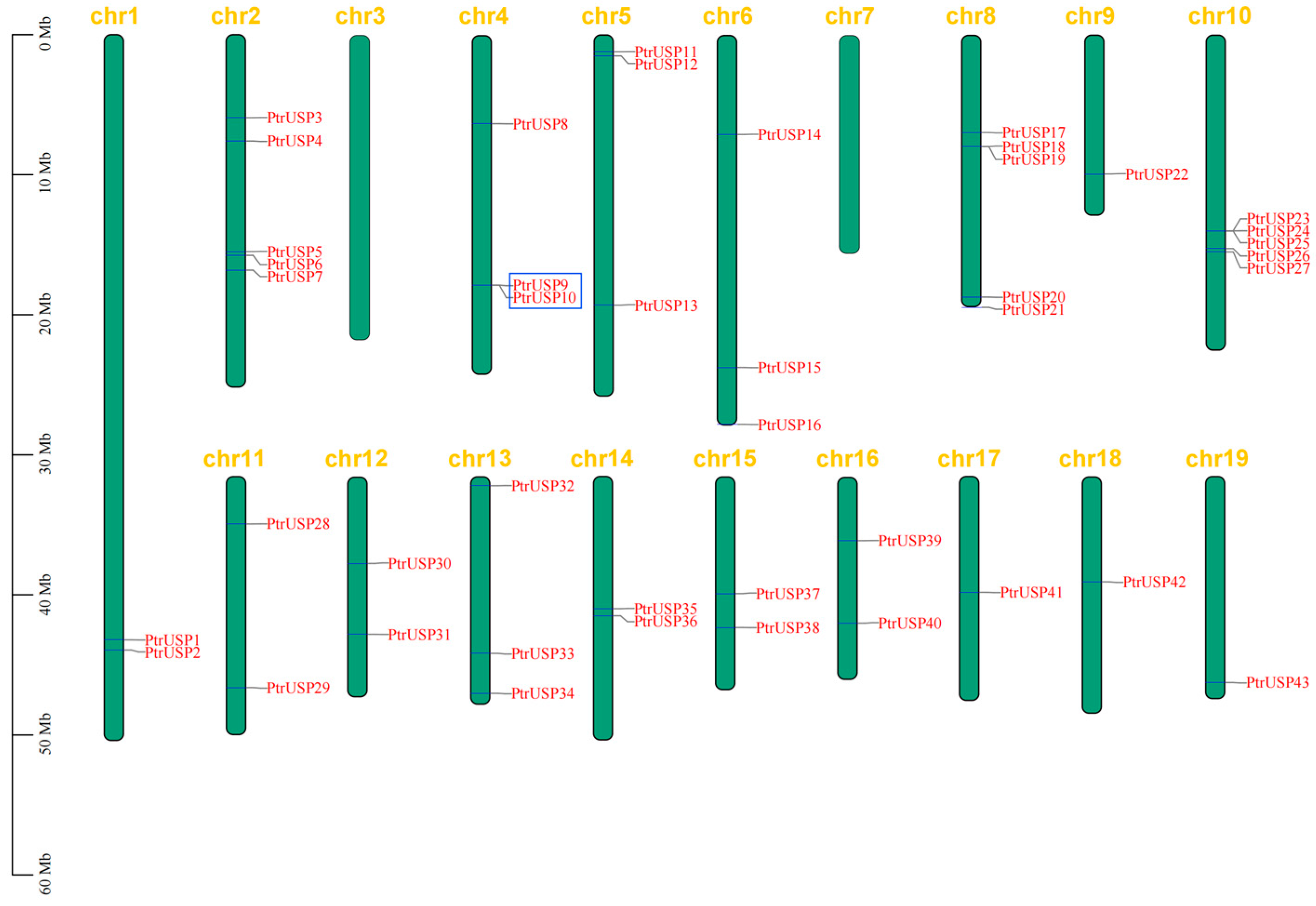
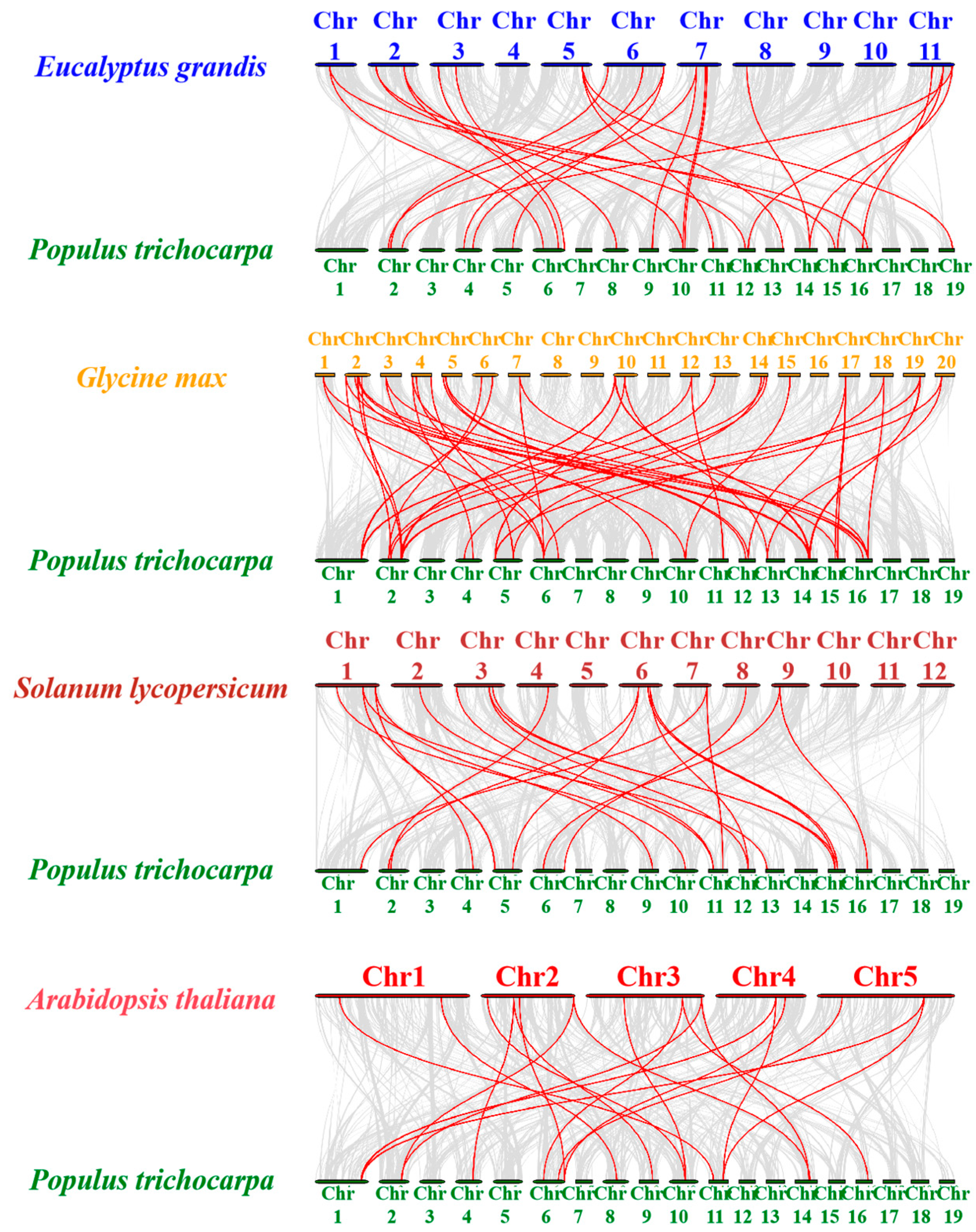
2.6. Chromosomal Localization and Collinearity Analysis of PtrUSPs
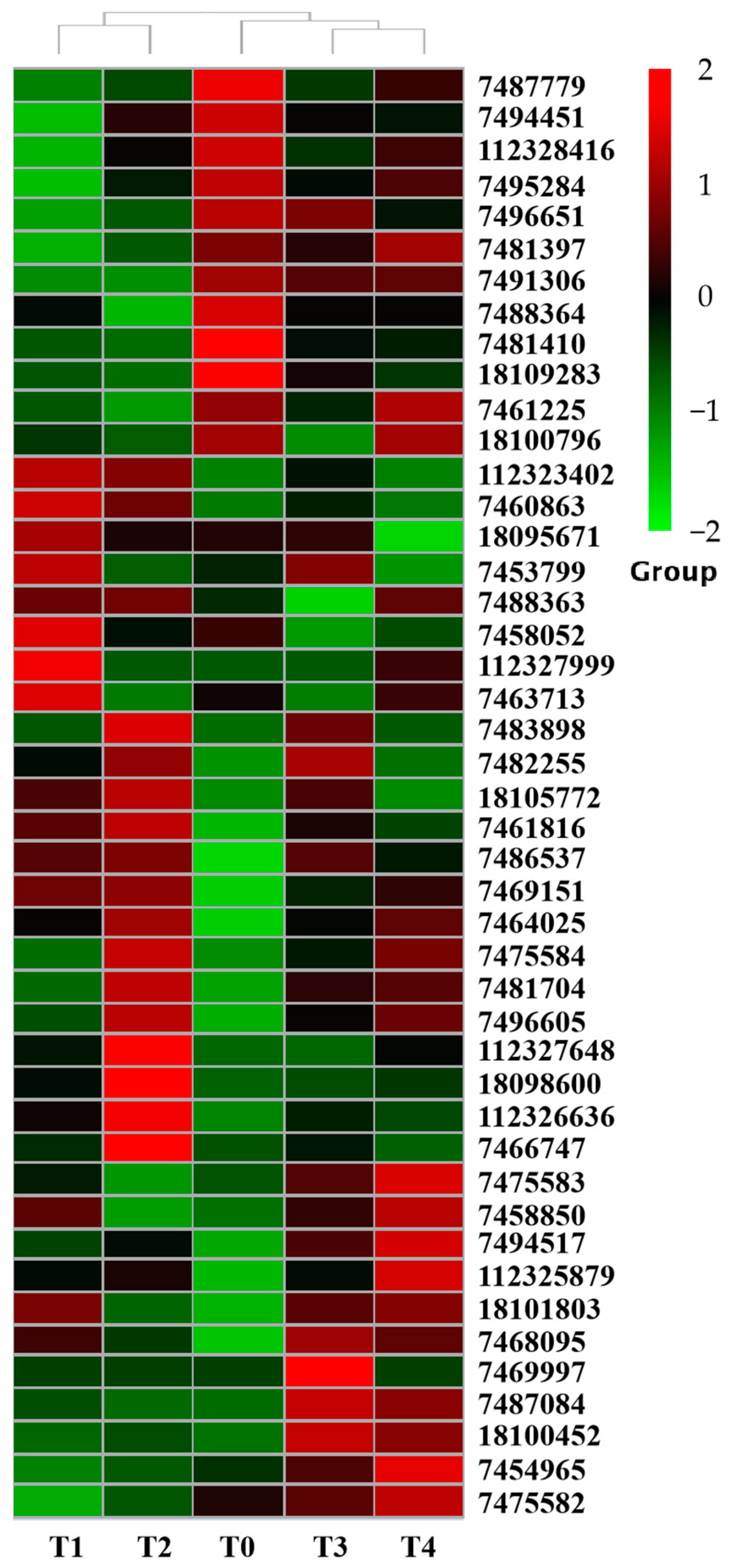
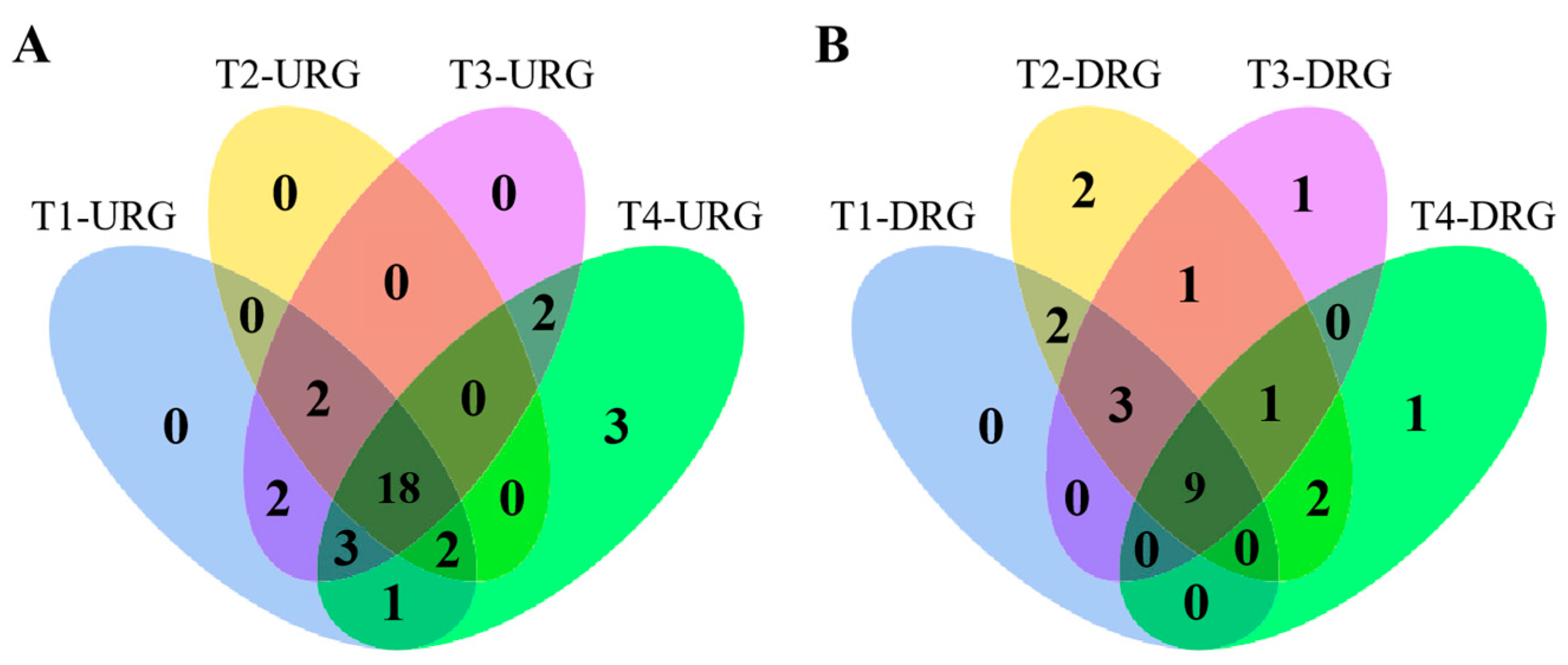
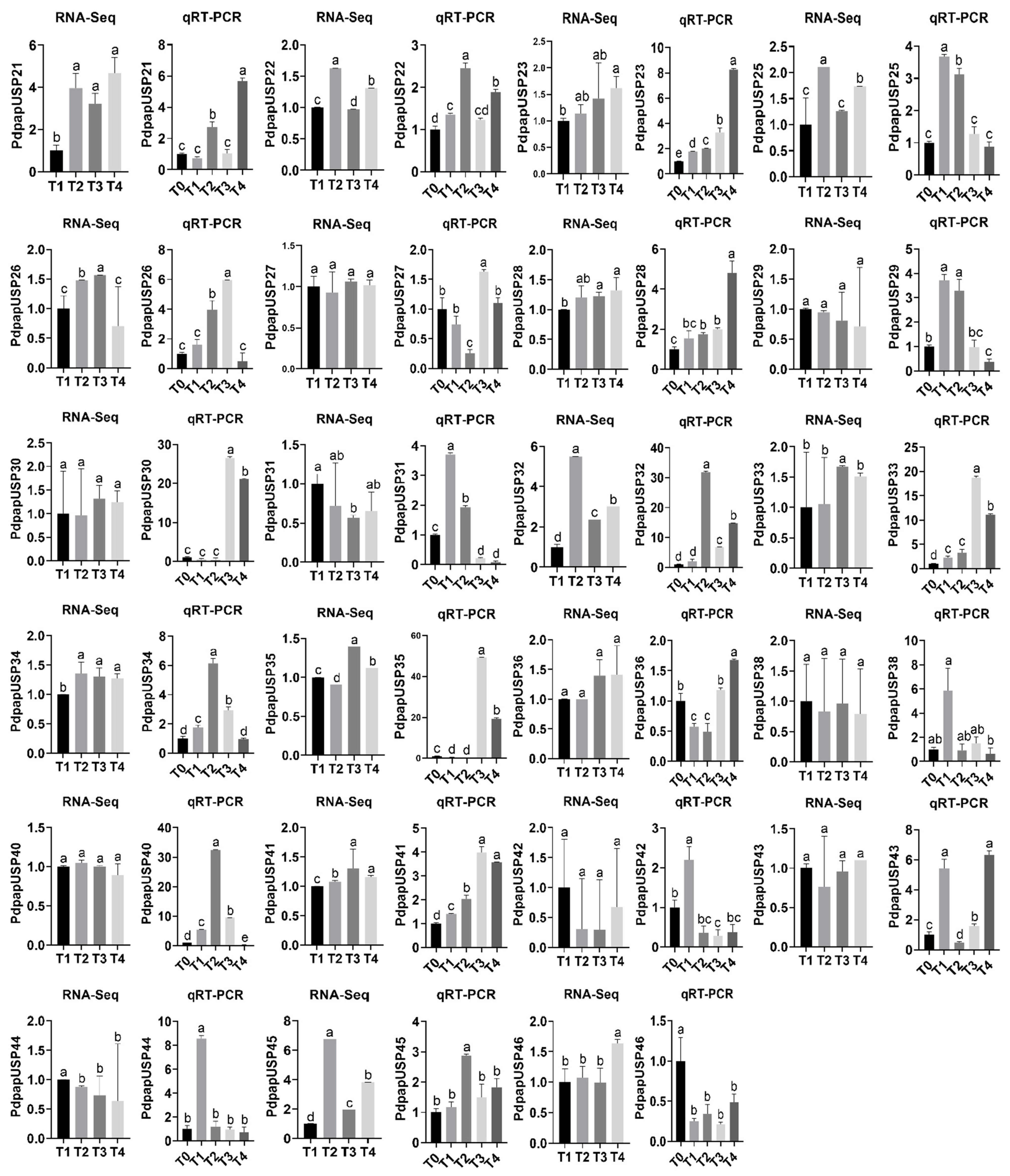
2.7. Expression Patterns of PdpapUSPs in Response to F. oxysporum
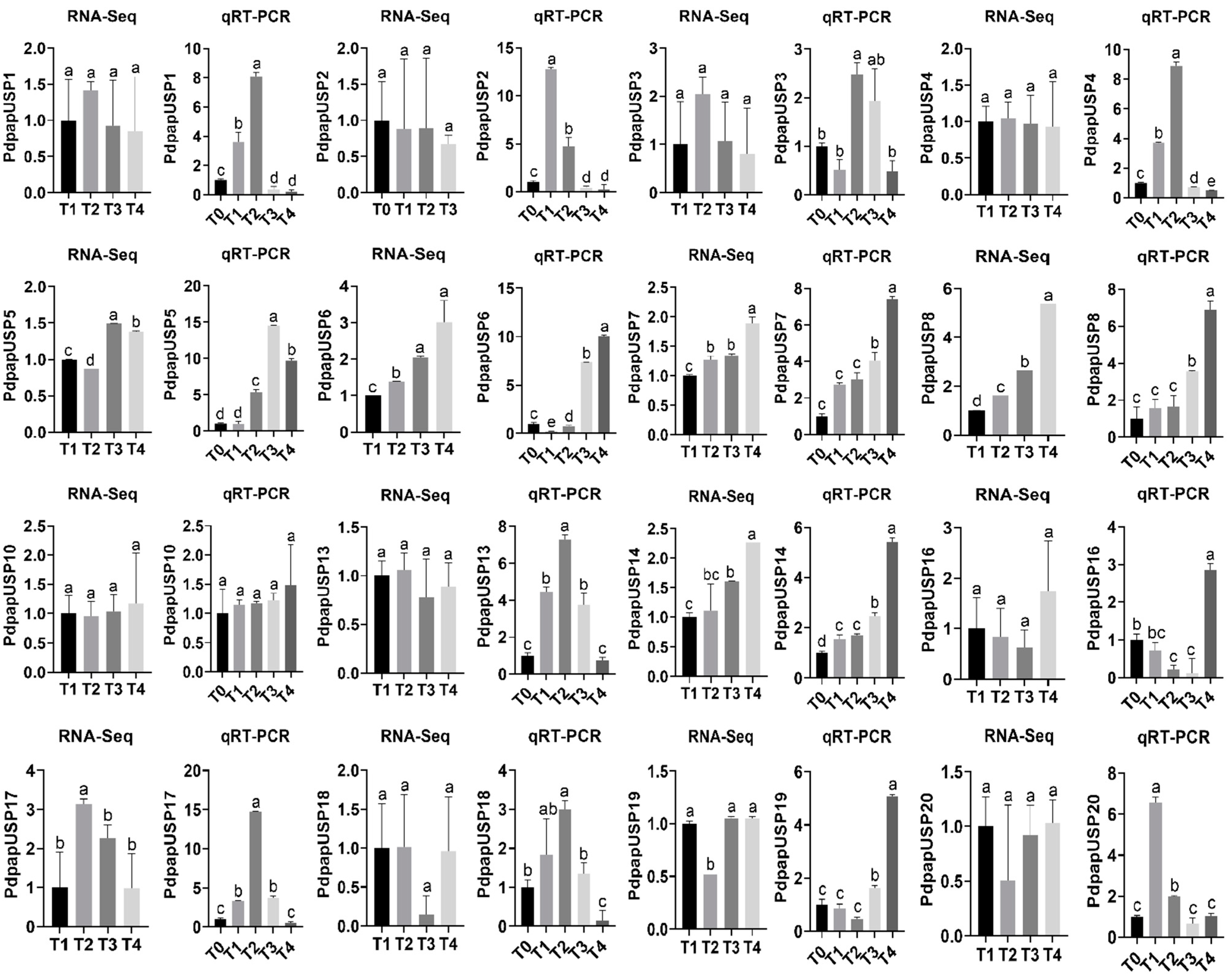
2.8. Validation of PtrUSP Expression Levels
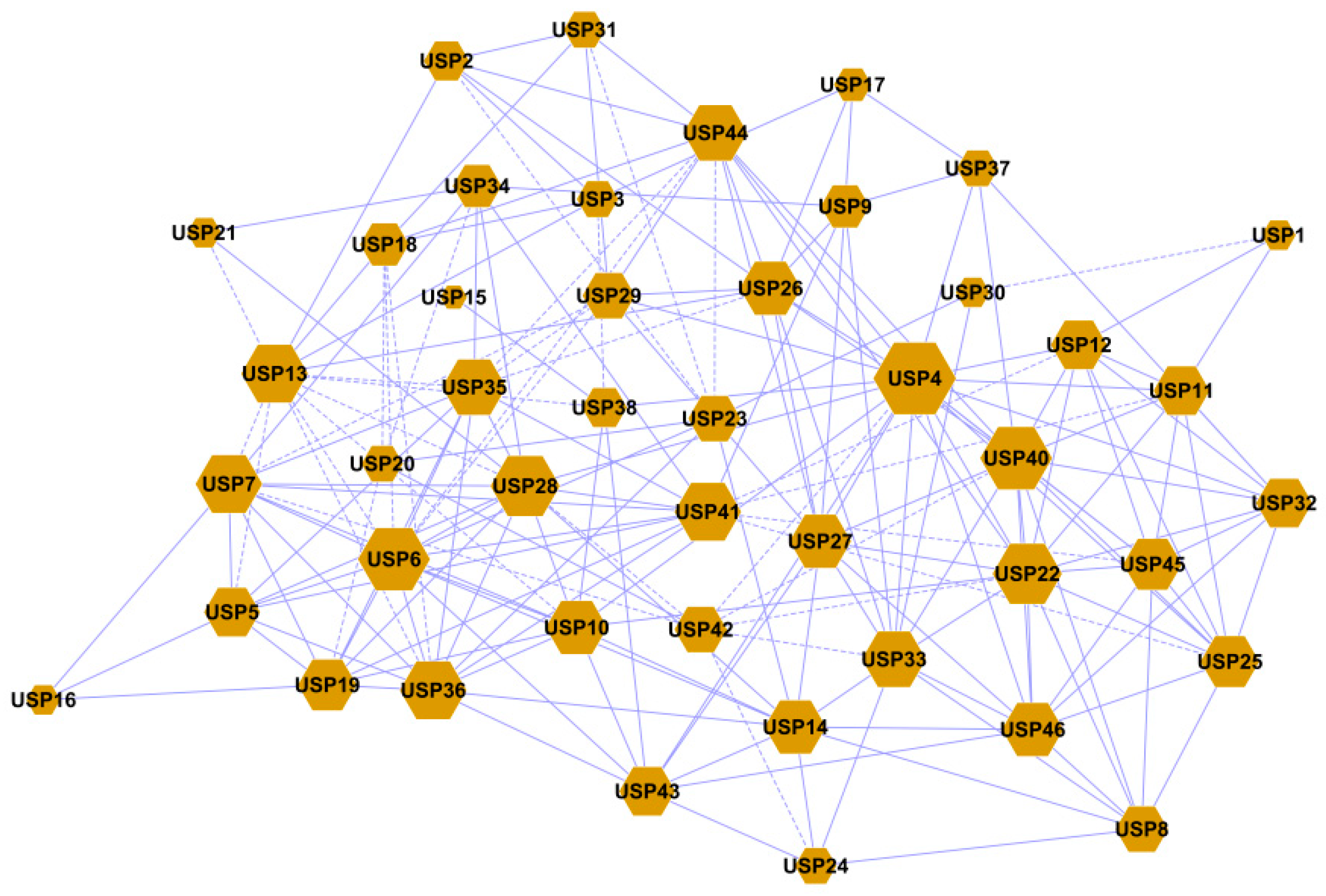
2.9. Gene Co-Expression Analysis
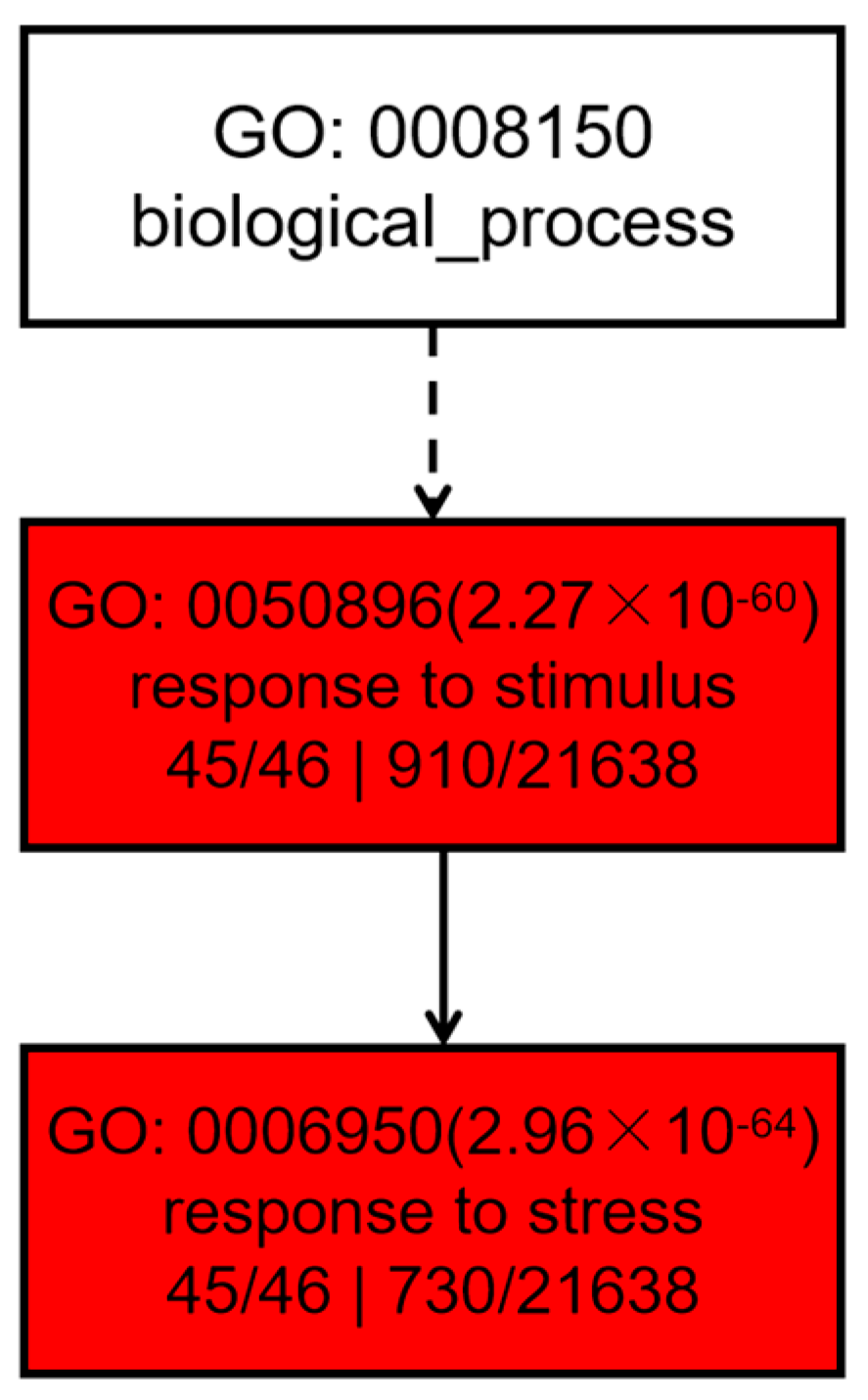
2.10. Gene Ontology (GO) Analysis
3. Discussion
4. Materials and Methods
4.1. Screening and Identification of USPs in P. trichocarpa
4.2. Evolutionary Relationship and Physicochemical Properties of USPs
4.3. Gene Structure and Protein Motif Analysis of PtrUSPs
4.4. Secondary and Tertiary Structure Analysis of PtrUSPs
4.5. Topological Heterogeneity Model and Subcellular Localization Prediction of PtrUSPs
4.6. Cis-Acting Element Analysis
4.7. Chromosome Distribution and Collinearity Analysis
4.8. Sample Preparation
4.9. Gene Expression Analysis
4.10. Co-Expression Network and Gene Ontology Annotation Analysis
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kvint, K.; Nachin, L.; Diez, A.; Nystrom, T. The bacterial universal stress protein: Function and regulation. Curr. Opin. Microbiol. 2003, 6, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.H.; Koo, S.S.; Oh, H.T.; Lee, E.S.; Park, J.H.; Phan, K.A.T.; Wi, S.D.; Bae, S.B.; Paeng, S.K.; Chae, H.B.; et al. The physiological functions of universal stress proteins and their molecular mechanism to protect plants from environmental stresses. Front. Plant Sci. 2019, 10, 750. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, A.; Berisio, R.; Lucey, M.; Schaffer, K.; Mcclean, S. A Universal Stress Protein upregulated by hypoxia may contribute to chronic lung colonisation and intramacrophage survival in cystic fibrosis. bioRxiv 2020. bioRxiv:2020.10.03.324806. [Google Scholar]
- Loukehaich, R.; Wang, T.T.; Ouyang, B.; Ziaf, K.; Li, H.X.; Zhang, J.H.; Lu, Y.E.; Ye, Z.B. SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 2012, 63, 5593–5606. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.Y.; Shen, J.; Ding, J.; Yuan, Y.; Gao, L.L.; Yang, Z.C.; Zhao, X. USP18 mitigates lipopolysaccharide-induced oxidative stress and inflammation in human pulmonary microvascular endothelial cells through the TLR4/NF-κB/ROS signaling. Toxicol. Vitr. 2021, 75, 105181. [Google Scholar] [CrossRef]
