Harnessing Fc/FcRn Affinity Data from Patents with Different Machine Learning Methods
Abstract
:1. Introduction
2. Results
2.1. Description of the Fc Variant Dataset and Creation of the Learning Sets
2.2. Algorithms and Tested Features
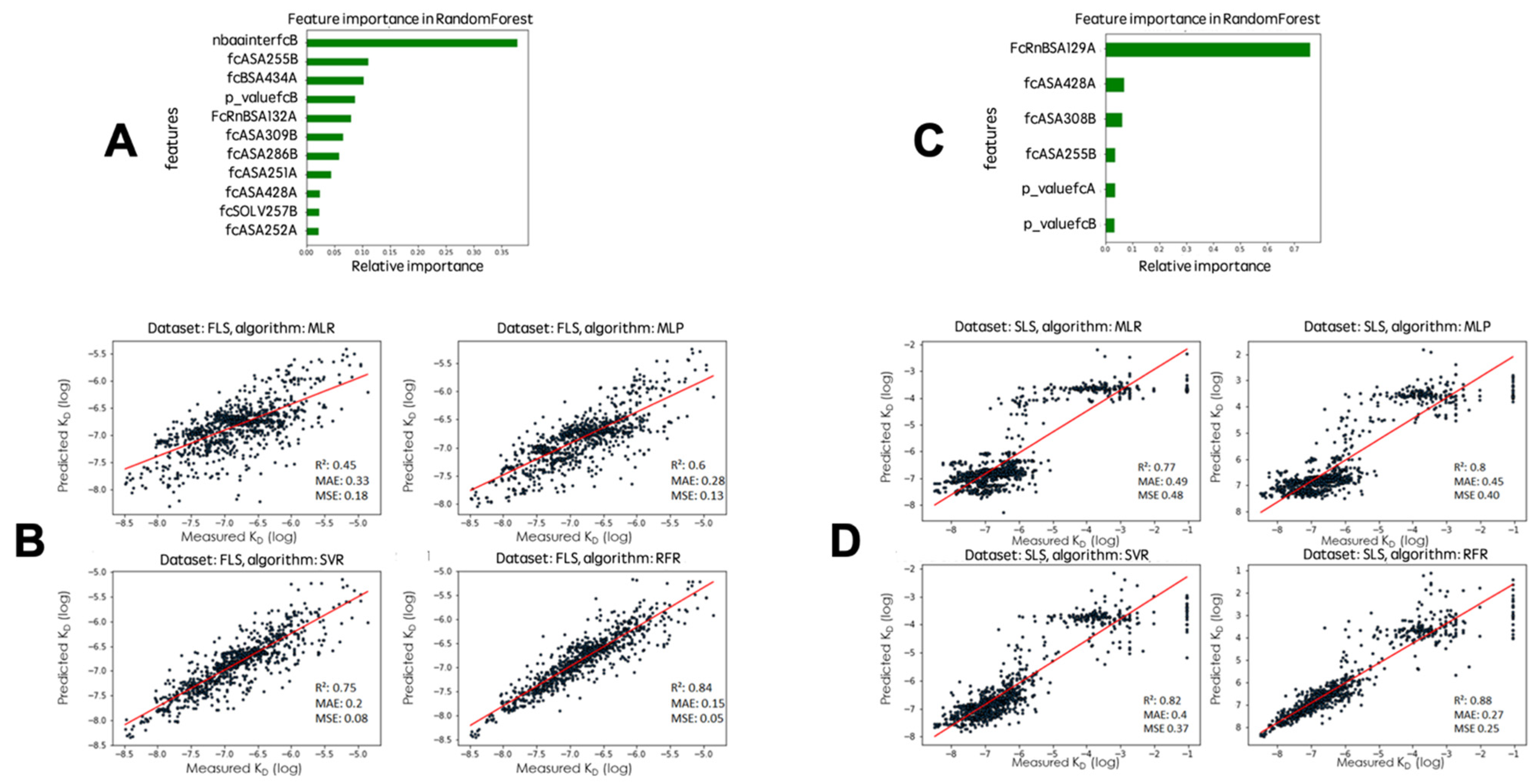
2.3. Randomly Generated Variants Predicted Affinity Comparison with the Four Algorithms
2.4. Experimental Validation
3. Discussion
3.1. Model FLS
3.2. Model SLS
3.3. Further Improvements
4. Materials and Methods
4.1. Antibody Expression and Purification
4.2. Surface Plasmon Resonance
4.3. Structure-Based Feature Extraction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| No. | Feature Name | Meaning |
|---|---|---|
| 1 | FcSOLV251A | Solvation effect of the 251 residue in Å2 |
| 2 | FcSOLV252A | Solvation effect of the 252 residue in Å2 |
| 3 | FcSOLV253A | Solvation effect of the 253 residue in Å2 |
| 4 | FcSOLV254A | Solvation effect of the 254 residue in Å2 |
| 5 | FcSOLV309A | Solvation effect of the 309 residue in Å2 |
| 6 | FcSOLV310A | Solvation effect of the 310 residue in Å2 |
| 7 | FcSOLV311A | Solvation effect of the 311 residue in Å2 |
| 8 | FcSOLV314A | Solvation effect of the 314 residue in Å2 |
| 9 | FcSOLV428A | Solvation effect of the 428 residue in Å2 |
| 10 | FcSOLV433A | Solvation effect of the 433 residue in Å2 |
| 11 | FcSOLV434A | Solvation effect of the 434 residue in Å2 |
| 12 | FcSOLV435A | Solvation effect of the 435 residue in Å2 |
| 13 | FcSOLV436A | Solvation effect of the 436 residue in Å2 |
| 14 | FcSOLV253B | Solvation effect of the 253 residue in Å2 |
| 15 | FcSOLV255B | Solvation effect of the 255 residue in Å2 |
| 16 | FcSOLV256B | Solvation effect of the 256 residue in Å2 |
| 17 | FcSOLV257B | Solvation effect of the 257 residue in Å2 |
| 18 | FcSOLV285B | Solvation effect of the 285 residue in Å2 |
| 19 | FcSOLV286B | Solvation effect of the 286 residue in Å2 |
| 20 | FcSOLV288B | Solvation effect of the 288 residue in Å2 |
| 21 | FcSOLV307B | Solvation effect of the 307 residue in Å2 |
| 22 | FcSOLV308 | Solvation effect of the 308 residue in Å2 |
| 23 | FcSOLV309B | Solvation effect of the 309 residue in Å2 |
| 24 | FcSOLV310B | Solvation effect of the 310 residue in Å2 |
| 25 | FcBSA251A | Buried surface of the 251 residue in Å2 |
| 26 | FcBSA252A | Buried surface of the 252 residue in Å2 |
| 27 | FcBSA253A | Buried surface of the 253 residue in Å2 |
| 28 | FcBSA254A | Buried surface of the 254 residue in Å2 |
| 29 | FcBSA309A | Buried surface of the 309 residue in Å2 |
| 30 | FcBSA310A | Buried surface of the 310 residue in Å2 |
| 31 | FcBSA311A | Buried surface of the 311 residue in Å2 |
| 32 | FcBSA314A | Buried surface of the 314 residue in Å2 |
| 33 | FcBSA428A | Buried surface of the 428 residue in Å2 |
| 34 | FcBSA433A | Buried surface of the 433 residue in Å2 |
| 35 | FcBSA434A | Buried surface of the 434 residue in Å2 |
| 36 | FcBSA435A | Buried surface of the 435 residue in Å2 |
| 37 | FcBSA436A | Buried surface of the 436 residue in Å2 |
| 38 | FcBSA253B | Buried surface of the 253 residue in Å2 |
| 39 | FcBSA255B | Buried surface of the 255 residue in Å2 |
| 40 | FcBSA256B | Buried surface of the 256 residue in Å2 |
| 41 | FcBSA257B | Buried surface of the 257 residue in Å2 |
| 42 | FcBSA285B | Buried surface of the 285 residue in Å2 |
| 43 | FcBSA286B | Buried surface of the 286 residue in Å2 |
| 44 | FcBSA288B | Buried surface of the 288 residue in Å2 |
| 45 | FcBSA307B | Buried surface of the 307 residue in Å2 |
| 46 | FcBSA308 | Buried surface of the 308 residue in Å2 |
| 47 | FcBSA309B | Buried surface of the 309 residue in Å2 |
| 48 | FcBSA310B | Buried surface of the 310 residue in Å2 |
| 49 | FcASA251A | Surface accessible to the solvent of the 251 residue in Å2 |
| 50 | FcASA252A | Surface accessible to the solvent of the 252 residue in Å2 |
| 51 | FcASA253A | Surface accessible to the solvent of the 253 residue in Å2 |
| 52 | FcASA254A | Surface accessible to the solvent of the 254 residue in Å2 |
| 53 | FcASA309A | Surface accessible to the solvent of the 309 residue in Å2 |
| 54 | FcASA310A | Surface accessible to the solvent of the 310 residue in Å2 |
| 55 | FcASA311A | Surface accessible to the solvent of the 311 residue in Å2 |
| 56 | FcASA314A | Surface accessible to the solvent of the 314 residue in Å2 |
| 57 | FcASA428A | Surface accessible to the solvent of the 428 residue in Å2 |
| 58 | FcASA433A | Surface accessible to the solvent of the 433 residue in Å2 |
| 59 | FcASA434A | Surface accessible to the solvent of the 434 residue in Å2 |
| 60 | FcASA435A | Surface accessible to the solvent of the 435 residue in Å2 |
| 61 | FcASA436A | Surface accessible to the solvent of the 436 residue in Å2 |
| 62 | FcASA253B | Surface accessible to the solvent of the 253 residue in Å2 |
| 63 | FcASA255B | Surface accessible to the solvent of the 255 residue in Å2 |
| 64 | FcASA256B | Surface accessible to the solvent of the 256 residue in Å2 |
| 65 | FcASA257B | Surface accessible to the solvent of the 257 residue in Å2 |
| 66 | FcASA285B | Surface accessible to the solvent of the 285 residue in Å2 |
| 67 | FcASA286B | Surface accessible to the solvent of the 286 residue in Å2 |
| 68 | FcASA288B | Surface accessible to the solvent of the 288 residue in Å2 |
| 69 | FcASA307B | Surface accessible to the solvent of the 307 residue in Å2 |
| 70 | FcASA308B | Surface accessible to the solvent of the 308 residue in Å2 |
| 71 | FcASA309B | Surface accessible to the solvent of the 309 residue in Å2 |
| 72 | FcASA310B | Surface accessible to the solvent of the 310 residue in Å2 |
| 73 | FcRnSOLV88A | Solvation effect of the 88 |
| 74 | FcRnSOLV112A | Solvation effect of the 112 |
| 75 | FcRnSOLV113A | Solvation effect of the 113 |
| 76 | FcRnSOLV114A | Solvation effect of the 114 |
| 77 | FcRnSOLV115A | Solvation effect of the 115 |
| 78 | FcRnSOLV116A | Solvation effect of the 116 |
| 79 | FcRnSOLV128A | Solvation effect of the 128 |
| 80 | FcRnSOLV129A | Solvation effect of the 129 |
| 81 | FcRnSOLV130A | Solvation effect of the 130 |
| 82 | FcRnSOLV131A | Solvation effect of the 131 |
| 83 | FcRnSOLV132A | Solvation effect of the 132 |
| 84 | FcRnSOLV133A | Solvation effect of the 133 |
| 85 | FcRnSOLV135A | Solvation effect of the 135 |
| 86 | FcRnSOLV1B | Solvation effect of the 1 |
| 87 | FcRnSOLV2B | Solvation effect of the 2 |
| 88 | FcRnSOLV3B | Solvation effect of the 3 |
| 89 | FcRnSOLV4B | Solvation effect of the 4 |
| 90 | FcRnSOLV85B | Solvation effect of the 85 |
| 91 | FcRnSOLV86B | Solvation effect of the 86 |
| 92 | FcRnBSA88A | Buried surface of the 88 |
| 93 | FcRnBSA112A | Buried surface of the 112 |
| 94 | FcRnBSA113A | Buried surface of the 113 |
| 95 | FcRnBSA114A | Buried surface of the 114 |
| 96 | FcRnBSA115A | Buried surface of the 115 |
| 97 | FcRnBSA116A | Buried surface of the 116 |
| 98 | FcRnBSA128A | Buried surface of the 128 |
| 99 | FcRnBSA129A | Buried surface of the 129 |
| 100 | FcRnBSA130A | Buried surface of the 130 |
| 101 | FcRnBSA131A | Buried surface of the 131 |
