Inhibition of Expression of the Circadian Clock Gene Cryptochrome 1 Causes Abnormal Glucometabolic and Cell Growth in Bombyx mori Cells
Abstract
1. Introduction
2. Results
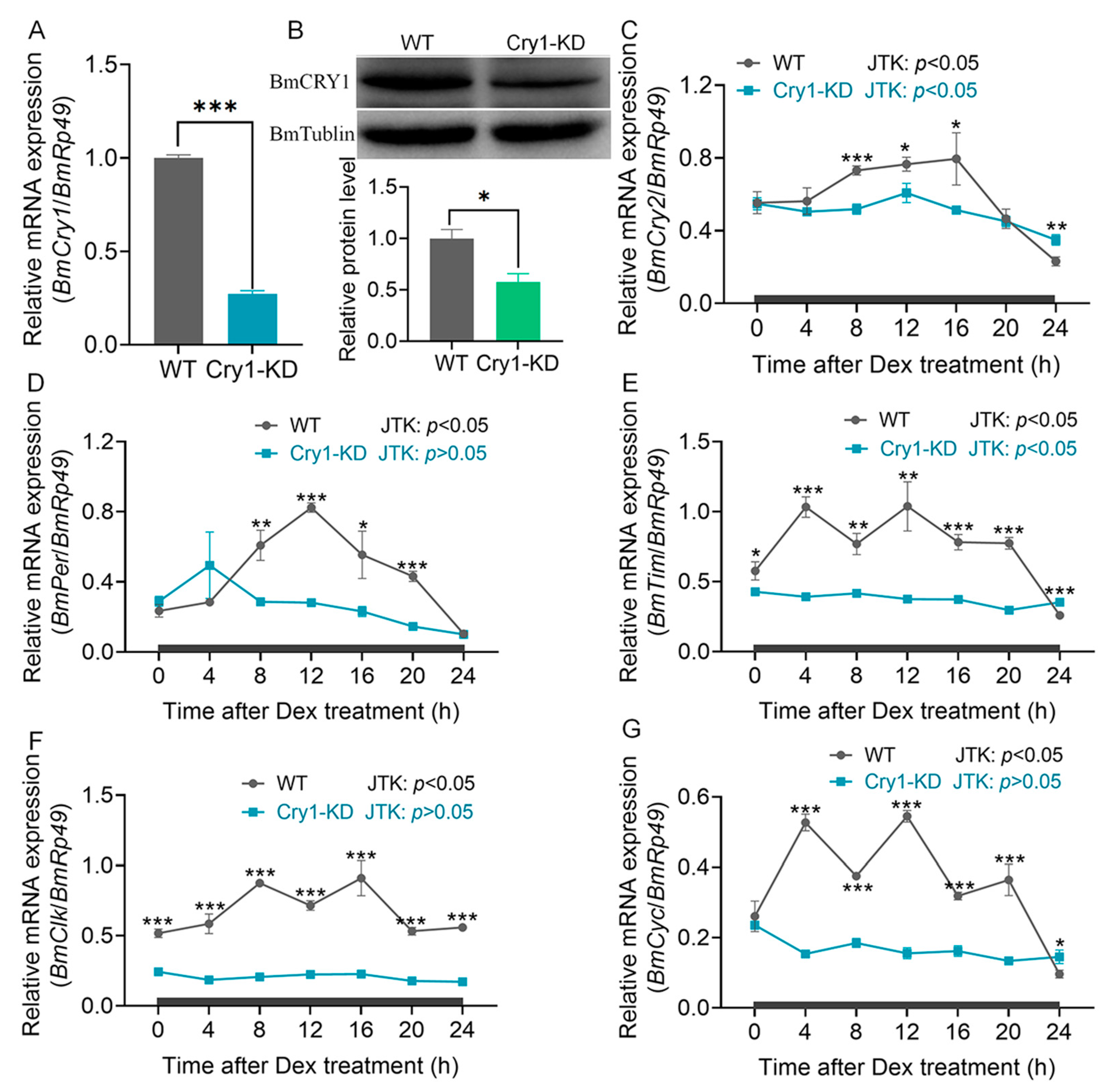
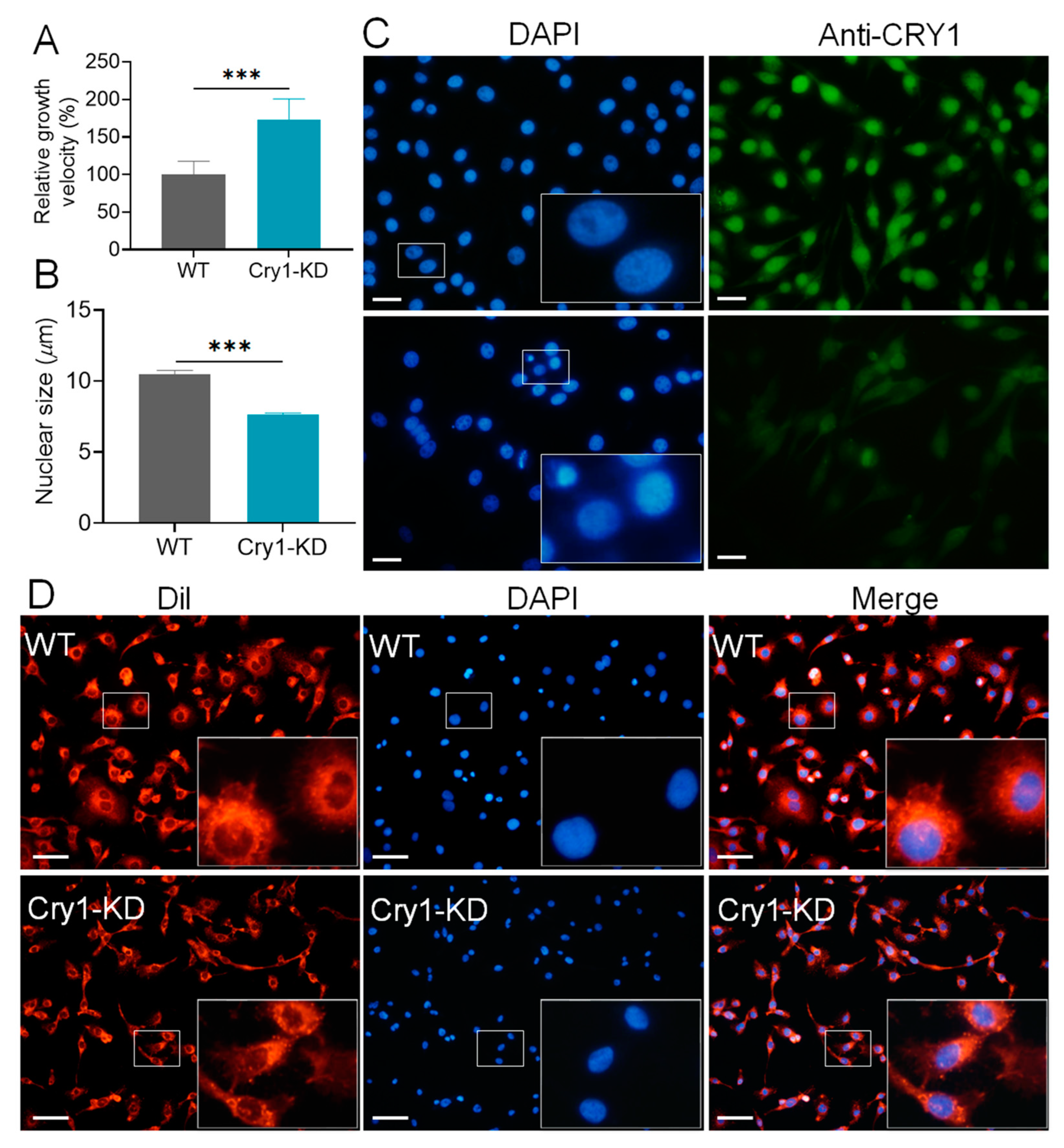
2.1. BmCry1 Knockdown Results in Accelerated Cell Growth and a Smaller Nucleus
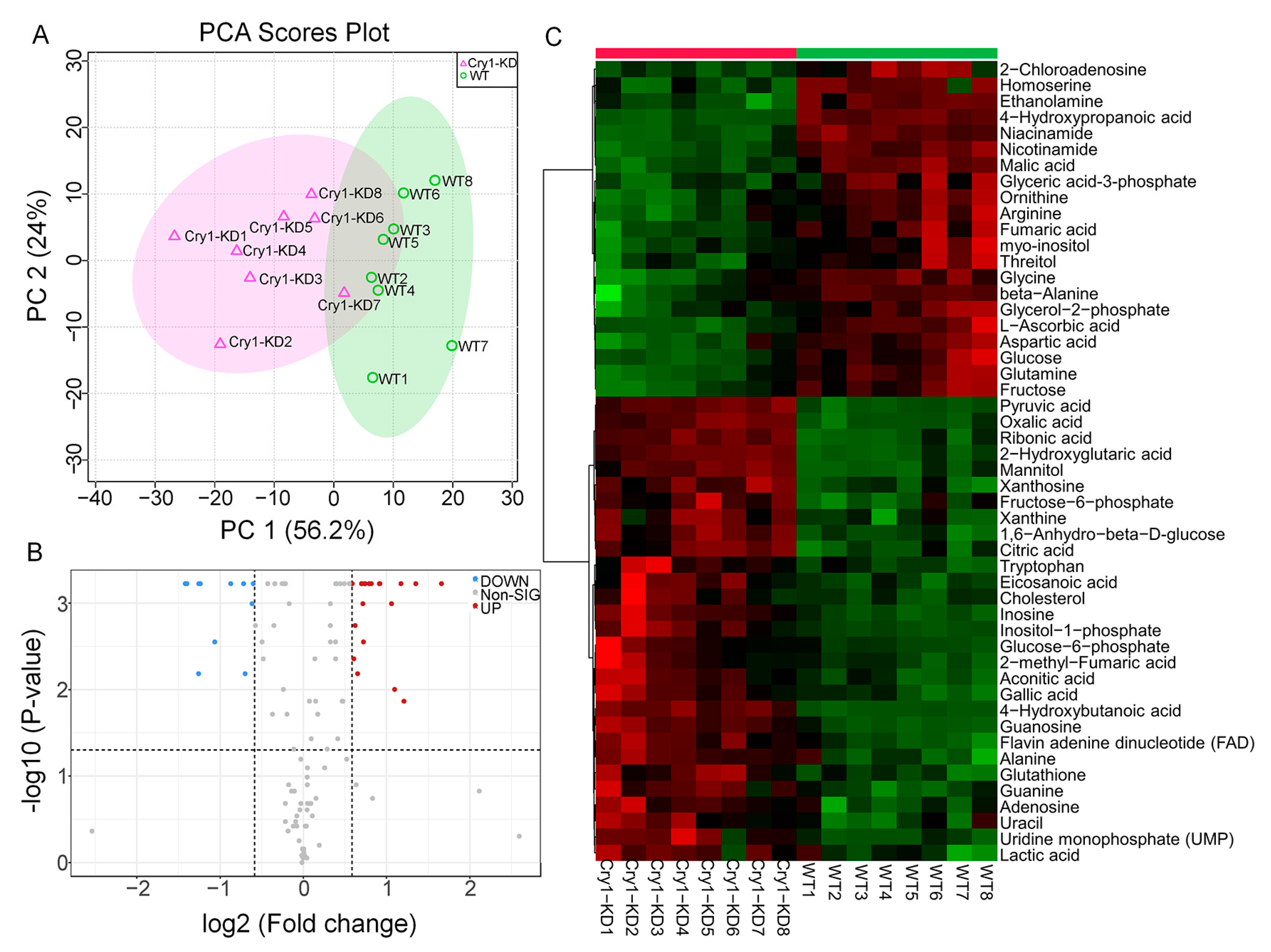
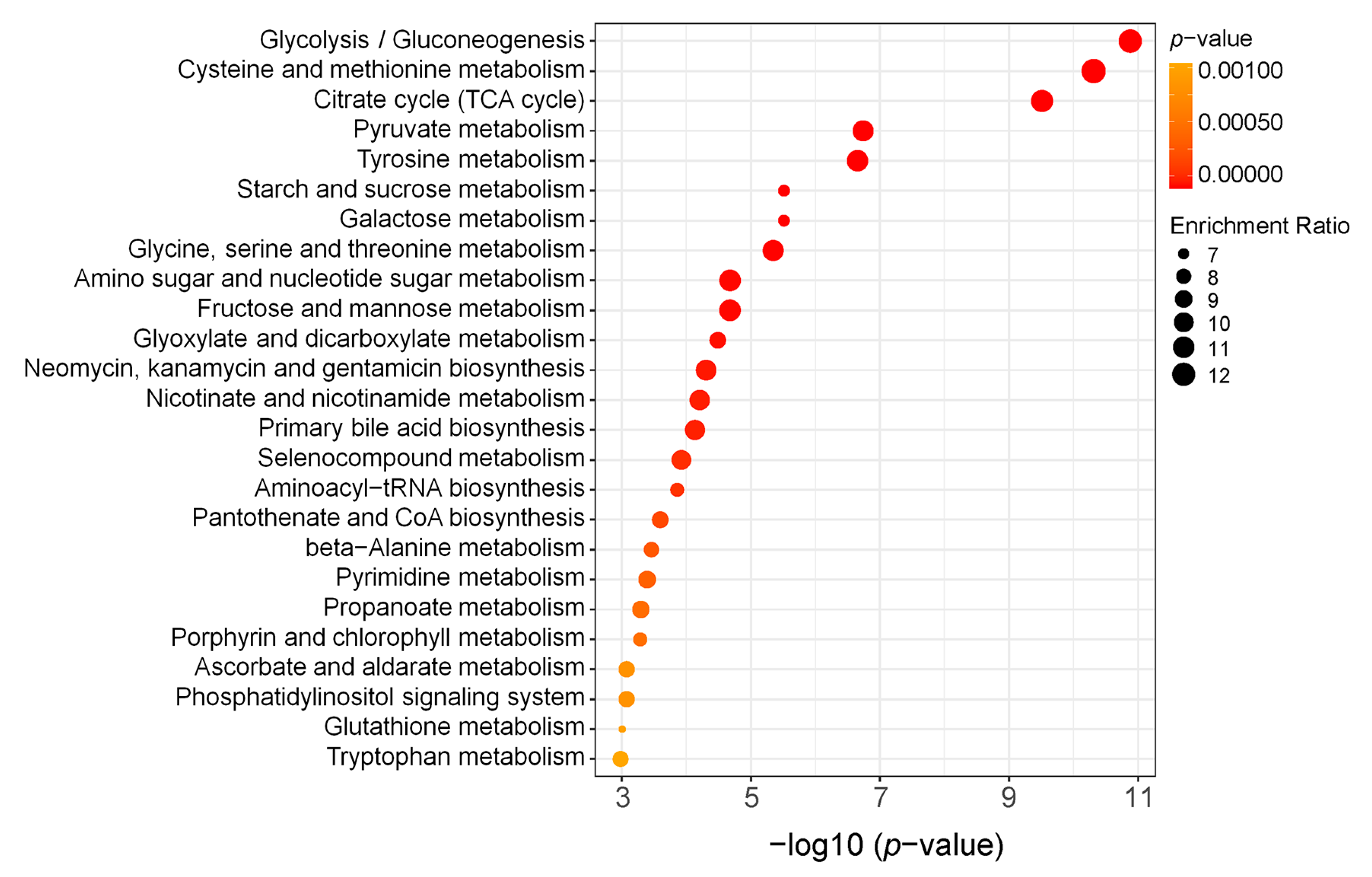
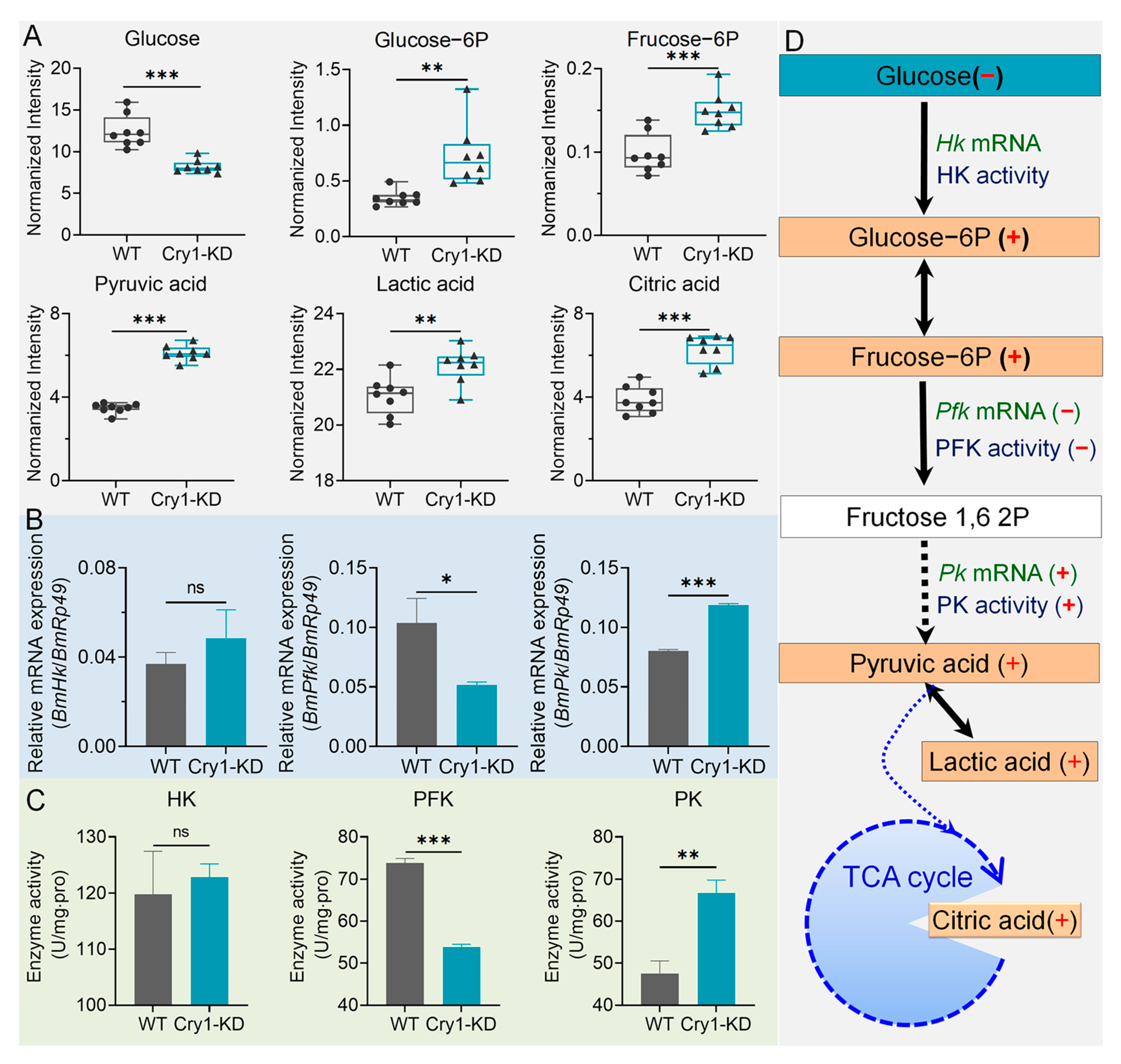
2.2. GC-MS and LC-MS/MS Assay Showed That the Knockdown of BmCry1 Caused Changes in Cell Metabolites
2.3. Glucose Metabolism Was Increased in BmCry1 Knockdown Cells
3. Discussion
3.1. Possible Functions of Cryptochrome in Bombyx mori
3.2. The Interaction of Circadian Clock, Cellular Glycometabolism and Cell Proliferation
3.3. Metabolic Difference between Knockdown BmCry1 and BmPer in BmN Cells
4. Materials and Methods
4.1. Preparation of the BmCry1 Knockdown (Cry1-KD) Cells
4.2. Cell Proliferation Assay
4.3. Cell Staining
4.4. Determination of Metabolomics
4.5. qRT-PCR Analysis
4.6. Determination of Enzyme Activity
4.7. Western Blotting
4.8. Immunohistochemistry
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ahmad, M.; Cashmore, A.R. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 1993, 366, 162–166. [Google Scholar] [CrossRef]
- Yuan, Q.; Metterville, D.; Briscoe, A.D.; Reppert, S.M. Insect Cryptochromes: Gene Duplication and Loss Define Diverse Ways to Construct Insect Circadian Clocks. Mol. Biol. Evol. 2007, 24, 948–955. [Google Scholar] [CrossRef]
- Ozturk, N. Light-dependent reactions of animal circadian photoreceptor cryptochrome. FEBS J. 2022, 289, 6622–6639. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.D.; Liang, H.; Zhu, X.S.; Tao, H.; Xu, L.; Sima, Y.H.; Xu, S.Q. Molecular cloning and bioinformatic analysis of circadian clock genes Bmcry1 and Bmcry2 in Bombyx mori. Acta Entomol. Sin. 2011, 54, 9–19. [Google Scholar]
- Zhu, H.; Sauman, I.; Yuan, Q.; Casselman, A.; Emery-Le, M.; Emery, P.; Reppert, S.M. Cryptochromes Define a Novel Circadian Clock Mechanism in Monarch Butterflies That May Underlie Sun Compass Navigation. PLoS Biol. 2008, 6, e4. [Google Scholar] [CrossRef]
- Rubin, E.B.; Shemesh, Y.; Cohen, M.; Elgavish, S.; Robertson, H.M.; Bloch, G. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 2006, 16, 1352–1365. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Hayden, A.N.; Iiams, S.E.; Merlin, C. Cryptochrome 1 mediates light-dependent inclination magnetosensing in monarch butterflies. Nat. Commun. 2021, 12, 771. [Google Scholar] [CrossRef] [PubMed]
- Emery, P.; So, W.V.; Kaneko, M.; Hall, J.C.; Rosbash, M. CRY, a Drosophila Clock and Light-Regulated Cryptochrome, Is a Major Contributor to Circadian Rhythm Resetting and Photosensitivity. Cell 1998, 95, 669–679. [Google Scholar] [CrossRef]