- Liu, W.T.; Karavolos, H.M.; Bulmer, M.D.; Allaoui, A.; Hormaeche, R.D.E.C.; Lee, J.J.; Khan, C.M.A. Role of the Universal Stress Protein UspA of Salmonella in growth arrest, stress and virulence. Microb. Pathog. 2007, 42, 2–10. [Google Scholar] [CrossRef]
- Wang, X.F.; Su, J.; Yang, N.; Zhang, H.; Cao, X.Y.; Kang, J.F. Functional characterization of selected Universal Stress Protein from Salvia miltiorrhiza (SmUSP) in Escherichia coli. Genes 2017, 8, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.; Does, V.D.C.; Albers, S.V. SaUspA, the Universal Stress Protein of Sulfolobus acidocaldarius stimulates the activity of the PP2A phosphatase and is involved in growth at high salinity. Front. Microbiol. 2020, 11, 598821. [Google Scholar] [CrossRef] [PubMed]
- Siegele, D.D. Universal stress proteins in Escherichia coli. J. Bacteriol. 2005, 187, 6253–6254. [Google Scholar] [CrossRef] [Green Version]
- Tkaczuk, L.K.; Shumilin, A.I.; Chruszcz, M.; Evdokimova, E.; Savchenko, A.; Minor, W. Structural and functional insight into the universal stress protein family. Evol. Appl. 2013, 6, 434–449. [Google Scholar] [CrossRef]
- Maqbool, A.; Zahur, M.; Husnain, T.; Riazuddin, S. GUSP1 and GUSP2, two drought-responsive genes in Gossypium arboreum have homology to universal stress proteins. Plant Mol. Biol. Rep. 2009, 27, 109–114. [Google Scholar] [CrossRef]
- Nachin, L.; Nannmark, U.; Nyström, T. Differential roles of the Universal Stress Proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 2005, 187, 6265–6272. [Google Scholar] [CrossRef] [Green Version]
- Hingley-Wilson, S.M.; Lougheed, K.E.A.; Ferguson, K.; Leiva, S.; Williams, H.D. Individual Mycobacterium tuberculosis Universal Stress Protein homologues are dispensable in vitro. Tuberculosis 2010, 90, 236–244. [Google Scholar] [CrossRef] [Green Version]
- Weber, A.; Jung, K. Biochemical properties of UspG, a Universal Stress Protein of Escherichia coli. Biochemistry 2006, 45, 1620–1628. [Google Scholar] [CrossRef] [PubMed]
- Li, W.T.; Wei, Y.M.; Wang, J.R.; Liu, C.J.; Lan, X.J.; Jiang, Q.T.; Pu, Z.E.; Zheng, Y.L. Identifification, localization, and characterization of putative USP genes in barley. Theor. Appl. Genet. 2010, 121, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Kerk, D.; Bulgrien, J.; Smith, W.D.; Gribskov, M. Arabidopsis proteins containing similarity to the universal stress protein domain of bacteria. Plant Physiol. 2003, 131, 1209–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Liu, H.M. Recent advances in the development of ubiquitin-specific-processing protease 7 (USP7) inhibitors. Eur. J. Med. Chem. 2020, 191, 112107. [Google Scholar] [CrossRef]