| 102 | FcRnBSA132A | Buried surface of the 132 |
| 103 | FcRnBSA133A | Buried surface of the 133 |
| 104 | FcRnBSA135A | Buried surface of the 135 |
| 105 | FcRnBSA1B | Buried surface of the 1 |
| 106 | FcRnBSA2B | Buried surface of the 2 |
| 107 | FcRnBSA3B | Buried surface of the 3 |
| 108 | FcRnBSA4B | Buried surface of the 4 |
| 109 | FcRnBSA85B | Buried surface of the 85 |
| 110 | FcRnBSA86B | Buried surface of the 86 |
| 111 | FcRnASA88A | Surface accessible to the solvent of the 88 |
| 112 | FcRnASA112A | Surface accessible to the solvent of the 112 |
| 113 | FcRnASA113A | Surface accessible to the solvent of the 113 |
| 114 | FcRnASA114A | Surface accessible to the solvent of the 114 |
| 115 | FcRnASA115A | Surface accessible to the solvent of the 115 |
| 116 | FcRnASA116A | Surface accessible to the solvent of the 116 |
| 117 | FcRnASA128A | Surface accessible to the solvent of the 128 |
| 118 | FcRnASA129A | Surface accessible to the solvent of the 129 |
| 119 | FcRnASA130A | Surface accessible to the solvent of the 130 |
| 120 | FcRnASA131A | Surface accessible to the solvent of the 131 |
| 121 | FcRnASA132A | Surface accessible to the solvent of the 132 |
| 122 | FcRnASA133A | Surface accessible to the solvent of the 133 |
| 123 | FcRnASA135A | Surface accessible to the solvent of the 135 |
| 124 | FcRnASA1B | Surface accessible to the solvent of the 1 |
| 125 | FcRnASA2B | Surface accessible to the solvent of the 2 |
| 126 | FcRnASA3B | Surface accessible to the solvent of the 3 |
| 127 | FcRnASA4B | Surface accessible to the solvent of the 4 |
| 128 | FcRnASA85B | Surface accessible to the solvent of the 85 |
| 129 | FcRnASA86B | Surface accessible to the solvent of the 86 |
| 130 | nbaainterFcA | Number of atoms interacting between Fc and the FcRns alpha chain |
| 131 | nbaainterFcB | Number of atoms interacting between Fc and the FcRns beta chain |
| 132 | nbliaiHFcA | Number of hydrogen bonds between Fc and the FcRns alpha chain |
| 133 | nbliaiHFcB | Number of hydrogen bonds between Fc and the FcRns beta chain |
| 134 | nbsaltFcA | Number of salt bridges between Fc and the FcRns alpha chain |
| 135 | nbsaltFcB | Number of salt bridges between Fc and the FcRns beta chain |
| 136 | interFace_solv_en_FcA | Solvation energy gain score calculated by PISA between Fc and the FcRns alpha chain |
| 137 | interface_solv_en_FcB | Solvation energy gain score calculated by PISA between Fc and the FcRns beta chain |
| 138 | p_valueFcA | Hydrophobic score calculated by PISA between Fc and the FcRns alpha chain |
| 139 | p_valueFcB | Hydrophobic score calculated by PISA between Fc and the FcRns beta chain |
| 140 | delta_g_theoriqueFcA | Theoretical binding energy score calculated by PISA between Fc and the FcRns alpha chain |
| 141 | delta_g_theoriqueFcB | Theoretical binding energy score calculated by PISA between Fc and the FcRns beta chain |
| 142 | Bond Strength | Average distance between bonds |
| 143 | paired hydrophilic | Number of paired hydrophilic amino acids |
| 144 | pH | pH |
| 145 | nbr_bounds_h | Total number of hydrogen bonds |
| 146 | nbr_ bounds _s | Total number of salt bridges |
| 147 | nbr_ bounds _c | Total number of contacts between amino acids’ cα atoms |
| Variant # | KD pH 7 Predicted | KD pH 7 Computed | KD pH 6 Measured |
|---|---|---|---|
| RFR | |||
| T256E/T307Q | 4.06 × 10−5 | 1.58 × 10−5 | 2.32 × 10−7 |
| T256D/T307W | 1.78 × 10−4 | 1.15 × 10−5 | 1.69 × 10−7 |
| M252Y/T256D | 3.23 × 10−5 | 6.39 × 10−6 | 9.40 × 10−8 |
| M252Y/T256E | 1.87 × 10−6 | 8.70 × 10−6 | 1.28 × 10−7 |
| M252Y/T307W | 2.31 × 10−5 | 8.02 × 10−6 | 1.18 × 10−7 |
| M252Y/T256D/T307Q | 5.48 × 10−7 | 7.82 × 10−6 | 1.15 × 10−7 |
| M252Y/T256E/T307Q | 6.69 × 10−7 | 1.48 × 10−5 | 2.18 × 10−7 |
| MLP | |||
| T256E/T307Q | 4.45 × 10−5 | 1.58 × 10−5 | 2.32 × 10−7 |
| T256D/T307W | 4.50 × 10−5 | 1.15 × 10−5 | 1.69 × 10−7 |
| M252Y/T256D | 8.66 × 10−6 | 6.39 × 10−6 | 9.40 × 10−8 |
| M252Y/T256E | 8.74 × 10−6 | 8.70 × 10−6 | 1.28 × 10−7 |
| M252Y/T307W | 1.33 × 10−6 | 8.02 × 10−6 | 1.18 × 10−7 |
| M252Y/T256D/T307Q | 1.46 × 10−6 | 7.82 × 10−6 | 1.15 × 10−7 |
| M252Y/T256E/T307Q | 1.48 × 10−6 | 1.48 × 10−5 | 2.18 × 10−7 |
| MLR | |||
| T256E/T307Q | 9.25 × 10−5 | 1.58 × 10−5 | 2.32 × 10−7 |
| T256D/T307W | 1.03 × 10−4 | 1.15 × 10−5 | 1.69 × 10−7 |
| M252Y/T256D | 7.33 × 10−5 | 6.39 × 10−6 | 9.40 × 10−8 |
| M252Y/T256E | 6.92 × 10−5 | 8.70 × 10−6 | 1.28 × 10−7 |
| M252Y/T307W | 3.00 × 10−5 | 8.02 × 10−6 | 1.18 × 10−7 |
| M252Y/T256D/T307Q | 2.81 × 10−5 | 7.82 × 10−6 | 1.15 × 10−7 |
| M252Y/T256E/T307Q | 2.72 × 10−5 | 1.48 × 10−5 | 2.18 × 10−7 |
| SVR | |||
| T256E/T307Q | 9.25 × 10−5 | 1.58 × 10−5 | 2.32 × 10−7 |
| T256D/T307W | 1.03 × 10−4 | 1.15 × 10−5 | 1.69 × 10−7 |
| M252Y/T256D | 7.33 × 10−5 | 6.39 × 10−6 | 9.40 × 10−8 |
| M252Y/T256E | 6.92 × 10−5 | 8.70 × 10−6 | 1.28 × 10−7 |
| M252Y/T307W | 3.00 × 10−5 | 8.02 × 10−6 | 1.18 × 10−7 |
| M252Y/T256D/T307Q | 2.81 × 10−5 | 7.82 × 10−6 | 1.15 × 10−7 |
| M252Y/T256E/T307Q | 2.72 × 10−5 | 1.48 × 10−5 | 2.18 × 10−7 |
| Model FL | ||
|---|---|---|
| RFR Learning_Set | ||
| Variant # | Mutations | KD |
| 833 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 3.46 × 10−9 |
| 831 | 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 3.61 × 10−9 |
| 802 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 3.70 × 10−9 |
| 829 | 235K, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 3.70 × 10−9 |
| 832 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 3.77 × 10−9 |
| 800 | 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 3.87 × 10−9 |
| 801 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 4.06 × 10−9 |
| 828 | 235K, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 4.06 × 10−9 |
| 568 | 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y | 4.24 × 10−9 |
| 1 | 239K, 252Y, 270F, 286E, 307Q, 308P, 311A, 428I, 434Y | 4.55 × 10−9 |
| 2 | 239K, 252W, 286E, 308P, 428Y, 434Y | 6.50 × 10−9 |
| 3 | 239K, 252W, 256E, 286E, 308P, 428Y, 434Y | 6.51 × 10−9 |
| 567 | 252Y, 286E, 307Q, 308P, 311A, 434Y | 6.78 × 10−9 |
| 527 | 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y | 7.15 × 10−9 |
| 5 | 239K, 252Y, 270F, 286E, 307Q, 308P, 428I, 434Y | 7.45 × 10−9 |
| 7 | 239K, 252Y, 270F, 286E, 308P, 387E, 428I, 434Y | 7.47 × 10−9 |
| 8 | 239K, 252Y, 270F, 286E, 308P, 428I, 434Y | 7.63 × 10−9 |
| 4 | 239K, 252Y, 270F, 286E, 308P, 311A, 428I, 434Y | 7.68 × 10−9 |
| 565 | 252Y, 286E, 308P, 428I, 434Y | 7.92 × 10−9 |
| 6 | 239K, 252Y, 286E, 308P, 428I, 434Y | 7.97 × 10−9 |
| MLP learning_set | ||
| Variant # | Mutations | KD |
| 568 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 7.65 × 10−9 |
| 1 | 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 7.65 × 10−9 |
| 29 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 7.98 × 10−9 |
| 5 | 235K, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 8.07 × 10−9 |
| 802 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 8.19 × 10−9 |
| 829 | 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 8.19 × 10−9 |
| 800 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 8.32 × 10−9 |