- Merlin, C.; Beaver, L.E.; Taylor, O.R.; Wolfe, S.A.; Reppert, S.M. Efficient targeted mutagenesis in the monarch butterfly using zinc-finger nucleases. Genome Res. 2013, 23, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Sehadova, H.; Markova, E.P.; Sehnal, F.; Takeda, M. Distribution of Circadian Clock-Related Proteins in the Cephalic Nervous System of the Silkworm, Bombyx Mori. J. Biol. Rhythm. 2004, 19, 466–482. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Liang, H.; Sima, Y.H.; Xu, S.Q. Effects of temperature and light rhythm on expression of clock genes Cry1 and Cry2 in Bombyx mori adult. Sci. Seric. 2013, 39, 453–459. [Google Scholar]
- Liang, H.; Xu, S.Q. Effect of diapause-inducing temperature and photoperiod on expression of major circadian clock feedback loop genes in embryos of silkworm, Bombyx mori. Sci. Seric. 2010, 36, 771–779. [Google Scholar]
- Chu, F.; Qiu, J.F.; Tao, H.; Li, X.; Shu, M.Y.; Liu, H.J.; Sima, Y.; Xu, S. Impact of cyclical changes in temperature on circadian clock genes expression in Bombyx BmN cells. Arch. Insect Biochem. Physiol. 2016, 91, 175–1886. [Google Scholar] [CrossRef]
- Sato, T.; Sassone-Corsi, P. Nutrition, metabolism, and epigenetics: Pathways of circadian reprogramming. EMBO Rep. 2022, 23, e52412. [Google Scholar] [CrossRef] [PubMed]
- Panda, S. Circadian physiology of metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef]
- Li, Y.; Li, G.; Görling, B.; Luy, B.; Du, J.; Yan, J. Integrative Analysis of Circadian Transcriptome and Metabolic Network Reveals the Role of De Novo Purine Synthesis in Circadian Control of Cell Cycle. PLoS Comput. Biol. 2015, 11, e1004086. [Google Scholar] [CrossRef]
- Pan, X.; Mota, S.; Zhang, B. Circadian clock regulation on lipid metabolism and metabolic diseases. In Lipid Transfer in Lipoprotein Metabolism and Cardiovascular Disease; Springer: Gateway East, Singapore, 2020; pp. 53–66. [Google Scholar]
- Chaix, A.; Lin, T.; Le, H.D.; Chang, M.W.; Panda, S. Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab. 2019, 29, 303–319.e4. [Google Scholar] [CrossRef]
- Mellor, J. The molecular basis of metabolic cycles and their relationship to circadian rhythms. Nat. Struct. Mol. Biol. 2016, 23, 1035–1044. [Google Scholar] [CrossRef]
- Wada, T.; Yamamoto, Y.; Takasugi, Y.; Ishii, H.; Uchiyama, T.; Saitoh, K.; Suzuki, M.; Uchiyama, M.; Yoshitane, H.; Fukada, Y.; et al. Adiponectin regulates the circadian rhythm of glucose and lipid metabolism. J. Endocrinol. 2022, 254, 121–133. [Google Scholar] [CrossRef]
- Yuan, P.; Yang, T.; Mu, J.; Zhao, J.; Yang, Y.; Yan, Z.; Hou, Y.; Chen, C.; Xing, J.; Zhang, H.; et al. Circadian clock gene NPAS2 promotes reprogramming of glucose metabolism in hepatocellular carcinoma cells. Cancer Lett. 2020, 469, 498–509. [Google Scholar] [CrossRef]