- VanBogelen, A.R.; Hutton, M.E.; Neidhardt, C.F. Gene-protein database of Escherichia coli K-12: Edition 3. Electrophoresis 1990, 11, 1131–1166. [Google Scholar] [CrossRef]
- Drumm, E.J.; Mi, K.X.; Bilder, P.; Sun, M.H.; Lim, J.; Bielefeldt-Ohmann, H.; Basaraba, R.; So, M.; Zhu, G.F.; Tufariello, M.J.; et al. Mycobacterium tuberculosis Universal Stress Protein Rv2623 regulates bacillary growth by ATP-Binding: Requirement for establishing chronic persistent infection. PLoS Pathog. 2009, 5, 1000460. [Google Scholar] [CrossRef]
- Sauter, M.; Rzewuski, G.; Marwedel, T.; Lorbiecke, R. The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 2002, 53, 2325–2331. [Google Scholar] [CrossRef] [Green Version]
- Verslues, E.P.; Lasky, R.J.; Juenger, E.T.; Liu, T.W.; Kumar, M.N. Genome-wide association mapping combined with reverse genetics identifies new effectors of low water potential-induced proline accumulation in Arabidopsis. Plant Physiol. 2014, 164, 144–159. [Google Scholar] [CrossRef] [Green Version]
- Gou, L.M.; Zhuo, C.L.; Lu, S.Y.; Guo, Z.F. A universal stress protein from Medicago falcata (MfUSP1) confers multiple stress tolerance by regulating antioxidant defense and proline accumulation. Environ. Exp. Bot. 2020, 178, 104168. [Google Scholar] [CrossRef]
- Jung, Y.J.; Melencion, S.M.B.; Lee, E.S.; Park, J.H.; Alinapon, C.V.; Oh, H.T.; Yun, D.J.; Chi, Y.H.; Lee, S.Y. Universal stress protein exhibits a redox-dependent chaperone function in Arabidopsis and enhances plant tolerance to heat shock and oxidative stress. Front. Plant Sci. 2015, 6, 1141. [Google Scholar] [CrossRef] [Green Version]
- Merkouropoulos, G.; Andreasson, E.; Hess, D.; Boller, T.; Peck, C.S. An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP kinases 3 and 6. J. Biol. Chem. 2008, 283, 10493–10499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenman, M.; Sörensson, C.; Andreasson, E. Enrichment of phosphoproteins and phosphopeptide derivatization identify universal stress proteins in elicitor-treated Arabidopsis. Mol. Plant Microbe Interact. 2008, 21, 1275–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.C.; Jung, Y.J.; Lee, Y.; Kim, I.R.; Seol, M.A.; Kim, E.J.; Jang, M.K.; Lee, J.R. Functional characterization of the Arabidopsis universal stress protein AtUSP with an antifungal activity. Biochem. Biophys. Res. Commun. 2017, 486, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Espinola, S.M.; Cancela, M.P.; Correa, L.B.; Zaha, A. Evolutionary fates of universal stress protein paralogs in Platyhelminthes. BMC Evol. Biol. 2018, 18, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzali, S.; Loreti, E.; Cardarelli, F.; Novi, G.; Parlanti, S.; Pucciariello, C.; Bassolino, L.; Banti, V.; Licausi, F.; Perata, P. Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants 2015, 1, 15151. [Google Scholar] [CrossRef] [PubMed]