| 801 | 235K, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 8.32 × 10−9 |
| 828 | 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y | 8.32 × 10−9 |
| 831 | 239K, 252Y, 270F, 286E, 307Q, 308P, 311A, 428I, 434Y | 8.66 × 10−9 |
| 832 | 239K, 252W, 286E, 308P, 428Y, 434Y | 8.66 × 10−9 |
| 19 | 239K, 252W, 256E, 286E, 308P, 428Y, 434Y | 8.69 × 10−9 |
| 47 | 252Y, 286E, 307Q, 308P, 311A, 434Y | 8.72 × 10−9 |
| 24 | 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y | 8.72 × 10−9 |
| 495 | 239K, 252Y, 270F, 286E, 307Q, 308P, 428I, 434Y | 8.72 × 10−9 |
| 833 | 239K, 252Y, 270F, 286E, 308P, 387E, 428I, 434Y | 9.13 × 10−9 |
| 567 | 239K, 252Y, 270F, 286E, 308P, 428I, 434Y | 9.44 × 10−9 |
| 527 | 239K, 252Y, 270F, 286E, 308P, 311A, 428I, 434Y | 9.44 × 10−9 |
| 3 | 252Y, 286E, 308P, 428I, 434Y | 1.02 × 10−8 |
| 570 | 239K, 252Y, 286E, 308P, 428I, 434Y | 1.09 × 10−8 |
| MLR learning_set | ||
| Variant # | Mutations | KD |
| 544 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 5.13 × 10−9 |
| 530 | 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 6.30 × 10−9 |
| 244 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 7.93 × 10−9 |
| 247 | 235K, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 8.08 × 10−9 |
| 343 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 8.10 × 10−9 |
| 543 | 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 9.51 × 10−9 |
| 568 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 9.56 × 10−9 |
| 1 | 235K, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 9.69 × 10−9 |
| 5 | 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y | 9.77 × 10−9 |
| 567 | 239K, 252Y, 270F, 286E, 307Q, 308P, 311A, 428I, 434Y | 1.10 × 10−8 |
| 527 | 239K, 252W, 286E, 308P, 428Y, 434Y | 1.11 × 10−8 |
| 29 | 239K, 252W, 256E, 286E, 308P, 428Y, 434Y | 1.11 × 10−8 |
| 47 | 252Y, 286E, 307Q, 308P, 311A, 434Y | 1.12 × 10−8 |
| 833 | 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y | 1.16 × 10−8 |
| 24 | 239K, 252Y, 270F, 286E, 307Q, 308P, 428I, 434Y | 1.18 × 10−8 |
| 495 | 239K, 252Y, 270F, 286E, 308P, 387E, 428I, 434Y | 1.18 × 10−8 |
| 19 | 239K, 252Y, 270F, 286E, 308P, 428I, 434Y | 1.27 × 10−8 |
| 802 | 239K, 252Y, 270F, 286E, 308P, 311A, 428I, 434Y | 1.31 × 10−8 |
| 829 | 252Y, 286E, 308P, 428I, 434Y | 1.31 × 10−8 |
| 536 | 239K, 252Y, 286E, 308P, 428I, 434Y | 1.45 × 10−8 |
| SVR learning_set | ||
| Variant # | Mutations | KD |
| 831 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 4.90 × 10−9 |
| 832 | 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 4.90 × 10−9 |
| 800 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 5.24 × 10−9 |
| 801 | 235K, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 5.24 × 10−9 |
| 828 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 5.24 × 10−9 |
| 833 | 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 5.87 × 10−9 |
| 2 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 6.92 × 10−9 |
| 3 | 235K, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 7.08 × 10−9 |
| 802 | 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y | 7.28 × 10−9 |
| 829 | 239K, 252Y, 270F, 286E, 307Q, 308P, 311A, 428I, 434Y | 7.28 × 10−9 |
| 5 | 239K, 252W, 286E, 308P, 428Y, 434Y | 8.13 × 10−9 |
| 565 | 239K, 252W, 256E, 286E, 308P, 428Y, 434Y | 8.71 × 10−9 |
| 6 | 252Y, 286E, 307Q, 308P, 311A, 434Y | 8.71 × 10−9 |
| 7 | 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y | 8.71 × 10−9 |
| 8 | 239K, 252Y, 270F, 286E, 307Q, 308P, 428I, 434Y | 8.71 × 10−9 |
| 763 | 239K, 252Y, 270F, 286E, 308P, 387E, 428I, 434Y | 1.02 × 10−8 |
| 818 | 239K, 252Y, 270F, 286E, 308P, 428I, 434Y | 1.02 × 10−8 |
| 783 | 239K, 252Y, 270F, 286E, 308P, 311A, 428I, 434Y | 1.02 × 10−8 |
| 567 | 252Y, 286E, 308P, 428I, 434Y | 1.20 × 10−8 |
| 527 | 239K, 252Y, 286E, 308P, 428I, 434Y | 1.20 × 10−8 |
| RFR 3mut | ||
| Variant # | Mutations | KD |
| 22,717 | L309Q, N434D, Y436L | 3.14 × 10−8 |
| 27,638 | H310N, H435G, Y436L | 3.72 × 10−8 |
| 21,828 | M252R, H310E, H433N | 4.01 × 10−8 |
| 23,131 | H310E, N434L, Y436K | 4.03 × 10−8 |
| 26,871 | H310R, N434F, Y436K | 4.18 × 10−8 |
| 22,177 | K288G, H310G, N434W | 5.50 × 10−8 |
| 27,282 | Q311R, M428F, N434F | 6.17 × 10−8 |
| 21,781 | K288G, H310G, H433T | 6.34 × 10−8 |
| 23,175 | M252W, I253D, N286R | 7.10 × 10−8 |
| 25,334 | M252R, K288S, H433S | 7.47 × 10−8 |
| 20,956 | M252R, I253G, H433G | 8.59 × 10−8 |
| 26,312 | I253D, H433A, Y436K | 8.70 × 10−8 |
| 25,958 | K288A, H310T, N434H | 8.87 × 10−8 |
| 23,441 | Q311R, M428N, N434F | 9.07 × 10−8 |
| 28,117 | K288R, H310D, H433Y | 9.16 × 10−8 |
| 27,621 | T256W, H435G, Y436N | 9.43 × 10−8 |
| 20,844 | P257V, N286D, T307Y | 9.79 × 10−8 |
| 27,672 | M252Q, T256N, H433Y | 9.80 × 10−8 |
| 20,219 | M252K, T256Y, H433N | 1.01 × 10−7 |
| 22,805 | N286R, T307R, Y436I | 1.02 × 10−7 |
| MLP 3mut | ||
| Variant # | Mutations | KD |
| 25,115 | L309Q, N434D, Y436L | 9.91 × 10−9 |
| 23,790 | H310N, H435G, Y436L | 1.18 × 10−8 |
| 27,256 | M252R, H310E, H433N | 1.30 × 10−8 |
| 27,568 | H310E, N434L, Y436K | 1.36 × 10−8 |
| 27,086 | H310R, N434F, Y436K | 1.40 × 10−8 |
| 22,280 | K288G, H310G, N434W | 1.42 × 10−8 |
| 21,044 | Q311R, M428F, N434F | 1.49 × 10−8 |
| 20,905 | K288G, H310G, H433T | 1.61 × 10−8 |
| 21,807 | M252W, I253D, N286R | 1.64 × 10−8 |
| 22,937 | M252R, K288S, H433S | 1.67 × 10−8 |
| 22,638 | M252R, I253G, H433G | 1.71 × 10−8 |
| 20,841 | I253D, H433A, Y436K | 1.74 × 10−8 |
| 25,914 | K288A, H310T, N434H | 1.76 × 10−8 |
| 21,608 | Q311R, M428N, N434F | 1.77 × 10−8 |
| 27,445 | K288R, H310D, H433Y | 1.79 × 10−8 |
| 26,619 | T256W, H435G, Y436N | 1.79 × 10−8 |
| 21,178 | P257V, N286D, T307Y | 1.83 × 10−8 |
| 24,744 | M252Q, T256N, H433Y | 1.84 × 10−8 |
| 21,756 | M252K, T256Y, H433N | 1.85 × 10−8 |
| 21,891 | N286R, T307R, Y436I | 1.85 × 10−8 |
| MLR 3mut | ||
| Variant # | Mutations | KD |
| 23,046 | L309Q, N434D, Y436L | 8.56 × 10−10 |
| 23,821 | H310N, H435G, Y436L | 1.90 × 10−9 |
| 20,005 | M252R, H310E, H433N | 1.90 × 10−9 |
| 21,325 | H310E, N434L, Y436K | 2.65 × 10−9 |
| 20,606 | H310R, N434F, Y436K | 2.72 × 10−9 |
| 25,146 | K288G, H310G, N434W | 3.56 × 10−9 |
| 23,660 | Q311R, M428F, N434F | 3.96 × 10−9 |
| 23,971 | K288G, H310G, H433T | 4.53 × 10−9 |
| 26,091 | M252W, I253D, N286R | 4.68 × 10−9 |
| 27,856 | M252R, K288S, H433S | 4.85 × 10−9 |
| 25,298 | M252R, I253G, H433G | 5.72 × 10−9 |
| 28,072 | I253D, H433A, Y436K | 5.80 × 10−9 |
| 28,288 | K288A, H310T, N434H | 5.88 × 10−9 |
| 27,058 | Q311R, M428N, N434F | 6.28 × 10−9 |
| 21,705 | K288R, H310D, H433Y | 6.42 × 10−9 |
| 25,221 | T256W, H435G, Y436N | 6.85 × 10−9 |
| 22,944 | P257V, N286D, T307Y | 6.95 × 10−9 |
| 22,506 | M252Q, T256N, H433Y | 7.37 × 10−9 |
| 25,804 | M252K, T256Y, H433N | 7.59 × 10−9 |
| 27,795 | N286R, T307R, Y436I | 7.70 × 10−9 |
| SVR 3mut | ||
| Variant # | Mutations | KD |
| 22,166 | L309Q, N434D, Y436L | 1.09 × 10−7 |
| 25,716 | H310N, H435G, Y436L | 1.16 × 10−7 |
| 26,932 | M252R, H310E, H433N | 1.26 × 10−7 |
| 20,339 | H310E, N434L, Y436K | 1.42 × 10−7 |
| 26,518 | H310R, N434F, Y436K | 1.44 × 10−7 |