- Hemmer, A.; Mareschal, J.; Dibner, C.; Pralong, J.A.; Dorribo, V.; Perrig, S.; Genton, L.; Pichard, C.; Collet, T.-H. The Effects of Shift Work on Cardio-Metabolic Diseases and Eating Patterns. Nutrients 2021, 13, 4178. [Google Scholar] [CrossRef]
- Marjot, T.; Ray, D.W.; Williams, F.R.; Tomlinson, J.W.; Armstrong, M.J. Sleep and liver disease: A bidirectional relationship. Lancet Gastroenterol. Hepatol. 2021, 6, 850–863. [Google Scholar] [CrossRef]
- McDonald, R.J.; Zelinski, E.L.; Keeley, R.J.; Sutherland, D.; Fehr, L.; Hong, N.S. Multiple effects of circadian dysfunction induced by photoperiod shifts: Alterations in context memory and food metabolism in the same subjects. Physiol. Behav. 2013, 118, 14–24. [Google Scholar] [CrossRef]
- Sahar, S.; Sassone-Corsi, P. Regulation of metabolism: The circadian clock dictates the time. Trends Endocrinol. Metab. 2012, 23, 1–8. [Google Scholar] [CrossRef]
- Jorgensen, P.; Tyers, M. How Cells Coordinate Growth and Division. Curr. Biol. 2004, 14, R1014–R1027. [Google Scholar] [CrossRef]
- Tu, B.P.; Kudlicki, A.; Rowicka, M.; McKnight, S.L. Logic of the Yeast Metabolic Cycle: Temporal Compartmentalization of Cellular Processes. Science 2005, 310, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Sutter, B.M.; Li, B.; Tu, B.P. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 2011, 42, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Rigaud, V.O.C.; Hoy, R.; Mohsin, S.; Khan, M. Stem Cell Metabolism: Powering Cell-Based Therapeutics. Cells 2020, 9, 2490. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef]
- Owusu-Ansah, E.; Yavari, A.; Mandal, S.; Banerjee, U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet. 2008, 40, 356–361. [Google Scholar] [CrossRef]
- Agathocleous, M.; Harris, W.A. Metabolism in physiological cell proliferation and differentiation. Trends Cell Biol. 2013, 23, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Moncada, S.; Higgs, E.A.; Colombo, S.L. Fulfilling the metabolic requirements for cell proliferation. Biochem. J. 2012, 446, 1–7. [Google Scholar] [CrossRef]
- Tao, H.; Li, X.; Qiu, J.-F.; Cui, W.-Z.; Sima, Y.-H.; Xu, S.-Q. Inhibition of expression of the circadian clock gene Period causes metabolic abnormalities including repression of glycometabolism in Bombyx mori cells. Sci. Rep. 2017, 7, 46258. [Google Scholar] [CrossRef]
- Manoogian, E.N.C.; Chow, L.S.; Taub, P.R.; Laferrère, B.; Panda, S. Time-restricted Eating for the Prevention and Management of Metabolic Diseases. Endocr. Rev. 2022, 43, 405–436. [Google Scholar] [CrossRef] [PubMed]
- Neves, A.R.; Albuquerque, T.; Quintela, T.; Costa, D. Circadian rhythm and disease: Relationship, new insights, and future perspectives. J. Cell. Physiol. 2022, 237, 3239–3256. [Google Scholar] [CrossRef] [PubMed]