- Melencion, S.M.B.; Chi, Y.H.; Pham, T.T.; Paeng, S.K.; Wi, S.D.; Lee, C.Y.; Ryu, S.W.; Koo, S.S.; Lee, S.Y. RNA chaperone function of a universal stress protein in Arabidopsis confers enhanced cold stress tolerance in plants. Int. J. Mol. Sci. 2017, 18, 2546. [Google Scholar] [CrossRef] [Green Version]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Isokpehi, D.R.; Simmons, S.S.; Cohly, H.P.H.; Ekunwe, I.N.S.; Begonia, B.G.; Ayensu, K.W. Identifification of drought-responsive universal stress proteins in Viridiplantae. Bioinform. Biol. Insights 2011, 5, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhao, F.A.; Fang, W.P.; Xie, D.Y.; Hou, J.N.; Yang, X.J.; Zhao, Y.M.; Tang, Z.J.; Nie, L.H.; Lv, S.P. Identification of early salt stress responsive proteins in seedling roots of upland cotton (Gossypium hirsutum L.) employing i TRAQ-based proteomic technique. Front. Plant Sci. 2015, 6, 732. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Beltrán, E.; Personat, J.M.; de la Torre, F.; del Pozo, O. A universal stress protein involved in oxidative stress is a phosphorylation target for protein kinase CIPK6. Plant Physiol. 2017, 173, 836–852. [Google Scholar] [CrossRef] [Green Version]
- Udawat, P.; Jha, K.R.; Sinha, D.; Mishra, A.; Jha, B. Overexpression of a cytosolic abiotic stress responsive universal stress protein (SbUSP) mitigates salt and osmotic stress in transgenic tobacco plants. Front. Plant Sci. 2016, 7, 518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, S.; Ahmad, A.; Batool, F.; Rashid, B.; Husnain, T. Genetic modifification of Gossypium arboreum universal stress protein (GUSP1) improves drought tolerance in transgenic cotton (Gossypium hirsutum). Physiol. Mol. Biol. Plants 2021, 27, 1779–1794. [Google Scholar] [CrossRef]
- Akram, A.; Arshad, K.; Hafeez, M.N.; Ahmad, A. Cloning and expression of universal stress protein 2 (USP2) gene in Escherichia coil. Biol. Clin. Sci. Res. J. 2021, 2021, e002. [Google Scholar] [CrossRef]
