Denervation Drives YAP/TAZ Activation in Muscular Fibro/Adipogenic Progenitors
Abstract
:1. Introduction
2. Results
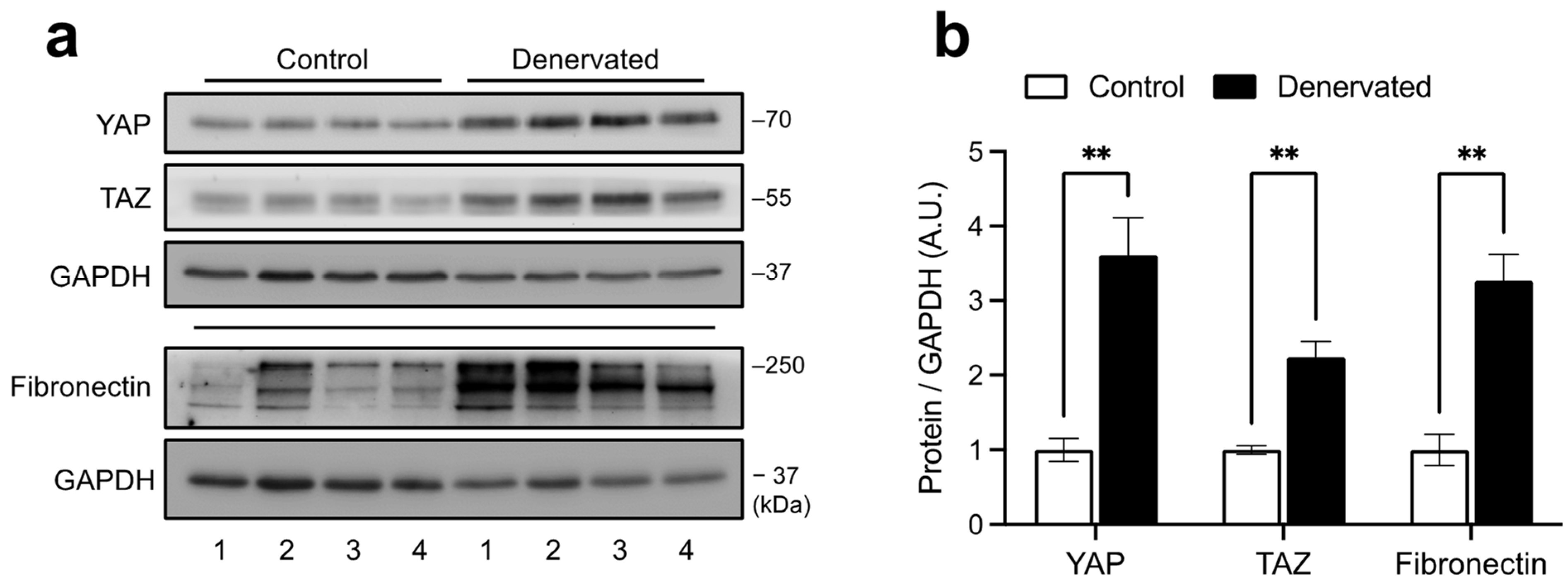
2.1. YAP and TAZ Are Increased in Denervated Skeletal Muscles
2.2. YAP/TAZ Activity Is Augmented in Denervated Skeletal Muscle
2.3. Expanded FAPs Accumulate YAP upon Denervation
2.4. Single-Cell Transcriptomics of Denervated Muscles Reveal Increased YAP/TAZ Activity in FAPs
3. Discussion
4. Materials and Methods
4.1. Animal Experiments
4.2. Protein Extraction and Western Blot
4.3. RNA Isolation and RT-qPCR
4.4. Immunofluorescence
4.5. Transcriptomics Analyses
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rebolledo, D.L.; Lipson, K.E.; Brandan, E. Driving fibrosis in neuromuscular diseases: Role and regulation of Connective tissue growth factor (CCN2/CTGF). Matrix Biol. Plus 2021, 11, 100059. [Google Scholar] [CrossRef] [PubMed]
- Sheets, K.; Overbey, J.; Ksajikian, A.; Bovid, K.; Kenter, K.; Li, Y. The pathophysiology and treatment of musculoskeletal fibrosis. J. Cell. Biochem. 2022, 123, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.; Shah, S.; Lovering, R. The Neuromuscular Junction: Roles in Aging and Neuromuscular Disease. Int. J. Mol. Sci. 2021, 22, 8058. [Google Scholar] [CrossRef] [PubMed]
- Accornero, F.; Kanisicak, O.; Tjondrokoesoemo, A.; Attia, A.C.; McNally, E.M.; Molkentin, J.D. Myofiber-specific inhibition of TGFβ signaling protects skeletal muscle from injury and dystrophic disease in mice. Hum. Mol. Genet. 2014, 23, 6903–6915. [Google Scholar] [CrossRef] [Green Version]
- Cohn, R.D.; van Erp, C.; Habashi, J.P.; Soleimani, A.; Klein, E.C.; Lisi, M.T.; Gamradt, M.; ap Rhys, C.M.; Holm, T.M.; Loeys, B.L.; et al. Angiotensin II type 1 receptor blockade attenuates TGF-β–induced failure of muscle regeneration in multiple myopathic states. Nat. Med. 2007, 13, 204–210. [Google Scholar] [CrossRef] [Green Version]
- Córdova-Casanova, A.; Cruz-Soca, M.; Chun, J.; Casar, J.C.; Brandan, E. Activation of the ATX/LPA/LPARs axis induces a fibrotic response in skeletal muscle. Matrix Biol. 2022, 109, 121–139. [Google Scholar] [CrossRef]
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From mechanisms to medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef]
- Contreras, O.; Rebolledo, D.L.; Oyarzún, J.E.; Olguín, H.C.; Brandan, E. Connective tissue cells expressing fibro/adipogenic progenitor markers increase under chronic damage: Relevance in fibroblast-myofibroblast differentiation and skeletal muscle fibrosis. Cell Tissue Res. 2016, 364, 647–660. [Google Scholar] [CrossRef]
- Contreras, O.; Soliman, H.; Theret, M.; Rossi, F.M.V.; Brandan, E. TGF-beta-driven downregulation of the tran-scription factor TCF7L2 affects Wnt/beta-catenin signaling in PDGFRalpha(+) fibroblasts. J. Cell Sci. 2020, 133, jcs242297. [Google Scholar] [CrossRef]