- Steele, T.A.; Louis, E.K.; Videnovic, A.; Auger, R.R. Circadian rhythm sleep-wake disorders: A contemporary review of neurobiology, treatment, and dysregulation in neurodegenerative disease. Neurotherapeutics 2021, 18, 53–74. [Google Scholar] [CrossRef]
- Yin, Z.; Klionsky, D.J. Intermittent time-restricted feeding promotes longevity through circadian autophagy. Autophagy 2022, 18, 471–472. [Google Scholar] [CrossRef] [PubMed]
- Froy, O. Circadian aspects of energy metabolism and aging. Ageing Res. Rev. 2013, 12, 931–940. [Google Scholar] [CrossRef]
- Tevy, M.F.; Giebultowicz, J.; Pincus, Z.; Mazzoccoli, G.; Vinciguerra, M. Aging signaling pathways and circadian clock-dependent metabolic derangements. Trends Endocrinol. Metab. 2013, 24, 229–237. [Google Scholar] [CrossRef]
- El Cheikh, R.; Bernard, S.; El Khatib, N. Modeling circadian clock–cell cycle interaction effects on cell population growth rates. J. Theor. Biol. 2014, 363, 318–331. [Google Scholar] [CrossRef]
- Lu, C.; Yang, Y.; Zhao, R.; Hua, B.; Bingxuan, H.; Xu, C.; Yan, Z.; Sun, N.; Qian, R. Role of circadian gene Clock during differentiation of mouse pluripotent stem cells. Protein Cell 2016, 7, 820–832. [Google Scholar] [CrossRef]
- Qiu, J.-F.; Li, X.; Cui, W.-Z.; Liu, X.-F.; Tao, H.; Yang, K.; Dai, T.-M.; Sima, Y.-H.; Xu, S.-Q. Inhibition of Period Gene Expression Causes Repression of Cell Cycle Progression and Cell Growth in the Bombyx mori Cells. Front. Physiol. 2019, 10, 537. [Google Scholar] [CrossRef] [PubMed]
- Crosby, P.; Partch, C.L. New insights into non-transcriptional regulation of mammalian core clock proteins. J. Cell Sci. 2020, 133, jcs241174. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-H.; Kim, S.-H.; Kim, H.-J.; Kim, W.; Lee, H.-R.; Jung, Y.; Choi, J.-H.; Hong, K.Y.; Jang, S.K.; Kim, K.-T. AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation. Nucleic Acids Res. 2014, 42, 3590–3606. [Google Scholar] [CrossRef]
- Lim, I.; Jung, Y.; Kim, D.-Y.; Kim, K.-T. HnRNP Q Has a Suppressive Role in the Translation of Mouse Cryptochrome1. PLoS ONE 2016, 11, e0159018. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Daimon, T.; Sezutsu, H.; Udaka, H.; Numata, H. Involvement of the Clock Gene period in the Circadian Rhythm of the Silkmoth Bombyx mori. J. Biol. Rhythm. 2019, 34, 283–292. [Google Scholar] [CrossRef]
- Cui, W.-Z.; Qiu, J.-F.; Dai, T.-M.; Chen, Z.; Li, J.-L.; Liu, K.; Wang, Y.-J.; Sima, Y.-H.; Xu, S.-Q. Circadian Clock Gene Period Contributes to Diapause via GABAeric-Diapause Hormone Pathway in Bombyx mori. Biology 2021, 10, 842. [Google Scholar] [CrossRef]