- Dhanyalakshmi, K.H.; Naika, B.N.M.; Sajeevan, R.S.; Mathew, K.O.; Shafifi, K.M.; Sowdhamini, R.; Nataraja, N.K. An approach to function annotation for Proteins of Unknown Function (PUFs) in the transcriptome of Indian mulberry. PLoS ONE 2016, 11, e0151323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahur, M.; Maqbool, A.; Irfan, M.; Barozai, M.Y.K.; Rashid, B.; Riazuddin, S.; Husnain, T. Isolation and functional analysis of cotton universal stress protein promoter in response to phytohormones and abiotic stresses. Cell. Mol. Biol. 2009, 43, 578–585. [Google Scholar] [CrossRef]
- Gorshkova, D.S.; Getman, I.A.; Voronkov, A.S.; Chizhova, S.I.; Kuznetsov, V.V.; Pojidaeva, E.S. The gene encoding the universal stress protein AtUSP is regulated by phytohormones and involved in seed germination of Arabidopsis thaliana. Biochem. Biophys. Mol. Biol. 2018, 479, 105–107. [Google Scholar] [CrossRef]
- Cia, M.C.; Guimaraes, A.C.R.; Medici, L.; Chabregas, S.M.; Azevedo, R.A. Antioxidant responses to water defificit by drought tolerant and sensitive sugarcane varieties. Ann. Appl. Biol. 2012, 161, 313–324. [Google Scholar] [CrossRef]
- Masamba, P.; Adenowo, A.F.; Oyinloye, B.E.; Kappo, A.P. Universal Stress Proteins as new targets for environmental and therapeutic interventions of schistosomiasis. Int. J. Environ. Res. Public Health 2016, 13, 972. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.B.; Guo, J.Y.; Jin, X.L.; Kim, J.S.; Ji, Y.; Fan, S.D.; Ha, N.C.; Quan, C.S. Crystal structure and functional implications of the tandem-type Universal Stress Protein UspE from Escherichia coli. BMC Struct. Biol. 2016, 16, 3. [Google Scholar] [CrossRef] [Green Version]
- Duprey, A.; Nasser, W.; Léonard, S.; Brochier-Armanet, C.; Reverchon, S. Transcriptional start site turnover in the evolution of bacterial paralogous genes: The pelE-pelD virulence genes in Dickeya. FEBS J. 2016, 283, 4192–4207. [Google Scholar] [CrossRef] [PubMed]
- Roy, W.S.; Penny, D. On the incidence of intron loss and gain in paralogous gene families. Mol. Biol. Evol. 2007, 24, 1579–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffares, C.D.; Penkett, J.C.; Bähler, J. Rapidly regulated genes are intron poor. Trend Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Prior, P.; Fujikawa, K.; Pratt, K.P. New insights into binding interfaces of coagulation factors V and VIII and their homologues-lessons from high resolution crystal structures. Curr. Protein Pept. Sci. 2002, 3, 313–339. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Khoshnoodi, J.; Takenaka, H.; Hosoyamada, M.; Nakajo, A.; Bessho, F.; Kudo, A.; Takahashi, S.; Arimura, Y.; Yamada, A.; et al. The effect of dexamethasone on defective nephrin transport caused by ER stress: A potential mechanism for the therapeutic action of glucocorticoids in the acquired glomerular diseases. Kidney Int. 2006, 69, 1350–1359. [Google Scholar] [CrossRef] [Green Version]