| 27,880 | K288G, H310G, N434W | 1.46 × 10−7 |
| 21,576 | Q311R, M428F, N434F | 1.51 × 10−7 |
| 27,672 | K288G, H310G, H433T | 1.51 × 10−7 |
| 22,597 | M252W, I253D, N286R | 1.58 × 10−7 |
| 20,333 | M252R, K288S, H433S | 1.64 × 10−7 |
| 22,168 | M252R, I253G, H433G | 1.64 × 10−7 |
| 23,757 | I253D, H433A, Y436K | 1.69 × 10−7 |
| 21,145 | K288A, H310T, N434H | 1.70 × 10−7 |
| 20,273 | Q311R, M428N, N434F | 1.71 × 10−7 |
| 27,350 | K288R, H310D, H433Y | 1.72 × 10−7 |
| 22,409 | T256W, H435G, Y436N | 1.73 × 10−7 |
| 24,464 | P257V, N286D, T307Y | 1.73 × 10−7 |
| 26,610 | M252Q, T256N, H433Y | 1.74 × 10−7 |
| 27,113 | M252K, T256Y, H433N | 1.76 × 10−7 |
| 23,602 | N286R, T307R, Y436I | 1.77 × 10−7 |
| RFR 5mut | ||
| Variant # | Mutations | KD |
| 31,995 | M252K, H285Q, T307D, L309K, H433Y | 1.24 × 10−8 |
| 32,898 | M252R, N286Q, H310R, M428I, H435N | 1.49 × 10−8 |
| 31,966 | M252W, I253H, N286D, Q311K, H433P | 1.67 × 10−8 |
| 37,526 | L251I, L309K, H433N, N434Q, Y436L | 1.72 × 10−8 |
| 36,771 | I253F, N286R, T307Q, M428F, H435E | 1.85 × 10−8 |
| 33,965 | L251A, M252K, L309K, L314S, H433Y | 1.94 × 10−8 |
| 37,863 | L309K, Q311E, H433G, N434H, Y436R | 2.09 × 10−8 |
| 32,948 | S254N, T307A, L309K, H433T, Y436R | 2.45 × 10−8 |
| 30,050 | T256E, N286Q, Q311N, N434H, Y436N | 2.53 × 10−8 |
| 35,099 | I253D, P257S, Q311E, M428Y, H433D | 2.64 × 10−8 |
| 38,056 | N286E, L309D, L314H, H435N, Y436N | 2.90 × 10−8 |
| 34,584 | L251D, M252Q, N286D, V308A, H433A | 2.97 × 10−8 |
| 32,714 | M252W, S254T, V308F, H310R, H435S | 2.98 × 10−8 |
| 35,697 | M252K, N286R, L309Q, H435N, Y436F | 3.14 × 10−8 |
| 31,821 | L251H, M252Q, N286H, Q311N, H433A | 3.34 × 10−8 |
| 37,707 | T256S, Q311K, L314R, M428W, N434W | 3.44 × 10−8 |
| 38,325 | K288T, V308A, L309K, H433G, Y436K | 3.68 × 10−8 |
| 30,121 | I253W, N286Y, L309V, M428F, H433D | 3.72 × 10−8 |
| 32,551 | M252F, P257W, H285E, Q311E, N434H | 3.84 × 10−8 |
| 34,867 | H285E, N286Y, L309Y, M428H, H433Y | 3.88 × 10−8 |
| MLP 5mut | ||
| Variant # | Mutations | KD |
| 36,622 | M252K, H285Q, T307D, L309K, H433Y | 5.13 × 10−9 |
| 38,413 | M252R, N286Q, H310R, M428I, H435N | 7.18 × 10−9 |
| 32,294 | M252W, I253H, N286D, Q311K, H433P | 7.20 × 10−9 |
| 34,399 | L251I, L309K, H433N, N434Q, Y436L | 7.91 × 10−9 |
| 35,394 | I253F, N286R, T307Q, M428F, H435E | 8.58 × 10−9 |
| 34,608 | L251A, M252K, L309K, L314S, H433Y | 8.68 × 10−9 |
| 31,958 | L309K, Q311E, H433G, N434H, Y436R | 8.91 × 10−9 |
| 34,236 | S254N, T307A, L309K, H433T, Y436R | 9.17 × 10−9 |
| 36,343 | T256E, N286Q, Q311N, N434H, Y436N | 9.26 × 10−9 |
| 35,234 | I253D, P257S, Q311E, M428Y, H433D | 9.47 × 10−9 |
| 38,030 | N286E, L309D, L314H, H435N, Y436N | 9.53 × 10−9 |
| 30,188 | L251D, M252Q, N286D, V308A, H433A | 9.83 × 10−9 |
| 35,109 | M252W, S254T, V308F, H310R, H435S | 9.98 × 10−9 |
| 32,632 | M252K, N286R, L309Q, H435N, Y436F | 1.00 × 10−8 |
| 30,914 | L251H, M252Q, N286H, Q311N, H433A | 1.04 × 10−8 |
| 35,398 | T256S, Q311K, L314R, M428W, N434W | 1.05 × 10−8 |
| 32,539 | K288T, V308A, L309K, H433G, Y436K | 1.05 × 10−8 |
| 35,860 | I253W, N286Y, L309V, M428F, H433D | 1.06 × 10−8 |
| 35,943 | M252F, P257W, H285E, Q311E, N434H | 1.07 × 10−8 |
| 37,387 | H285E, N286Y, L309Y, M428H, H433Y | 1.07 × 10−8 |
| MLR 5mut | ||
| Variant # | Mutations | KD |
| 36,120 | M252K, H285Q, T307D, L309K, H433Y | 4.79 × 10−10 |
| 31,116 | M252R, N286Q, H310R, M428I, H435N | 5.14 × 10−10 |
| 37,434 | M252W, I253H, N286D, Q311K, H433P | 1.40 × 10−9 |
| 33,162 | L251I, L309K, H433N, N434Q, Y436L | 1.44 × 10−9 |
| 35,517 | I253F, N286R, T307Q, M428F, H435E | 1.66 × 10−9 |
| 37,684 | L251A, M252K, L309K, L314S, H433Y | 1.88 × 10−9 |
| 37,301 | L309K, Q311E, H433G, N434H, Y436R | 1.88 × 10−9 |
| 30,930 | S254N, T307A, L309K, H433T, Y436R | 1.94 × 10−9 |
| 36,097 | T256E, N286Q, Q311N, N434H, Y436N | 1.95 × 10−9 |
| 38,430 | I253D, P257S, Q311E, M428Y, H433D | 1.97 × 10−9 |
| 38,202 | N286E, L309D, L314H, H435N, Y436N | 2.09 × 10−9 |
| 37,863 | L251D, M252Q, N286D, V308A, H433A | 2.09 × 10−9 |
| 30,545 | M252W, S254T, V308F, H310R, H435S | 2.15 × 10−9 |
| 31,317 | M252K, N286R, L309Q, H435N, Y436F | 2.25 × 10−9 |
| 34,813 | L251H, M252Q, N286H, Q311N, H433A | 2.34 × 10−9 |
| 36,045 | T256S, Q311K, L314R, M428W, N434W | 2.40 × 10−9 |
| 38,596 | K288T, V308A, L309K, H433G, Y436K | 2.55 × 10−9 |
| 33,006 | I253W, N286Y, L309V, M428F, H433D | 2.69 × 10−9 |
| 33,288 | M252F, P257W, H285E, Q311E, N434H | 2.72 × 10−9 |
| 33,871 | H285E, N286Y, L309Y, M428H, H433Y | 2.75 × 10−9 |
| SVR 5mut | ||
| Variant # | Mutations | KD |
| 31,131 | M252K, H285Q, T307D, L309K, H433Y | 6.12 × 10−8 |
| 37,573 | M252R, N286Q, H310R, M428I, H435N | 8.12 × 10−8 |
| 34,132 | M252W, I253H, N286D, Q311K, H433P | 8.81 × 10−8 |
| 32,677 | L251I, L309K, H433N, N434Q, Y436L | 9.49 × 10−8 |
| 38,342 | I253F, N286R, T307Q, M428F, H435E | 1.10 × 10−7 |
| 31,134 | L251A, M252K, L309K, L314S, H433Y | 1.12 × 10−7 |
| 37,613 | L309K, Q311E, H433G, N434H, Y436R | 1.23 × 10−7 |
| 36,014 | S254N, T307A, L309K, H433T, Y436R | 1.29 × 10−7 |
| 32,967 | T256E, N286Q, Q311N, N434H, Y436N | 1.48 × 10−7 |
| 30,390 | I253D, P257S, Q311E, M428Y, H433D | 1.52 × 10−7 |
| 31,621 | N286E, L309D, L314H, H435N, Y436N | 1.58 × 10−7 |
| 30,551 | L251D, M252Q, N286D, V308A, H433A | 1.63 × 10−7 |
| 32,946 | M252W, S254T, V308F, H310R, H435S | 1.68 × 10−7 |
| 32,204 | M252K, N286R, L309Q, H435N, Y436F | 1.74 × 10−7 |
| 30,254 | L251H, M252Q, N286H, Q311N, H433A | 1.75 × 10−7 |
| 31,168 | T256S, Q311K, L314R, M428W, N434W | 1.77 × 10−7 |
| 32,902 | K288T, V308A, L309K, H433G, Y436K | 1.78 × 10−7 |
| 30,243 | I253W, N286Y, L309V, M428F, H433D | 1.79 × 10−7 |
| 30,417 | M252F, P257W, H285E, Q311E, N434H | 1.83 × 10−7 |
| 30,211 | H285E, N286Y, L309Y, M428H, H433Y | 1.84 × 10−7 |
| RFR 8mut | ||
| Variant # | Mutations | KD |
| 30,849 | M252W, T256P, N286K, L309K, Q311A, M428F, Y436G | 5.67 × 10−9 |
| 30,198 | L251R, I253T, H285N, N286D, L309K, M428F, N434D | 1.21 × 10−8 |
| 30,864 | L251T, M252Y, I253P, N286K, V308F, L309R, H433G | 1.52 × 10−8 |
| 30,501 | M252Y, I253E, H285I, N286D, V308A, N434H | 1.73 × 10−8 |
| 30,454 | M252Y, N286Q, K288F, L309W, Q311L, N434Y | 1.77 × 10−8 |
| 30,390 | R255Y, P257N, H285D, V308A, L309K, M428W, N434H | 1.94 × 10−8 |
| 30,947 | M252W, I253Y, R255F, N286E, L309D, Q311K, H433P | 2.32 × 10−8 |
| 30,169 | L251T, I253D, R255S, T256S, Q311A, M428F, H433G | 2.60 × 10−8 |
| 30,358 | M252E, P257V, L309K, M428L, H433F, H435K | 2.85 × 10−8 |
| 30,582 | P257A, H285I, T307W, M428W, N434H | 2.85 × 10−8 |
| 30,338 | R255Q, N286K, T307Q, L309P, M428F, H433I, H435R | 3.57 × 10−8 |
| 30,211 | H285E, N286K, T307R, L309E, Q311K, M428W, N434F | 3.59 × 10−8 |
| 30,974 | I253S, T256S, H285D, N286E, V308A, M428L, H435E | 3.94 × 10−8 |
| 30,696 | M252D, N286W, L309R, Q311V, N434H, H435K | 4.33 × 10−8 |
| 30,401 | M252W, I253D, P257A, V308F, L309E, N434W | 4.42 × 10−8 |
| 30,416 | I253P, T256V, N286R, Q311K, M428W, N434F | 4.79 × 10−8 |
| 30,777 | M252W, P257T, N286H, T307F, L309G, H433L | 5.22 × 10−8 |
| 30,280 | M252V, R255Q, T256N, N286H, M428F, N434Y, Y436K | 5.47 × 10−8 |