- Nartey, M.A.; Sun, X.; Qin, S.; Hou, C.X.; Li, M.W. CRISPR/Cas9-based knockout reveals that the clock gene timeless is indispensable for regulating circadian behavioral rhythms in Bombyx mori. Insect Sci. 2021, 28, 1414–1425. [Google Scholar] [CrossRef]
- Qiu, J.-F.; Cui, W.-Z.; Zhang, Q.; Dai, T.-M.; Liu, K.; Li, J.-L.; Wang, Y.-J.; Sima, Y.-H.; Xu, S.-Q. Temporal transcriptome reveals that circadian clock is involved in the dynamic regulation of immune response to bacterial infection in Bombyx mori. Insect Sci. 2023, 30, 31–46. [Google Scholar] [CrossRef]
- Qiu, J.-F.; Dai, T.-M.; Luo, C.; Cui, W.-Z.; Liu, K.; Li, J.-L.; Sima, Y.-H.; Xu, S.-Q. Circadian clock regulates developmental time through ecdysone and juvenile hormones in Bombyx mori. Insect Mol. Biol. 2023. [Google Scholar] [CrossRef]
- Zhang, Y.; Markert, M.J.; Groves, S.C.; Hardin, P.E.; Merlin, C. Vertebrate-like CRYPTOCHROME 2 from monarch regulates circadian transcription via independent repression of CLOCK and BMAL1 activity. Proc. Natl. Acad. Sci. USA 2017, 114, E7516–E7525. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M. Circadian Rhythms, Metabolism, and Insulin Sensitivity: Transcriptional Networks in Animal Models. Curr. Diabetes Rep. 2013, 13, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Lazar, M.A. Interconnections between circadian clocks and metabolism. J. Clin. Investig. 2021, 131, e148278. [Google Scholar] [CrossRef]
- Zhang, E.E.; Liu, Y.; Dentin, R.; Pongsawakul, P.Y.; Liu, A.C.; Hirota, T.; Nusinow, D.A.; Sun, X.; Landais, S.; Kodama, Y.; et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluco-neogenesis. Nat. Med. 2010, 16, 1152–1156. [Google Scholar] [CrossRef]
- Lamia, K.A.; Papp, S.J.; Yu, R.T.; Barish, G.D.; Uhlenhaut, N.H.; Jonker, J.W.; Downes, M.; Evans, R.M. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 2011, 480, 552–556. [Google Scholar] [CrossRef]
- Okano, S.; Akashi, M.; Hayasaka, K.; Nakajima, O. Unusual circadian locomotor activity and pathophysiology in mutant CRY1 transgenic mice. Neurosci. Lett. 2009, 451, 246–251. [Google Scholar] [CrossRef]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef]
- Doi, M.; Takahashi, Y.; Komatsu, R.; Yamazaki, F.; Yamada, H.; Haraguchi, S.; Emoto, N.; Okuno, Y.; Tsujimoto, G.; Kanematsu, A.; et al. Salt-sensitive hypertension in circadian clock–deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat. Med. 2010, 16, 67–74. [Google Scholar] [CrossRef]
- Cal-Kayitmazbatir, S.; Kulkoyluoglu-Cotul, E.; Growe, J.; Selby, C.P.; Rhoades, S.D.; Malik, D.; Oner, H.; Asimgil, H.; Francey, L.J.; Sancar, A.; et al. CRY1-CBS binding regulates circadian clock function and metabolism. FEBS J. 2021, 288, 614–639. [Google Scholar] [CrossRef]
- Peek, C.B.; Levine, D.C.; Cedernaes, J.; Taguchi, A.; Kobayashi, Y.; Tsai, S.J.; Bonar, N.A.; McNulty, M.R.; Ramsey, K.M.; Bass, J. Circadian Clock Interaction with HIF1α Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab. 2017, 25, 86–92. [Google Scholar] [CrossRef]