- Kandel, M.B.; Yamamoto, S.; Midorikawa, R.; Morise, J.; Wakazono, Y.; Oka, S.; Takamiya, K. N-glycosylation of the AMPA-type glutamate receptor regulates cell surface expression and tetramer formation affecting channel function. J. Neurochem. 2019, 147, 730–747. [Google Scholar] [CrossRef] [Green Version]
- Li, L.B.; Chen, N.H.; Ramamoorthy, S.; Chi, L.M.; Cui, X.N.; Wang, L.J.C.; Reith, M.E.A. The role of N-glycosylation in function and surface trafficking of the human dopamine transporter. J. Biol. Chem. 2004, 279, 21012–21020. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.P.; Ye, Q.; Chen, M.; Li, A.M.; Mi, W.T.; Fang, Y.X.; Zaytseva, Y.Y.; O’Connor, K.L.; Kooi, C.W.V.; Liu, S.; et al. N-glycosylation-defective splice variants of neuropilin-1 promote metastasis by activating endosomal signals. Nat. Commun. 2019, 10, 3708. [Google Scholar] [CrossRef] [Green Version]
- Girin, T.; Lejay, L.; Wirth, J.; Widiez, T.; Palenchar, M.P.; Nazoa, P.; Touraine, B.; Gojon, A.; Lepetit, M. Identification of a 150 bp cis-acting element of the AtNRT2.1 promoter involved in the regulation of gene expression by the N and C status of the plant. Plant Cell Environ. 2007, 30, 1366–1380. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed] [Green Version]
- Wang, Y.; Salasini, C.B.; Khan, M.; Devi, B.; Bush, M.; Subramaniam, R.; Hepworth, R.S. Clade I TGACG-motif binding basic leucine zipper transcription factors mediate BLADE-ON-PETIOLE-dependent regulation of development. Plant Physiol. 2019, 180, 937–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, H.Z.; Landherr, L.L.; Frohlich, W.M.; Leebens-Mack, J.; Ma, H.; DePamphilis, W.C. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef]
- Cannon, B.S.; Mitra, A.; Baumgarten, A.; Young, D.N.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Flagel, E.L.; Wendel, F.J. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pan, F.; Chen, D.M.; Chu, W.Y.; Liu, H.L.; Xiang, Y. Genome-wide identification and analysis of the Populus trichocarpa TIFY gene family. Plant Physiol. Bioch. 2017, 115, 360–371. [Google Scholar] [CrossRef]
- Xu, T.; Chen, T.; Zhang, T.; Shen, L.; Chen, Z.; Xu, Y.; Wu, Y.; Yang, J. Genome-Wide Identification and Characterization of USP Gene Family in Grapes (Vitis vinifera L.). Horticulturae 2022, 8, 1024. [Google Scholar] [CrossRef]
- Cui, X.Y.; Zhang, P.Y.; Hu, Y.F.; Chen, C.C.; Liu, Q.Y.; Guan, P.Y.; Zhang, J.X. Genome-wide analysis of the Universal stress protein A gene family in Vitis and expression in response to abiotic stress. Plant Physiol. Bioch. 2021, 165, 57–70. [Google Scholar] [CrossRef]