- Joe, A.W.; Yi, L.; Natarajan, A.; Le Grand, F.; So, L.; Wang, J.; Rudnicki, M.A.; Rossi, F. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010, 12, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Mathew, S.; Hansen, J.M.; Merrell, A.J.; Murphy, M.M.; Lawson, J.A.; Hutcheson, D.A.; Hansen, M.S.; Angus-Hill, M.; Kardon, G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 2011, 138, 371–384. [Google Scholar] [CrossRef] [Green Version]
- Murphy, M.M.; Lawson, J.A.; Mathew, S.J.; Hutcheson, D.A.; Kardon, G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 2011, 138, 3625–3637. [Google Scholar] [CrossRef] [Green Version]
- Uezumi, A.; Fukada, S.-I.; Yamamoto, N.; Takeda, S.; Tsuchida, K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010, 12, 143–152. [Google Scholar] [CrossRef]
- Uezumi, A.; Ito, T.; Morikawa, D.; Shimizu, N.; Yoneda, T.; Segawa, M.; Yamaguchi, M.; Ogawa, R.; Matev, M.M.; Miyagoe-Suzuki, Y.; et al. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J. Cell Sci. 2011, 124, 3654–3664. [Google Scholar] [CrossRef] [Green Version]
- González, D.; Contreras, O.; Rebolledo, D.L.; Espinoza, J.P.; van Zundert, B.; Brandan, E. ALS skeletal muscle shows enhanced TGF-β signaling, fibrosis and induction of fibro/adipogenic progenitor markers. PLoS ONE 2017, 12, e0177649. [Google Scholar] [CrossRef] [Green Version]
- Madaro, L.; Passafaro, M.; Sala, D.; Etxaniz, U.; Lugarini, F.; Proietti, D.; Alfonsi, M.V.; Nicoletti, C.; Gatto, S.; De Bardi, M.; et al. Denervation-activated STAT3–IL-6 signalling in fibro-adipogenic progenitors promotes myofibres atrophy and fibrosis. Nat. Cell Biol. 2018, 20, 917–927. [Google Scholar] [CrossRef]
- Rebolledo, D.L.; González, D.; Faundez-Contreras, J.; Contreras, O.; Vio, C.P.; Murphy-Ullrich, J.E.; Lipson, K.E.; Brandan, E. Denervation-induced skeletal muscle fibrosis is mediated by CTGF/CCN2 independently of TGF-β. Matrix Biol. 2019, 82, 20–37. [Google Scholar] [CrossRef]
- Chen, J.; Wang, X.; He, Q.; Bulus, N.; Fogo, A.B.; Zhang, M.-Z.; Harris, R.C. YAP Activation in Renal Proximal Tubule Cells Drives Diabetic Renal Interstitial Fibrogenesis. Diabetes 2020, 69, 2446–2457. [Google Scholar] [CrossRef]
- Liu, F.; Lagares, D.; Choi, K.M.; Stopfer, L.; Marinković, A.; Vrbanac, V.; Probst, C.K.; Hiemer, S.E.; Sisson, T.H.; Horowitz, J.C.; et al. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L344–L357. [Google Scholar] [CrossRef] [Green Version]
- Mannaerts, I.; Leite, S.B.; Verhulst, S.; Claerhout, S.; Eysackers, N.; Thoen, L.F.; Hoorens, A.; Reynaert, H.; Halder, G.; van Grunsven, L.A. The Hippo pathway effector YAP controls mouse hepatic stellate cell activation. J. Hepatol. 2015, 63, 679–688. [Google Scholar] [CrossRef]