| 30,913 | L251P, T256P, N286Q, L309K, Q311I, Y436G | 5.66 × 10−8 |
| 30,948 | L251G, T256N, N286E, V308A, L309K, H433L, N434T | 5.66 × 10−8 |
| MLP 8mut | ||
| Variant # | Mutations | KD |
| 30,126 | M252W, T256P, N286K, L309K, Q311A, M428F, Y436G | 5.75 × 10−9 |
| 30,501 | L251R, I253T, H285N, N286D, L309K, M428F, N434D | 6.73 × 10−9 |
| 30,603 | L251T, M252Y, I253P, N286K, V308F, L309R, H433G | 7.09 × 10−9 |
| 30,947 | M252Y, I253E, H285I, N286D, V308A, N434H | 8.43 × 10−9 |
| 30,070 | M252Y, N286Q, K288F, L309W, Q311L, N434Y | 9.86 × 10−9 |
| 30,582 | R255Y, P257N, H285D, V308A, L309K, M428W, N434H | 1.09 × 10−8 |
| 30,198 | M252W, I253Y, R255F, N286E, L309D, Q311K, H433P | 1.14 × 10−8 |
| 30,822 | L251T, I253D, R255S, T256S, Q311A, M428F, H433G | 1.18 × 10−8 |
| 30,554 | M252E, P257V, L309K, M428L, H433F, H435K | 1.37 × 10−8 |
| 30,950 | P257A, H285I, T307W, M428W, N434H | 1.40 × 10−8 |
| 30,154 | R255Q, N286K, T307Q, L309P, M428F, H433I, H435R | 1.46 × 10−8 |
| 30,942 | H285E, N286K, T307R, L309E, Q311K, M428W, N434F | 1.47 × 10−8 |
| 30,842 | I253S, T256S, H285D, N286E, V308A, M428L, H435E | 1.49 × 10−8 |
| 30,454 | M252D, N286W, L309R, Q311V, N434H, H435K | 1.52 × 10−8 |
| 30,259 | M252W, I253D, P257A, V308F, L309E, N434W | 1.55 × 10−8 |
| 30,042 | I253P, T256V, N286R, Q311K, M428W, N434F | 1.59 × 10−8 |
| 30,782 | M252W, P257T, N286H, T307F, L309G, H433L | 1.60 × 10−8 |
| 30,241 | M252V, R255Q, T256N, N286H, M428F, N434Y, Y436K | 1.62 × 10−8 |
| 30,171 | L251P, T256P, N286Q, L309K, Q311I, Y436G | 1.64 × 10−8 |
| 30,365 | L251G, T256N, N286E, V308A, L309K, H433L, N434T | 1.68 × 10−8 |
| MLR 8mut | ||
| Variant # | Mutations | KD |
| 30,835 | M252W, T256P, N286K, L309K, Q311A, M428F, Y436G | 2.34 × 10-11 |
| 30,259 | L251R, I253T, H285N, N286D, L309K, M428F, N434D | 8.60 × 10-11 |
| 30,184 | L251T, M252Y, I253P, N286K, V308F, L309R, H433G | 3.28 × 10−10 |
| 30,558 | M252Y, I253E, H285I, N286D, V308A, N434H | 4.17 × 10−10 |
| 30,317 | M252Y, N286Q, K288F, L309W, Q311L, N434Y | 5.83 × 10−10 |
| 30,395 | R255Y, P257N, H285D, V308A, L309K, M428W, N434H | 6.03 × 10−10 |
| 30,787 | M252W, I253Y, R255F, N286E, L309D, Q311K, H433P | 6.18 × 10−10 |
| 30,968 | L251T, I253D, R255S, T256S, Q311A, M428F, H433G | 1.34 × 10−9 |
| 30,762 | M252E, P257V, L309K, M428L, H433F, H435K | 1.36 × 10−9 |
| 30,253 | P257A, H285I, T307W, M428W, N434H | 1.68 × 10−9 |
| 30,500 | R255Q, N286K, T307Q, L309P, M428F, H433I, H435R | 2.17 × 10−9 |
| 30,926 | H285E, N286K, T307R, L309E, Q311K, M428W, N434F | 2.26 × 10−9 |
| 30,023 | I253S, T256S, H285D, N286E, V308A, M428L, H435E | 2.27 × 10−9 |
| 30,515 | M252D, N286W, L309R, Q311V, N434H, H435K | 2.49 × 10−9 |
| 30,087 | M252W, I253D, P257A, V308F, L309E, N434W | 2.51 × 10−9 |
| 30,209 | I253P, T256V, N286R, Q311K, M428W, N434F | 3.25 × 10−9 |
| 30,832 | M252W, P257T, N286H, T307F, L309G, H433L | 3.36 × 10−9 |
| 30,947 | M252V, R255Q, T256N, N286H, M428F, N434Y, Y436K | 3.49 × 10−9 |
| 30,179 | L251P, T256P, N286Q, L309K, Q311I, Y436G | 3.51 × 10−9 |
| 30,577 | L251G, T256N, N286E, V308A, L309K, H433L, N434T | 3.74 × 10−9 |
| SVR 8mut | ||
| Variant # | Mutations | KD |
| 30,401 | M252W, T256P, N286K, L309K, Q311A, M428F, Y436G | 1.53 × 10−7 |
| 30,245 | L251R, I253T, H285N, N286D, L309K, M428F, N434D | 1.71 × 10−7 |
| 30,105 | L251T, M252Y, I253P, N286K, V308F, L309R, H433G | 1.84 × 10−7 |
| 30,625 | M252Y, I253E, H285I, N286D, V308A, N434H | 1.91 × 10−7 |
| 30,022 | M252Y, N286Q, K288F, L309W, Q311L, N434Y | 1.92 × 10−7 |
| 30,142 | R255Y, P257N, H285D, V308A, L309K, M428W, N434H | 1.96 × 10−7 |
| 30,501 | M252W, I253Y, R255F, N286E, L309D, Q311K, H433P | 2.02 × 10−7 |
| 30,097 | L251T, I253D, R255S, T256S, Q311A, M428F, H433G | 2.03 × 10−7 |
| 30,974 | M252E, P257V, L309K, M428L, H433F, H435K | 2.04 × 10−7 |
| 30,684 | P257A, H285I, T307W, M428W, N434H | 2.04 × 10−7 |
| 30,186 | R255Q, N286K, T307Q, L309P, M428F, H433I, H435R | 2.04 × 10−7 |
| 30,955 | H285E, N286K, T307R, L309E, Q311K, M428W, N434F | 2.04 × 10−7 |
| 30,582 | I253S, T256S, H285D, N286E, V308A, M428L, H435E | 2.05 × 10−7 |
| 30,012 | M252D, N286W, L309R, Q311V, N434H, H435K | 2.05 × 10−7 |
| 30,905 | M252W, I253D, P257A, V308F, L309E, N434W | 2.05 × 10−7 |
| 30,502 | I253P, T256V, N286R, Q311K, M428W, N434F | 2.05 × 10−7 |
| 30,785 | M252W, P257T, N286H, T307F, L309G, H433L | 2.05 × 10−7 |
| 30,685 | M252V, R255Q, T256N, N286H, M428F, N434Y, Y436K | 2.06 × 10−7 |
| 30,261 | L251P, T256P, N286Q, L309K, Q311I, Y436G | 2.06 × 10−7 |
| 30,206 | L251G, T256N, N286E, V308A, L309K, H433L, N434T | 2.06 × 10−7 |
| Model FLS | ||
| RFR Learning_Set | ||
| Variant # | Mutations | KD |
| 833 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 3.66 × 10−9 |
| 831 | 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 3.79 × 10−9 |
| 832 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 3.92 × 10−9 |
| 802 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 4.09 × 10−9 |
| 829 | 235K, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 4.09 × 10−9 |
| 800 | 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 4.24 × 10−9 |
| 801 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 4.39 × 10−9 |
| 828 | 235K, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 4.39 × 10−9 |
| 568 | 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y | 4.71 × 10−9 |
| 1 | 239K, 252Y, 270F, 286E, 307Q, 308P, 311A, 428I, 434Y | 5.40 × 10−9 |
| 567 | 252Y, 286E, 307Q, 308P, 311A, 434Y | 7.07 × 10−9 |
| 5 | 239K, 252Y, 270F, 286E, 307Q, 308P, 428I, 434Y | 7.27 × 10−9 |
| 2 | 239K, 252W, 286E, 308P, 428Y, 434Y | 7.34 × 10−9 |
| 7 | 239K, 252Y, 270F, 286E, 308P, 387E, 428I, 434Y | 7.60 × 10−9 |
| 527 | 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y | 7.90 × 10−9 |
| 6 | 239K, 252Y, 286E, 308P, 428I, 434Y | 7.95 × 10−9 |
| 8 | 239K, 252Y, 270F, 286E, 308P, 428I, 434Y | 8.12 × 10−9 |
| 4 | 239K, 252Y, 270F, 286E, 308P, 311A, 428I, 434Y | 8.41 × 10−9 |
| 3 | 239K, 252W, 256E, 286E, 308P, 428Y, 434Y | 8.59 × 10−9 |
| 565 | 252Y, 286E, 308P, 428I, 434Y | 1.03 × 10−8 |
| MLP learning_set | ||
| Variant # | Mutations | KD |
| 20 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 2.96 × 10−8 |
| 90 | 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 3.42 × 10−8 |
| 40 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 3.42 × 10−8 |
| 566 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 3.51 × 10−8 |
| 570 | 235K, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 3.51 × 10−8 |
| 569 | 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 3.51 × 10−8 |
| 204 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 3.51 × 10−8 |
| 119 | 235K, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 3.51 × 10−8 |
| 110 | 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y | 3.51 × 10−8 |
| 44 | 239K, 252Y, 270F, 286E, 307Q, 308P, 311A, 428I, 434Y | 3.51 × 10−8 |
| 131 | 252Y, 286E, 307Q, 308P, 311A, 434Y | 3.51 × 10−8 |
| 581 | 239K, 252Y, 270F, 286E, 307Q, 308P, 428I, 434Y | 3.54 × 10−8 |
| 23 | 239K, 252W, 286E, 308P, 428Y, 434Y | 3.54 × 10−8 |