- Mathias, J.T.; Natarajan, K.; Shukla, S.; Doshi, A.K.; Singh, N.Z.; Ambudkar, V.S.; Baer, R.M. The FLT3 and PDGFR inhibitor crenolanib is a substrate of the multidrug resistance protein ABCB1 but does not inhibit transport function at pharmacologically relevant concentrations. Invest. New Drug 2015, 33, 300–309. [Google Scholar] [CrossRef]
- Goodstein, M.D.; Shu, S.Q.; Howson, R.; Neupane, R.; Hayes, D.R.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for green Plant Genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, D.R.; Eddy, R.S.; Bateman, A.; Punta, M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.R.; Williams, L.; Qureshi, M.; Salazar, A.G.; Sonnhammer, L.L.E.; Tosatto, S.; Paladin, L.; Raj, S.; Richardson, J.L.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using Maximum Likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a Maximum-Likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, D.R.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.J.; Xia, R.; Chen, H.; He, Y.H. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. Mol. Plant 2018, 3, 1–7. [Google Scholar]
- Bailey, L.T.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identififier. Nucleic Acids Res. 2005, 33, 116–120. [Google Scholar] [CrossRef] [Green Version]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Torsten, S.; Jürgen, K.; Nicolas, G.; Peitsch, C.M. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar]
- Ulrich, O.; Christian, H.A.; Sebastian, M.; Bernd, W. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; de Peer, V.Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, D.J.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny andcollinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, H.B.; Bowers, E.J.; Wang, X.Y.; Ming, R.; Alam, M.; Paterson, H.A. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Kang, P.Z.; Bai, X.J.; Zhang, L.R.; Du, Y.N.; Zhang, Z.K. Biological characteristics of Fusarium oxysporium (Schl.) f. sp. Cucumerinum Owen. N. Horticult. 2018, 22, 65–69. [Google Scholar]
- Diao, J.; Wang, J.Q.; Zhang, P.; Hao, X.; Wang, Y.; Liang, L.W.; Zhang, Y.; Ma, W.; Ma, L. Transcriptome analysis reveals the important role of WRKY28 in Fusarium Oxysporum resistance. Front. Plant Sci. 2021, 12, 1723. [Google Scholar] [CrossRef]
- Zhai, Z.H.; Chen, X.N.; Wang, J. Primer design with Primer Premier 5.0. Adv. Med. Educ. Pract. 2008, 16, 695–698. [Google Scholar]