- Szeto, S.G.; Narimatsu, M.; Lu, M.; He, X.; Sidiqi, A.M.; Tolosa, M.F.; Chan, L.; De Freitas, K.; Bialik, J.F.; Majumder, S.; et al. YAP/TAZ Are Mechanoregulators of TGF-β-Smad Signaling and Renal Fibrogenesis. J. Am. Soc. Nephrol. 2016, 27, 3117–3128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Totaro, A.; Panciera, T.; Piccolo, S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 2018, 20, 888–899. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, C.-Y.; Zha, Z.-Y.; Zhao, B.; Yao, J.; Zhao, S.; Xiong, Y.; Lei, Q.-Y.; Guan, K.-L. TEAD Transcription Factors Mediate the Function of TAZ in Cell Growth and Epithelial-Mesenchymal Transition. J. Biol. Chem. 2009, 284, 13355–13362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Yu, J.; Lin, J.D.; Wang, C.-Y.; Chinnaiyan, A.M.; et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piersma, B.; Bank, R.A.; Boersema, M. Signaling in Fibrosis: TGF-β, WNT, and YAP/TAZ Converge. Front. Med. 2015, 2, 59. [Google Scholar] [CrossRef]
- Labibi, B.; Bashkurov, M.; Wrana, J.L.; Attisano, L. Modeling the Control of TGF-β/Smad Nuclear Accumulation by the Hippo Pathway Effectors, Taz/Yap. Iscience 2020, 23, 101416. [Google Scholar] [CrossRef]
- Wang, Y.; Tu, K.; Liu, D.; Guo, L.; Chen, Y.; Li, Q.; Maiers, J.L.; Liu, Z.; Shah, V.H.; Dou, C.; et al. p300 Acetyltransferase Is a Cytoplasm-to-Nucleus Shuttle for SMAD2/3 and TAZ Nuclear Transport in Transforming Growth Factor β–Stimulated Hepatic Stellate Cells. Hepatology 2019, 70, 1409–1423. [Google Scholar] [CrossRef] [Green Version]
- Azzolin, L.; Panciera, T.; Soligo, S.; Enzo, E.; Bicciato, S.; Dupont, S.; Bresolin, S.; Frasson, C.; Basso, G.; Guzzardo, V.; et al. YAP/TAZ Incorporation in the β-Catenin Destruction Complex Orchestrates the Wnt Response. Cell 2014, 158, 157–170. [Google Scholar] [CrossRef] [Green Version]
- Azzolin, L.; Zanconato, F.; Bresolin, S.; Forcato, M.; Basso, G.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Role of TAZ as Mediator of Wnt Signaling. Cell 2012, 151, 1443–1456. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.-X.; Zhao, B.; Panupinthu, N.; Jewell, J.L.; Lian, I.; Wang, L.H.; Zhao, J.; Yuan, H.; Tumaneng, K.; Li, H.; et al. Regulation of the Hippo-YAP Pathway by G-Protein-Coupled Receptor Signaling. Cell 2012, 150, 780–791. [Google Scholar] [CrossRef] [Green Version]
- Panciera, T.; Azzolin, L.; Cordenonsi, M.; Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 758–770. [Google Scholar] [CrossRef]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef] [Green Version]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef]
- Scott, K.E.; Fraley, S.I.; Rangamani, P. A spatial model of YAP/TAZ signaling reveals how stiffness, dimensionality, and shape contribute to emergent outcomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2021571118. [Google Scholar] [CrossRef]
- Fischer, M.; Rikeit, P.; Knaus, P.; Coirault, C. YAP-Mediated Mechanotransduction in Skeletal Muscle. Front. Physiol. 2016, 7, 41. [Google Scholar] [CrossRef] [Green Version]
- Silver, J.S.; Günay, K.A.; Cutler, A.A.; Vogler, T.O.; Brown, T.E.; Pawlikowski, B.T.; Bednarski, O.J.; Bannister, K.L.; Rogowski, C.J.; Mckay, A.G.; et al. Injury-mediated stiffening persistently activates muscle stem cells through YAP and TAZ mechanotransduction. Sci. Adv. 2021, 7, eabe4501. [Google Scholar] [CrossRef]