- Lamia, K.A.; Sachdeva, U.M.; Di Tacchio, L.; Williams, E.C.; Alvarez, J.G.; Egan, D.F.; Vasquez, D.S.; Juguilon, H.; Panda, S.; Shaw, R.J.; et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 2009, 326, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Onuma, S.; Kinoshita, S.; Shimba, S.; Ozono, K.; Michigami, T.; Kawai, M. The Lack of Bmal1, a Core Clock Gene, in the Intestine Decreases Glucose Absorption in Mice. Endocrinology 2022, 163, bqac119. [Google Scholar] [CrossRef]
- Barca-Mayo, O.; López, M. Astrocyte Clocks and Glucose Homeostasis. Front. Endocrinol. 2021, 12, 662017. [Google Scholar] [CrossRef] [PubMed]
- Kohsaka, A.; Das, P.; Hashimoto, I.; Nakao, T.; Deguchi, Y.; Gouraud, S.S.; Waki, H.; Muragaki, Y.; Maeda, M. The Circadian Clock Maintains Cardiac Function by Regulating Mitochondrial Metabolism in Mice. PLoS ONE 2014, 9, e112811. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhou, Z.; Chen, Y.; Wu, Y.; Liu, Y. RBP4 functions as a hepatokine in the regulation of glucose metabolism by the circadian clock in mice. Diabetologia 2016, 59, 354–362. [Google Scholar] [CrossRef]
- Neufeld-Cohen, A.; Robles, M.S.; Aviram, R.; Manella, G.; Adamovich, Y.; Ladeuix, B.; Nir, D.; Rousso-Noori, L.; Kuperman, Y.; Golik, M.; et al. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by PERIOD proteins. Proc. Natl. Acad. Sci. USA 2016, 113, E1673–E1682. [Google Scholar] [CrossRef] [PubMed]
- Farshadi, E.; van der Horst, G.T.J.; Chaves, I. Molecular Links between the Circadian Clock and the Cell Cycle. J. Mol. Biol. 2020, 432, 3515–3524. [Google Scholar] [CrossRef]
- Qu, M.; Zhang, G.; Qu, H.; Vu, A.; Wu, R.; Tsukamoto, H.; Jia, Z.; Huang, W.; Lenz, H.J.; Rich, J.N.; et al. Circadian regulator BMAL1::CLOCK promotes cell proliferation in hepatocellular carcinoma by controlling apoptosis and cell cycle. Proc. Natl. Acad. Sci. USA 2023, 120, e2214829120. [Google Scholar] [CrossRef]
- Wang, Z.; Su, G.; Dai, Z.; Meng, M.; Zhang, H.; Fan, F.; Liu, Z.; Zhang, L.; Weygant, N.; He, F.; et al. Circadian clock genes promote glioma progression by affecting tumour immune infiltration and tumour cell proliferation. Cell Prolif. 2021, 54, e12988. [Google Scholar] [CrossRef]
- Miller, B.H.; McDearmon, E.L.; Panda, S.; Hayes, K.R.; Zhang, J.; Andrews, J.L.; Antoch, M.P.; Walker, J.R.; Esser, K.A.; Hogenesch, J.B.; et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. USA 2007, 104, 3342–3347. [Google Scholar] [CrossRef]
- Stokes, K.; Nunes, M.; Trombley, C.; Flôres, D.E.F.L.; Wu, G.; Taleb, Z.; Alkhateeb, A.; Banskota, S.; Harris, C.; Love, O.P.; et al. The Circadian Clock Gene, Bmal1, Regulates Intestinal Stem Cell Signaling and Represses Tumor Initiation. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1847–1872.e0. [Google Scholar] [CrossRef]