- Guo, R.T.; Ji, S.D.; Wang, Z.Y.; Zhang, H.F.; Wang, Y.C.; Liu, Z.H. Trichoderma asperellum xylanases promote growth and induce resistance in poplar. Microbiol. Res. 2021, 248, 126767. [Google Scholar] [CrossRef]
- Livak, J.K.; Schmittgen, D.T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Damian, S.; Andrea, F.; Michael, K.; Milan, S.; Alexander, R.; Pablo, M.; Tobias, D.; Manuel, S.; Jean, M.; Peer, B.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, 561–568. [Google Scholar]
- Smoot, E.M.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, T.; Liu, Y.; Yan, H.Y.; You, Q.; Yi, X.; Du, Z.; Xu, W.Y.; Su, Z. AgriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, 122–129. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K. SPSS for applied sciences: Basic statistical testing. J. R. Stat. Soc. 2014, 4, 990. [Google Scholar] [CrossRef]
- Choi, K.J.; Kim, H.B.; Park, S.H. The comparison of postoperative pain: Total laparoscopic hysterectomy versus vaginal hysterectomy. Korean J. Obstet. Gynecol. 2012, 55, 384–391. [Google Scholar] [CrossRef] [Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diao, J.; Gu, W.; Jiang, Z.; Wang, J.; Zou, H.; Zong, C.; Ma, L. Comprehensive Analysis of Universal Stress Protein Family Genes and Their Expression in Fusarium oxysporum Response of Populus davidiana × P. alba var. pyramidalis Louche Based on the Transcriptome. Int. J. Mol. Sci. 2023, 24, 5405. https://doi.org/10.3390/ijms24065405
Diao J, Gu W, Jiang Z, Wang J, Zou H, Zong C, Ma L. Comprehensive Analysis of Universal Stress Protein Family Genes and Their Expression in Fusarium oxysporum Response of Populus davidiana × P. alba var. pyramidalis Louche Based on the Transcriptome. International Journal of Molecular Sciences. 2023; 24(6):5405. https://doi.org/10.3390/ijms24065405
Chicago/Turabian StyleDiao, Jian, Wei Gu, Zhehui Jiang, Jiaqi Wang, Hongfei Zou, Cheng Zong, and Ling Ma. 2023. "Comprehensive Analysis of Universal Stress Protein Family Genes and Their Expression in Fusarium oxysporum Response of Populus davidiana × P. alba var. pyramidalis Louche Based on the Transcriptome" International Journal of Molecular Sciences 24, no. 6: 5405. https://doi.org/10.3390/ijms24065405
APA StyleDiao, J., Gu, W., Jiang, Z., Wang, J., Zou, H., Zong, C., & Ma, L. (2023). Comprehensive Analysis of Universal Stress Protein Family Genes and Their Expression in Fusarium oxysporum Response of Populus davidiana × P. alba var. pyramidalis Louche Based on the Transcriptome. International Journal of Molecular Sciences, 24(6), 5405. https://doi.org/10.3390/ijms24065405