| 19 | 239K, 252Y, 270F, 286E, 308P, 387E, 428I, 434Y | 3.55 × 10−8 |
| 568 | 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y | 3.57 × 10−8 |
| 1 | 239K, 252Y, 286E, 308P, 428I, 434Y | 3.57 × 10−8 |
| 5 | 239K, 252Y, 270F, 286E, 308P, 428I, 434Y | 3.59 × 10−8 |
| 53 | 239K, 252Y, 270F, 286E, 308P, 311A, 428I, 434Y | 3.80 × 10−8 |
| 98 | 239K, 252W, 256E, 286E, 308P, 428Y, 434Y | 3.80 × 10−8 |
| 59 | 252Y, 286E, 308P, 428I, 434Y | 3.84 × 10−8 |
| MLR learning_set | ||
| Variant # | Mutations | KD |
| 684 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 9.96 × 10−9 |
| 163 | 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 1.79 × 10−8 |
| 167 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 2.03 × 10−8 |
| 216 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 2.03 × 10−8 |
| 182 | 235K, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 2.55 × 10−8 |
| 128 | 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 2.60 × 10−8 |
| 94 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 2.64 × 10−8 |
| 231 | 235K, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 2.64 × 10−8 |
| 120 | 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y | 2.68 × 10−8 |
| 495 | 239K, 252Y, 270F, 286E, 307Q, 308P, 311A, 428I, 434Y | 2.69 × 10−8 |
| 24 | 252Y, 286E, 307Q, 308P, 311A, 434Y | 2.69 × 10−8 |
| 192 | 239K, 252Y, 270F, 286E, 307Q, 308P, 428I, 434Y | 2.84 × 10−8 |
| 127 | 239K, 252W, 286E, 308P, 428Y, 434Y | 2.84 × 10−8 |
| 145 | 239K, 252Y, 270F, 286E, 308P, 387E, 428I, 434Y | 2.85 × 10−8 |
| 496 | 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y | 2.89 × 10−8 |
| 235 | 239K, 252Y, 286E, 308P, 428I, 434Y | 2.89 × 10−8 |
| 130 | 239K, 252Y, 270F, 286E, 308P, 428I, 434Y | 2.89 × 10−8 |
| 77 | 239K, 252Y, 270F, 286E, 308P, 311A, 428I, 434Y | 2.89 × 10−8 |
| 82 | 239K, 252W, 256E, 286E, 308P, 428Y, 434Y | 2.89 × 10−8 |
| 107 | 252Y, 286E, 308P, 428I, 434Y | 2.89 × 10−8 |
| SVR learning_set | ||
| Variant # | Mutations | KD |
| 243 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 7.25 × 10−9 |
| 276 | 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 7.26 × 10−9 |
| 208 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y, 436V | 1.29 × 10−8 |
| 8 | 235R, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 1.31 × 10−8 |
| 7 | 235K, 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 1.31 × 10−8 |
| 6 | 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 1.31 × 10−8 |
| 565 | 235R, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 1.31 × 10−8 |
| 4 | 235K, 239K, 250V, 252Y, 286E, 307Q, 308P, 311A, 434Y, 436V | 1.31 × 10−8 |
| 800 | 252Y, 286E, 307Q, 308P, 311A, 428I, 434Y | 1.34 × 10−8 |
| 828 | 239K, 252Y, 270F, 286E, 307Q, 308P, 311A, 428I, 434Y | 1.34 × 10−8 |
| 801 | 252Y, 286E, 307Q, 308P, 311A, 434Y | 1.34 × 10−8 |
| 802 | 239K, 252Y, 270F, 286E, 307Q, 308P, 428I, 434Y | 1.34 × 10−8 |
| 829 | 239K, 252W, 286E, 308P, 428Y, 434Y | 1.34 × 10−8 |
| 633 | 239K, 252Y, 270F, 286E, 308P, 387E, 428I, 434Y | 1.36 × 10−8 |
| 59 | 239K, 252Y, 286E, 307Q, 308P, 311A, 434Y | 1.42 × 10−8 |
| 43 | 239K, 252Y, 286E, 308P, 428I, 434Y | 1.67 × 10−8 |
| 38 | 239K, 252Y, 270F, 286E, 308P, 428I, 434Y | 1.67 × 10−8 |
| 37 | 239K, 252Y, 270F, 286E, 308P, 311A, 428I, 434Y | 1.67 × 10−8 |
| 42 | 239K, 252W, 256E, 286E, 308P, 428Y, 434Y | 1.67 × 10−8 |
| 41 | 252Y, 286E, 308P, 428I, 434Y | 1.67 × 10−8 |
| RFR 3mut | ||
| Variant # | Mutations | KD |
| 24,936 | H310L, N434H, Y436Q | 3.32 × 10−8 |
| 23,681 | T307K, H310D, N434F | 3.80 × 10−8 |
| 22,900 | P257H, H310G, N434F | 4.03 × 10−8 |
| 22,177 | K288G, H310G, N434W | 4.59 × 10−8 |
| 24,569 | H310V, Q311Y, N434F | 4.84 × 10−8 |
| 23,303 | H310D, H433S, N434F | 4.85 × 10−8 |
| 24,652 | H310E, Q311G, N434F | 4.88 × 10−8 |
| 22,597 | H285W, H310E, N434W | 4.98 × 10−8 |
| 20,285 | P257I, T307G, N434F | 5.05 × 10−8 |
| 23,152 | I253E, H310A, N434W | 5.38 × 10−8 |
| 27,018 | H310A, M428K, N434F | 6.27 × 10−8 |
| 25,958 | K288A, H310T, N434H | 6.53 × 10−8 |
| 26,256 | R255K, T307K, N434F | 8.46 × 10−8 |
| 23,826 | S254A, T307D, N434F | 9.36 × 10−8 |
| 26,389 | R255K, T307F, N434W | 9.95 × 10−8 |
| 21,973 | H285T, L309N, N434F | 1.04 × 10−7 |
| 27,024 | M252F, L309D, N434H | 1.17 × 10−7 |
| 26,949 | T256L, V308P, N434W | 1.26 × 10−7 |
| 20,052 | L309G, M428L, N434F | 1.27 × 10−7 |
| 23,029 | R255Y, T307S, N434H | 1.28 × 10−7 |
| MLP 3mut | ||
| Variant # | Mutations | KD |
| 20,285 | H310L, N434H, Y436Q | 2.53 × 10−8 |
| 23,242 | T307K, H310D, N434F | 2.71 × 10−8 |
| 26,256 | P257H, H310G, N434F | 3.19 × 10−8 |
| 22,900 | K288G, H310G, N434W | 3.77 × 10−8 |
| 20,052 | H310V, Q311Y, N434F | 4.56 × 10−8 |
| 26,389 | H310D, H433S, N434F | 5.99 × 10−8 |
| 23,681 | H310E, Q311G, N434F | 6.05 × 10−8 |
| 22,715 | H285W, H310E, N434W | 6.82 × 10−8 |
| 23,826 | P257I, T307G, N434F | 6.94 × 10−8 |
| 21,576 | I253E, H310A, N434W | 7.11 × 10−8 |
| 20,293 | H310A, M428K, N434F | 7.80 × 10−8 |
| 21,093 | K288A, H310T, N434H | 8.15 × 10−8 |
| 22,166 | R255K, T307K, N434F | 8.20 × 10−8 |
| 21,460 | S254A, T307D, N434F | 9.15 × 10−8 |
| 27,689 | R255K, T307F, N434W | 1.10 × 10−7 |
| 20,625 | H285T, L309N, N434F | 1.12 × 10−7 |
| 26,125 | M252F, L309D, N434H | 1.15 × 10−7 |
| 22,533 | T256L, V308P, N434W | 1.15 × 10−7 |
| 26,616 | L309G, M428L, N434F | 1.16 × 10−7 |
| 20,670 | R255Y, T307S, N434H | 1.21 × 10−7 |
| MLR 3mut | ||
| Variant # | Mutations | KD |
| 20,285 | H310L, N434H, Y436Q | 2.00 × 10−8 |
| 23,242 | T307K, H310D, N434F | 2.28 × 10−8 |
| 26,256 | P257H, H310G, N434F | 3.15 × 10−8 |
| 22,900 | K288G, H310G, N434W | 3.73 × 10−8 |
| 21,460 | H310V, Q311Y, N434F | 5.02 × 10−8 |
| 20,052 | H310D, H433S, N434F | 5.07 × 10−8 |
| 23,681 | H310E, Q311G, N434F | 5.34 × 10−8 |
| 23,826 | H285W, H310E, N434W | 6.64 × 10−8 |
| 20,625 | P257I, T307G, N434F | 6.95 × 10−8 |
| 26,616 | I253E, H310A, N434W | 7.54 × 10−8 |
| 20,293 | H310A, M428K, N434F | 7.62 × 10−8 |
| 21,093 | K288A, H310T, N434H | 7.68 × 10−8 |
| 23,441 | R255K, T307K, N434F | 7.70 × 10−8 |
| 27,689 | S254A, T307D, N434F | 8.09 × 10−8 |
| 25,977 | R255K, T307F, N434W | 8.61 × 10−8 |
| 21,576 | H285T, L309N, N434F | 8.84 × 10−8 |
| 26,389 | M252F, L309D, N434H | 8.84 × 10−8 |
| 22,715 | T256L, V308P, N434W | 9.07 × 10−8 |
| 22,166 | L309G, M428L, N434F | 9.34 × 10−8 |
| 27,485 | R255Y, T307S, N434H | 1.02 × 10−7 |
| SVR 3mut | ||
| Variant # | Mutations | KD |
| 22,166 | H310L, N434H, Y436Q | 3.15 × 10−8 |
| 22,050 | T307K, H310D, N434F | 3.63 × 10−8 |
| 26,109 | P257H, H310G, N434F | 5.30 × 10−8 |
| 20,052 | K288G, H310G, N434W | 1.15 × 10−7 |
| 23,242 | H310V, Q311Y, N434F | 1.21 × 10−7 |
| 23,889 | H310D, H433S, N434F | 1.28 × 10−7 |
| 25,411 | H310E, Q311G, N434F | 1.40 × 10−7 |
| 22,409 | H285W, H310E, N434W | 1.66 × 10−7 |
| 25,263 | P257I, T307G, N434F | 1.69 × 10−7 |
| 21,432 | I253E, H310A, N434W | 1.70 × 10−7 |
| 22,743 | H310A, M428K, N434F | 1.78 × 10−7 |
| 22,378 | K288A, H310T, N434H | 1.82 × 10−7 |
| 20,285 | R255K, T307K, N434F | 1.88 × 10−7 |
| 23,826 | S254A, T307D, N434F | 1.93 × 10−7 |
| 24,481 | R255K, T307F, N434W | 2.00 × 10−7 |