- Fu, L.; Pelicano, H.; Liu, J.; Huang, P.; Lee, C.C. The circadian gene period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 2002, 111, 41–50. [Google Scholar] [CrossRef]
- Yang, W.S.; Stockwell, B.R. Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. 2008, 9, R92. [Google Scholar] [CrossRef]
- Rosbash, M.; Takahashi, J.S. Circadian rhythms: The cancer connection. Nature 2002, 420, 373–374. [Google Scholar] [CrossRef]
- Shostak, A.; Ruppert, B.; Ha, N.; Bruns, P.; Toprak, U.H.; Lawerenz, C.; Lichter, P.; Radlwimmer, B.; Eils, J.; Brors, B.; et al. MYC/MIZ1-dependent gene repression inversely coordinates the circadian clock with cell cycle and proliferation. Nat. Commun. 2016, 7, 11807. [Google Scholar] [CrossRef]
- Neufeld, T.P.; de la Cruz, A.F.A.; Johnston, L.A.; Edgar, B.A. Coordination of Growth and Cell Division in the Drosophila Wing. Cell 1998, 93, 1183–1193. [Google Scholar] [CrossRef]
- Harris, W.A.; Hartenstein, V. Neuronal determination without cell division in xenopus embryos. Neuron 1991, 6, 499–515. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011, 13, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Lin, L.; Lin, H.; Chen, W.; Chen, L.; Chen, X.; Chen, S.; Lin, Q.; Xu, Y.; Zeng, Y. KCNK3 inhibits proliferation and glucose metabolism of lung adenocarcinoma via activation of AMPK-TXNIP pathway. Cell Death Discov. 2022, 8, 360. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.L.; Fan, X.G.; Wu, J.; Wang, Y.; Hu, X.W.; Pei, S.Y.; Zheng, Y.X.; Chen, J.; Huang, Y.; Li, N.; et al. CB2R agonist GW405833 alleviates acute liver failure in mice via inhibiting HIF-1α-mediated reprogramming of glycometabolism and macrophage proliferation. Acta Pharmacol. Sin 2023. [Google Scholar] [CrossRef]
- Zhang, Y.; Mou, Y.; Liang, C.; Zhu, S.; Liu, S.; Shao, P.; Li, J.; Wang, Z. Promoting cell proliferation, cell cycle progression, and glycolysis: Glycometabolism-related genes act as prognostic signatures for prostate cancer. Prostate 2021, 81, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, R.; Tsuchiya, Y.; Tokuda, I.; Matsuo, T.; Sato, M.; Node, K.; Nishida, E.; Akashi, M. The Mammalian Circadian Clock Protein Period Counteracts Cryptochrome in Phosphorylation Dynamics of Circadian Locomotor Output Cycles Kaput (CLOCK). J. Biol. Chem. 2014, 289, 32064–32072. [Google Scholar] [CrossRef] [PubMed]
- Mahesh, G.; Jeong, E.; Ng, F.S.; Liu, Y.; Gunawardhana, K.; Houl, J.H.; Yildirim, E.; Amunugama, R.; Jones, R.; Allen, D.L.; et al. Phosphorylation of the Transcription Activator CLOCK Regulates Progression through a ~24-h Feedback Loop to Influence the Circadian Period in Drosophila. J. Biol. Chem. 2014, 289, 19681–19693. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.; Selby, C.P.; Chiou, Y.-Y.; Ozkan-Dagliyan, I.; Gaddameedhi, S.; Sancar, A. Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev. 2014, 28, 1989–1998. [Google Scholar] [CrossRef] [PubMed]