- Watt, K.I.; Turner, B.J.; Hagg, A.; Zhang, X.; Davey, J.R.; Qian, H.; Beyer, C.; Winbanks, C.E.; Harvey, K.F.; Gregorevic, P. The Hippo pathway effector YAP is a critical regulator of skeletal muscle fibre size. Nat. Commun. 2015, 6, 6048. [Google Scholar] [CrossRef] [Green Version]
- Ehmsen, J.T.; Kawaguchi, R.; Mi, R.; Coppola, G.; Höke, A. Longitudinal RNA-Seq analysis of acute and chronic neurogenic skeletal muscle atrophy. Sci. Data 2019, 6, 179. [Google Scholar] [CrossRef] [Green Version]
- Zanconato, F.; Forcato, M.; Battilana, G.; Azzolin, L.; Quaranta, E.; Bodega, B.; Rosato, A.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015, 17, 1218–1227. [Google Scholar] [CrossRef]
- Contreras, O.; Cruz-Soca, M.; Theret, M.; Soliman, H.; Tung, L.W.; Groppa, E.; Rossi, F.M.; Brandan, E. Cross-talk between TGF-beta and PDGFRalpha signaling pathways regulates the fate of stromal fibro-adipogenic progenitors. J. Cell Sci. 2019, 132, jcs232157. [Google Scholar] [CrossRef]
- Hamilton, T.G.; Klinghoffer, R.A.; Corrin, P.D.; Soriano, P. Evolutionary Divergence of Platelet-Derived Growth Factor Alpha Receptor Signaling Mechanisms. Mol. Cell. Biol. 2003, 23, 4013–4025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Ma, X.; Sun, Y.; Peng, H.; Wang, Y.; Thomas, S.S.; Hu, Z. Decoding the transcriptome of denervated muscle at single-nucleus resolution. J. Cachex-Sarcopenia Muscle 2022, 13, 2102–2117. [Google Scholar] [CrossRef] [PubMed]
- Liu-Chittenden, Y.; Huang, B.; Shim, J.S.; Chen, Q.; Lee, S.-J.; Anders, R.A.; Liu, J.O.; Pan, D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012, 26, 1300–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitani, A.; Nagase, T.; Fukuchi, K.; Aburatani, H.; Makita, R.; Kurihara, H. Transcriptional Coactivator with PDZ-binding Motif Is Essential for Normal Alveolarization in Mice. Am. J. Respir. Crit. Care Med. 2009, 180, 326–338. [Google Scholar] [CrossRef]
- Judson, R.N.; Gray, S.R.; Walker, C.; Carroll, A.M.; Itzstein, C.; Lionikas, A.; Zammit, P.S.; De Bari, C.; Wackerhage, H. Constitutive Expression of Yes-Associated Protein (Yap) in Adult Skeletal Muscle Fibres Induces Muscle Atrophy and Myopathy. PLoS ONE 2013, 8, e59622. [Google Scholar] [CrossRef] [Green Version]
- Vita, G.L.; Polito, F.; Oteri, R.; Arrigo, R.; Ciranni, A.M.; Musumeci, O.; Messina, S.; Rodolico, C.; Di Giorgio, R.M.; Vita, G.; et al. Hippo signaling pathway is altered in Duchenne muscular dystrophy. PLoS ONE 2018, 13, e0205514. [Google Scholar] [CrossRef] [Green Version]
- Liang, M.; Yu, M.; Xia, R.; Song, K.; Wang, J.; Luo, J.; Chen, G.; Cheng, J. Yap/Taz Deletion in Gli+ Cell-Derived Myofibroblasts Attenuates Fibrosis. J. Am. Soc. Nephrol. 2017, 28, 3278–3290. [Google Scholar] [CrossRef] [Green Version]
- Piersma, B.; de Rond, S.; Werker, P.M.; Boo, S.; Hinz, B.; van Beuge, M.M.; Bank, R.A. YAP1 Is a Driver of Myofibroblast Differentiation in Normal and Diseased Fibroblasts. Am. J. Pathol. 2015, 185, 3326–3337. [Google Scholar] [CrossRef] [Green Version]