| 21,447 | H285T, L309N, N434F | 2.01 × 10−7 |
| 23,303 | M252F, L309D, N434H | 2.05 × 10−7 |
| 28,002 | T256L, V308P, N434W | 2.15 × 10−7 |
| 26,447 | L309G, M428L, N434F | 2.32 × 10−7 |
| 25,009 | R255Y, T307S, N434H | 2.35 × 10−7 |
| RFR 5mut | ||
| Variant # | Mutations | KD |
| 37,435 | S254G, T256Q, H310I, Q311W, N434H | 1.07 × 10−8 |
| 35,309 | M252Q, R255K, L309N, N434H, Y436R | 1.16 × 10−8 |
| 35,463 | H285Y, N286R, H310N, H433Y, N434H | 1.36 × 10−8 |
| 37,379 | S254T, K288G, H310S, M428V, N434F | 2.91 × 10−8 |
| 38,616 | M252R, P257E, T307Q, V308L, N434H | 4.50 × 10−8 |
| 31,621 | I253E, V308Y, H310D, L314I, N434Y | 5.33 × 10−8 |
| 30,182 | I253T, K288N, V308T, H310D, N434W | 5.58 × 10−8 |
| 37,088 | R255K, T256Q, V308H, L314S, N434Y | 5.96 × 10−8 |
| 31,104 | S254A, V308D, H310I, M428Q, N434H | 6.29 × 10−8 |
| 37,174 | M252P, T307Y, V308R, Q311E, N434F | 6.62 × 10−8 |
| 33,328 | S254G, H285S, N286F, L309A, N434Y | 6.77 × 10−8 |
| 33,978 | M252F, R255A, N286R, H310N, N434H | 7.29 × 10−8 |
| 31,320 | V308I, L309N, M428S, N434W, H435D | 8.29 × 10−8 |
| 33,091 | R255K, T307W, Q311D, H433Q, N434F | 8.31 × 10−8 |
| 31,232 | I253P, P257G, T307E, L309Y, N434H | 8.36 × 10−8 |
| 33,342 | I253D, S254D, T256V, T307G, N434F | 8.43 × 10−8 |
| 33,591 | M252T, I253V, N286G, H310L, N434H | 8.67 × 10−8 |
| 30,984 | P257K, T307A, V308F, M428H, N434Y | 8.69 × 10−8 |
| 30,585 | M252T, P257H, V308Y, L309F, N434F | 8.74 × 10−8 |
| 38,371 | M252E, S254F, P257Y, T307A, N434H | 8.88 × 10−8 |
| MLP 5mut | ||
| Variant # | Mutations | KD |
| 33,091 | S254G, T256Q, H310I, Q311W, N434H | 3.22 × 10−8 |
| 37,254 | M252Q, R255K, L309N, N434H, Y436R | 3.28 × 10−8 |
| 33,646 | H285Y, N286R, H310N, H433Y, N434H | 3.42 × 10−8 |
| 34,469 | S254T, K288G, H310S, M428V, N434F | 4.33 × 10−8 |
| 34,320 | M252R, P257E, T307Q, V308L, N434H | 4.47 × 10−8 |
| 32,501 | I253E, V308Y, H310D, L314I, N434Y | 4.61 × 10−8 |
| 34,132 | I253T, K288N, V308T, H310D, N434W | 5.00 × 10−8 |
| 30,984 | R255K, T256Q, V308H, L314S, N434Y | 5.02 × 10−8 |
| 32,098 | S254A, V308D, H310I, M428Q, N434H | 5.06 × 10−8 |
| 34,494 | M252P, T307Y, V308R, Q311E, N434F | 5.83 × 10−8 |
| 34,889 | S254G, H285S, N286F, L309A, N434Y | 5.97 × 10−8 |
| 33,342 | M252F, R255A, N286R, H310N, N434H | 6.06 × 10−8 |
| 31,505 | V308I, L309N, M428S, N434W, H435D | 6.15 × 10−8 |
| 35,586 | R255K, T307W, Q311D, H433Q, N434F | 6.60 × 10−8 |
| 37,174 | I253P, P257G, T307E, L309Y, N434H | 6.85 × 10−8 |
| 31,465 | I253D, S254D, T256V, T307G, N434F | 7.08 × 10−8 |
| 36,149 | M252T, I253V, N286G, H310L, N434H | 7.21 × 10−8 |
| 33,080 | P257K, T307A, V308F, M428H, N434Y | 7.22 × 10−8 |
| 37,661 | M252T, P257H, V308Y, L309F, N434F | 7.23 × 10−8 |
| 30,906 | M252E, S254F, P257Y, T307A, N434H | 7.32 × 10−8 |
| MLR 5mut | ||
| Variant # | Mutations | KD |
| 30,906 | S254G, T256Q, H310I, Q311W, N434H | 1.22 × 10−8 |
| 37,580 | M252Q, R255K, L309N, N434H, Y436R | 1.33 × 10−8 |
| 33,885 | H285Y, N286R, H310N, H433Y, N434H | 1.39 × 10−8 |
| 32,098 | S254T, K288G, H310S, M428V, N434F | 1.75 × 10−8 |
| 33,646 | M252R, P257E, T307Q, V308L, N434H | 3.23 × 10−8 |
| 34,320 | I253E, V308Y, H310D, L314I, N434Y | 3.36 × 10−8 |
| 34,469 | I253T, K288N, V308T, H310D, N434W | 3.62 × 10−8 |
| 33,091 | R255K, T256Q, V308H, L314S, N434Y | 3.74 × 10−8 |
| 37,954 | S254A, V308D, H310I, M428Q, N434H | 4.06 × 10−8 |
| 30,984 | M252P, T307Y, V308R, Q311E, N434F | 4.59 × 10−8 |
| 37,254 | S254G, H285S, N286F, L309A, N434Y | 4.88 × 10−8 |
| 34,889 | M252F, R255A, N286R, H310N, N434H | 5.60 × 10−8 |
| 33,342 | V308I, L309N, M428S, N434W, H435D | 5.62 × 10−8 |
| 31,505 | R255K, T307W, Q311D, H433Q, N434F | 6.39 × 10−8 |
| 33,080 | I253P, P257G, T307E, L309Y, N434H | 6.61 × 10−8 |
| 34,248 | I253D, S254D, T256V, T307G, N434F | 6.68 × 10−8 |
| 35,586 | M252T, I253V, N286G, H310L, N434H | 6.86 × 10−8 |
| 33,509 | P257K, T307A, V308F, M428H, N434Y | 6.93 × 10−8 |
| 37,174 | M252T, P257H, V308Y, L309F, N434F | 7.15 × 10−8 |
| 34,132 | M252E, S254F, P257Y, T307A, N434H | 7.67 × 10−8 |
| SVR 5mut | ||
| Variant # | Mutations | KD |
| 31,465 | S254G, T256Q, H310I, Q311W, N434H | 9.50 × 10−9 |
| 33,646 | M252Q, R255K, L309N, N434H, Y436R | 2.70 × 10−8 |
| 30,585 | H285Y, N286R, H310N, H433Y, N434H | 2.83 × 10−8 |
| 31,612 | S254T, K288G, H310S, M428V, N434F | 3.98 × 10−8 |
| 30,423 | M252R, P257E, T307Q, V308L, N434H | 4.17 × 10−8 |
| 31,505 | I253E, V308Y, H310D, L314I, N434Y | 4.66 × 10−8 |
| 38,216 | I253T, K288N, V308T, H310D, N434W | 6.22 × 10−8 |
| 34,132 | R255K, T256Q, V308H, L314S, N434Y | 7.10 × 10−8 |
| 32,098 | S254A, V308D, H310I, M428Q, N434H | 7.50 × 10−8 |
| 37,379 | M252P, T307Y, V308R, Q311E, N434F | 9.68 × 10−8 |
| 33,080 | S254G, H285S, N286F, L309A, N434Y | 1.03 × 10−7 |
| 36,338 | M252F, R255A, N286R, H310N, N434H | 1.03 × 10−7 |
| 31,469 | V308I, L309N, M428S, N434W, H435D | 1.14 × 10−7 |
| 37,661 | R255K, T307W, Q311D, H433Q, N434F | 1.23 × 10−7 |
| 36,149 | I253P, P257G, T307E, L309Y, N434H | 1.30 × 10−7 |
| 37,777 | I253D, S254D, T256V, T307G, N434F | 1.37 × 10−7 |
| 34,998 | M252T, I253V, N286G, H310L, N434H | 1.37 × 10−7 |
| 38,029 | P257K, T307A, V308F, M428H, N434Y | 1.44 × 10−7 |
| 34,712 | M252T, P257H, V308Y, L309F, N434F | 1.49 × 10−7 |
| 32,754 | M252E, S254F, P257Y, T307A, N434H | 1.51 × 10−7 |
| RFR 8mut | ||
| Variant # | Mutations | KD |
| 30,401 | M252W, I253D, P257A, V308F, L309E, N434W | 3.06 × 10−8 |
| 30,320 | L251Q, P257S, N286P, V308W, L309E, Q311A, N434H | 5.89 × 10−8 |
| 30,747 | M252G, T256A, L309D, N434W, H435E | 6.07 × 10−8 |
| 30,663 | I253S, P257V, K288G, T307G, N434H, Y436S | 7.74 × 10−8 |
| 30,083 | L251P, P257T, K288N, T307R, V308P, L309K, N434H | 8.02 × 10−8 |
| 30,582 | P257A, H285I, T307W, M428W, N434H | 9.00 × 10−8 |
| 30,549 | T256G, H285D, T307Y, L309T, N434F, H435T | 9.50 × 10−8 |
| 30,647 | M252W, T256P, P257A, K288L, T307S, M428I, N434H | 9.59 × 10−8 |
| 30,596 | L251Q, K288F, T307I, L309K, Q311T, M428I, N434W | 1.01 × 10−7 |
| 30,548 | L251R, M252H, V308R, L309D, N434H, Y436G | 1.06 × 10−7 |
| 30,915 | K288E, T307E, V308N, L309W, M428W, N434Y | 1.07 × 10−7 |
| 30,501 | M252Y, I253E, H285I, N286D, V308A, N434H | 1.10 × 10−7 |
| 30,848 | M252V, T256A, L309G, H433S, N434W, H435K | 1.11 × 10−7 |
| 30,912 | R255S, P257N, H285R, L309D, M428I, N434Y | 1.14 × 10−7 |
| 30,116 | M252I, I253S, N434W, H435P, Y436K | 1.17 × 10−7 |
| 30,780 | I253T, N286Q, V308P, Q311A, N434Y, Y436S | 1.21 × 10−7 |
| 30,625 | T256N, N286L, K288P, T307P, Q311A, M428L, N434Y | 1.23 × 10−7 |
| 30,245 | M252E, P257T, H285N, V308P, Q311L, N434Y | 1.24 × 10−7 |
| 30,045 | P257Y, Q311T, M428L, N434Y, H435P | 1.36 × 10−7 |
| 30,560 | P257A, K288T, T307F, Q311V, N434H, H435K | 1.38 × 10−7 |
| MLP 8mut | ||
| Variant # | Mutations | KD |
| 30,829 | M252W, I253D, P257A, V308F, L309E, N434W | 4.39 × 10−8 |
| 30,549 | L251Q, P257S, N286P, V308W, L309E, Q311A, N434H | 4.75 × 10−8 |
| 30,625 | M252G, T256A, L309D, N434W, H435E | 6.72 × 10−8 |
| 30,061 | I253S, P257V, K288G, T307G, N434H, Y436S | 7.36 × 10−8 |
| 30,860 | L251P, P257T, K288N, T307R, V308P, L309K, N434H | 8.15 × 10−8 |