- Bae, K.; Jin, X.; Maywood, E.S.; Hastings, M.H.; Reppert, S.M.; Weaver, D.R. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 2001, 30, 525–536. [Google Scholar] [CrossRef]
- Zheng, B.; Albrecht, U.; Kaasik, K.; Sage, M.; Lu, W.; Vaishnav, S.; Li, Q.; Sun, Z.S.; Eichele, G.; Bradley, A.; et al. Non-redundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 2001, 105, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Kaasik, K.; Lee, C.C. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 2004, 430, 467–471. [Google Scholar] [CrossRef]
- Akashi, M.; Okamoto, A.; Tsuchiya, Y.; Todo, T.; Nishida, E.; Node, K. A Positive Role for PERIOD in Mammalian Circadian Gene Expression. Cell Rep. 2014, 7, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.; All, S.; Skopis, G.; Cheng, K.-Y.; Compton, B.; Srialluri, N.; Stow, L.; Jeffers, L.A.; Gumz, M.L. Opposing actions of Per1 and Cry2 in the regulation of Per1 target gene expression in the liver and kidney. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 305, R735–R747. [Google Scholar] [CrossRef] [PubMed]
- Sellick, C.A.; Hansen, R.; Stephens, G.M.; Goodacre, R.; Dickson, A.J. Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nat. Protoc. 2011, 6, 1241–1249. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, J.; Dai, T.; Tao, H.; Li, X.; Luo, C.; Sima, Y.; Xu, S. Inhibition of Expression of the Circadian Clock Gene Cryptochrome 1 Causes Abnormal Glucometabolic and Cell Growth in Bombyx mori Cells. Int. J. Mol. Sci. 2023, 24, 5435. https://doi.org/10.3390/ijms24065435
Qiu J, Dai T, Tao H, Li X, Luo C, Sima Y, Xu S. Inhibition of Expression of the Circadian Clock Gene Cryptochrome 1 Causes Abnormal Glucometabolic and Cell Growth in Bombyx mori Cells. International Journal of Molecular Sciences. 2023; 24(6):5435. https://doi.org/10.3390/ijms24065435
Chicago/Turabian StyleQiu, Jianfeng, Taiming Dai, Hui Tao, Xue Li, Cheng Luo, Yanghu Sima, and Shiqing Xu. 2023. "Inhibition of Expression of the Circadian Clock Gene Cryptochrome 1 Causes Abnormal Glucometabolic and Cell Growth in Bombyx mori Cells" International Journal of Molecular Sciences 24, no. 6: 5435. https://doi.org/10.3390/ijms24065435
APA StyleQiu, J., Dai, T., Tao, H., Li, X., Luo, C., Sima, Y., & Xu, S. (2023). Inhibition of Expression of the Circadian Clock Gene Cryptochrome 1 Causes Abnormal Glucometabolic and Cell Growth in Bombyx mori Cells. International Journal of Molecular Sciences, 24(6), 5435. https://doi.org/10.3390/ijms24065435