- Shi-Wen, X.; Racanelli, M.; Ali, A.; Simon, A.; Quesnel, K.; Stratton, R.J.; Leask, A. Verteporfin inhibits the persistent fibrotic phenotype of lesional scleroderma dermal fibroblasts. J. Cell Commun. Signal. 2021, 15, 71–80. [Google Scholar] [CrossRef]
- Morales, M.G.; Cabello-Verrugio, C.; Santander, C.; Cabrera, D.; Goldschmeding, R.; Brandan, E. CTGF/CCN-2 over-expression can directly induce features of skeletal muscle dystrophy. J. Pathol. 2011, 225, 490–501. [Google Scholar] [CrossRef]
- Morales, M.G.; Gutierrez, J.; Cabello-Verrugio, C.; Cabrera, D.; Lipson, K.; Goldschmeding, R.; Brandan, E. Reducing CTGF/CCN2 slows down mdx muscle dystrophy and improves cell therapy. Hum. Mol. Genet. 2013, 22, 4938–4951. [Google Scholar] [CrossRef] [Green Version]
- Dey, A.; Varelas, X.; Guan, K.-L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 2020, 19, 480–494. [Google Scholar] [CrossRef]
- Engler, A.; Griffin, M.A.; Sen, S.; Bonnemann, C.G.; Sweeney, H.L.; Discher, D.E. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J. Cell Biol. 2004, 166, 877–887. [Google Scholar] [CrossRef] [Green Version]
- Pavan, P.; Monti, E.; Bondí, M.; Fan, C.; Stecco, C.; Narici, M.; Reggiani, C.; Marcucci, L. Alterations of Extracellular Matrix Mechanical Properties Contribute to Age-Related Functional Impairment of Human Skeletal Muscles. Int. J. Mol. Sci. 2020, 21, 3992. [Google Scholar] [CrossRef]
- Wood, L.K.; Kayupov, E.; Gumucio, J.P.; Mendias, C.; Claflin, D.; Brooks, S.V. Intrinsic stiffness of extracellular matrix increases with age in skeletal muscles of mice. J. Appl. Physiol. 2014, 117, 363–369. [Google Scholar] [CrossRef] [Green Version]
- Loomis, T.; Hu, L.-Y.; Wohlgemuth, R.P.; Chellakudam, R.R.; Muralidharan, P.D.; Smith, L.R. Matrix stiffness and architecture drive fibro-adipogenic progenitors’ activation into myofibroblasts. Sci. Rep. 2022, 12, 13582. [Google Scholar] [CrossRef]
- Kurimoto, S.; Jung, J.; Tapadia, M.; Lengfeld, J.; Agalliu, D.; Waterman, M.; Mozaffar, T.; Gupta, R. Activation of the Wnt/β-catenin signaling cascade after traumatic nerve injury. Neuroscience 2015, 294, 101–108. [Google Scholar] [CrossRef] [Green Version]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallardo, F.S.; Córdova-Casanova, A.; Bock-Pereda, A.; Rebolledo, D.L.; Ravasio, A.; Casar, J.C.; Brandan, E. Denervation Drives YAP/TAZ Activation in Muscular Fibro/Adipogenic Progenitors. Int. J. Mol. Sci. 2023, 24, 5585. https://doi.org/10.3390/ijms24065585
Gallardo FS, Córdova-Casanova A, Bock-Pereda A, Rebolledo DL, Ravasio A, Casar JC, Brandan E. Denervation Drives YAP/TAZ Activation in Muscular Fibro/Adipogenic Progenitors. International Journal of Molecular Sciences. 2023; 24(6):5585. https://doi.org/10.3390/ijms24065585
Chicago/Turabian StyleGallardo, Felipe S., Adriana Córdova-Casanova, Alexia Bock-Pereda, Daniela L. Rebolledo, Andrea Ravasio, Juan Carlos Casar, and Enrique Brandan. 2023. "Denervation Drives YAP/TAZ Activation in Muscular Fibro/Adipogenic Progenitors" International Journal of Molecular Sciences 24, no. 6: 5585. https://doi.org/10.3390/ijms24065585