| 30,721 | P257A, H285I, T307W, M428W, N434H | 8.68 × 10−8 |
| 30,045 | T256G, H285D, T307Y, L309T, N434F, H435T | 9.13 × 10−8 |
| 30,234 | M252W, T256P, P257A, K288L, T307S, M428I, N434H | 9.13 × 10−8 |
| 30,852 | L251Q, K288F, T307I, L309K, Q311T, M428I, N434W | 9.38 × 10−8 |
| 30,063 | L251R, M252H, V308R, L309D, N434H, Y436G | 9.75 × 10−8 |
| 30,022 | K288E, T307E, V308N, L309W, M428W, N434Y | 1.09 × 10−7 |
| 30,565 | M252Y, I253E, H285I, N286D, V308A, N434H | 1.09 × 10−7 |
| 30,490 | M252V, T256A, L309G, H433S, N434W, H435K | 1.10 × 10−7 |
| 30,669 | R255S, P257N, H285R, L309D, M428I, N434Y | 1.13 × 10−7 |
| 30,401 | M252I, I253S, N434W, H435P, Y436K | 1.13 × 10−7 |
| 30,583 | I253T, N286Q, V308P, Q311A, N434Y, Y436S | 1.17 × 10−7 |
| 30,245 | T256N, N286L, K288P, T307P, Q311A, M428L, N434Y | 1.21 × 10−7 |
| 30,211 | M252E, P257T, H285N, V308P, Q311L, N434Y | 1.26 × 10−7 |
| 30,596 | P257Y, Q311T, M428L, N434Y, H435P | 1.42 × 10−7 |
| 30,683 | P257A, K288T, T307F, Q311V, N434H, H435K | 1.43 × 10−7 |
| MLR 8mut | ||
| Variant # | Mutations | KD |
| 30,829 | M252W, I253D, P257A, V308F, L309E, N434W | 2.17 × 10−8 |
| 30,669 | L251Q, P257S, N286P, V308W, L309E, Q311A, N434H | 2.84 × 10−8 |
| 30,549 | M252G, T256A, L309D, N434W, H435E | 4.85 × 10−8 |
| 30,583 | I253S, P257V, K288G, T307G, N434H, Y436S | 7.34 × 10−8 |
| 30,022 | L251P, P257T, K288N, T307R, V308P, L309K, N434H | 7.90 × 10−8 |
| 30,721 | P257A, H285I, T307W, M428W, N434H | 8.09 × 10−8 |
| 30,063 | T256G, H285D, T307Y, L309T, N434F, H435T | 8.84 × 10−8 |
| 30,100 | M252W, T256P, P257A, K288L, T307S, M428I, N434H | 1.05 × 10−7 |
| 30,625 | L251Q, K288F, T307I, L309K, Q311T, M428I, N434W | 1.05 × 10−7 |
| 30,401 | L251R, M252H, V308R, L309D, N434H, Y436G | 1.11 × 10−7 |
| 30,683 | K288E, T307E, V308N, L309W, M428W, N434Y | 1.11 × 10−7 |
| 30,565 | M252Y, I253E, H285I, N286D, V308A, N434H | 1.12 × 10−7 |
| 30,225 | M252V, T256A, L309G, H433S, N434W, H435K | 1.13 × 10−7 |
| 30,045 | R255S, P257N, H285R, L309D, M428I, N434Y | 1.24 × 10−7 |
| 30,605 | M252I, I253S, N434W, H435P, Y436K | 1.30 × 10−7 |
| 30,860 | I253T, N286Q, V308P, Q311A, N434Y, Y436S | 1.36 × 10−7 |
| 30,061 | T256N, N286L, K288P, T307P, Q311A, M428L, N434Y | 1.40 × 10−7 |
| 30,245 | M252E, P257T, H285N, V308P, Q311L, N434Y | 1.52 × 10−7 |
| 30,211 | P257Y, Q311T, M428L, N434Y, H435P | 1.66 × 10−7 |
| 30,085 | P257A, K288T, T307F, Q311V, N434H, H435K | 1.67 × 10−7 |
| SVR 8mut | ||
| Variant # | Mutations | KD |
| 30,501 | M252W, I253D, P257A, V308F, L309E, N434W | 3.20 × 10−8 |
| 30,401 | L251Q, P257S, N286P, V308W, L309E, Q311A, N434H | 4.26 × 10−8 |
| 30,848 | M252G, T256A, L309D, N434W, H435E | 8.11 × 10−8 |
| 30,479 | I253S, P257V, K288G, T307G, N434H, Y436S | 1.19 × 10−7 |
| 30,829 | L251P, P257T, K288N, T307R, V308P, L309K, N434H | 1.26 × 10−7 |
| 30,045 | P257A, H285I, T307W, M428W, N434H | 1.27 × 10−7 |
| 30,397 | T256G, H285D, T307Y, L309T, N434F, H435T | 1.69 × 10−7 |
| 30,116 | M252W, T256P, P257A, K288L, T307S, M428I, N434H | 1.70 × 10−7 |
| 30,157 | L251Q, K288F, T307I, L309K, Q311T, M428I, N434W | 1.74 × 10−7 |
| 30,336 | L251R, M252H, V308R, L309D, N434H, Y436G | 1.86 × 10−7 |
| 30,061 | K288E, T307E, V308N, L309W, M428W, N434Y | 1.88 × 10−7 |
| 30,560 | M252Y, I253E, H285I, N286D, V308A, N434H | 1.89 × 10−7 |
| 30,549 | M252V, T256A, L309G, H433S, N434W, H435K | 2.00 × 10−7 |
| 30,891 | R255S, P257N, H285R, L309D, M428I, N434Y | 2.26 × 10−7 |
| 30,228 | M252I, I253S, N434W, H435P, Y436K | 2.27 × 10−7 |
| 30,911 | I253T, N286Q, V308P, Q311A, N434Y, Y436S | 2.31 × 10−7 |
| 30,386 | T256N, N286L, K288P, T307P, Q311A, M428L, N434Y | 2.50 × 10−7 |
| 30,605 | M252E, P257T, H285N, V308P, Q311L, N434Y | 2.62 × 10−7 |
| 30,150 | P257Y, Q311T, M428L, N434Y, H435P | 2.76 × 10−7 |
| 30,924 | P257A, K288T, T307F, Q311V, N434H, H435K | 2.79 × 10−7 |



References
- Zalevsky, J.; Chamberlain, A.K.; Horton, H.M.; Karki, S.; Leung, I.W.L.; Sproule, T.J.; Lazar, G.A.; Roopenian, D.C.; Desjarlais, J.R. Enhanced Antibody Half-Life Improves in Vivo Activity. Nat. Biotechnol. 2010, 28, 157–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, S.-Y.; Pegu, A.; Rudicell, R.S.; Yang, Z.; Joyce, M.G.; Chen, X.; Wang, K.; Bao, S.; Kraemer, T.D.; Rath, T.; et al. Enhanced Neonatal Fc Receptor Function Improves Protection against Primate SHIV Infection. Nature 2014, 514, 642–645. [Google Scholar] [CrossRef] [Green Version]
- Ramdani, Y.; Lamamy, J.; Watier, H.; Gouilleux-Gruart, V. Monoclonal Antibody Engineering and Design to Modulate FcRn Activities: A Comprehensive Review. Int. J. Mol. Sci. 2022, 23, 9604. [Google Scholar] [CrossRef] [PubMed]
- Liu, L. Pharmacokinetics of Monoclonal Antibodies and Fc-Fusion Proteins. Protein Cell 2018, 9, 15–32. [Google Scholar] [CrossRef] [Green Version]
- Ternant, D.; Arnoult, C.; Pugnière, M.; Dhommée, C.; Drocourt, D.; Perouzel, E.; Passot, C.; Baroukh, N.; Mulleman, D.; Tiraby, G.; et al. IgG1 Allotypes Influence the Pharmacokinetics of Therapeutic Monoclonal Antibodies through FcRn Binding. J. Immunol. 2016, 196, 607–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG Subclasses and Allotypes: From Structure to Effector Functions. Front. Immunol. 2014, 5, 520. [Google Scholar] [CrossRef] [Green Version]
- Dall’Acqua, W.F.; Kiener, P.A.; Wu, H. Properties of Human IgG1s Engineered for Enhanced Binding to the Neonatal Fc Receptor (FcRn). J. Biol. Chem. 2006, 281, 23514–23524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumet, C.; Pottier, J.; Gouilleux-Gruart, V.; Watier, H. Insights into the IgG Heavy Chain Engineering Patent Landscape as Applied to IgG4 Antibody Development. mAbs 2019, 11, 1341–1350. [Google Scholar] [CrossRef] [Green Version]
- Yeung, Y.A.; Leabman, M.K.; Marvin, J.S.; Qiu, J.; Adams, C.W.; Lien, S.; Starovasnik, M.A.; Lowman, H.B. Engineering Human IgG1 Affinity to Human Neonatal Fc Receptor: Impact of Affinity Improvement on Pharmacokinetics in Primates. J. Immunol. 2009, 182, 7663–7671. [Google Scholar] [CrossRef] [Green Version]
- Deng, R.; Loyet, K.M.; Lien, S.; Iyer, S.; DeForge, L.E.; Theil, F.-P.; Lowman, H.B.; Fielder, P.J.; Prabhu, S. Pharmacokinetics of Humanized Monoclonal Anti-Tumor Necrosis Factor-α Antibody and Its Neonatal Fc Receptor Variants in Mice and Cynomolgus Monkeys. Drug Metab. Dispos.: Biol. Fate Chem. 2010, 38, 600–605. [Google Scholar] [CrossRef] [Green Version]
- Ward, E.S.; Ober, R.J. Targeting FcRn to Generate Antibody-Based Therapeutics. Trends Pharmacol. Sci. 2018, 39, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.L.; Namenuk, A.K.; Hong, K.; Meng, Y.G.; Rae, J.; Briggs, J.; Xie, D.; Lai, J.; Stadlen, A.; Li, B.; et al. High Resolution Mapping of the Binding Site on Human IgG1 for Fc Gamma RI, Fc Gamma RII, Fc Gamma RIII, and FcRn and Design of IgG1 Variants with Improved Binding to the Fc Gamma R. J. Biol. Chem. 2001, 276, 6591–6604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oganesyan, V.; Damschroder, M.M.; Cook, K.E.; Li, Q.; Gao, C.; Wu, H.; Dall’Acqua, W.F. Structural Insights into Neonatal Fc Receptor-Based Recycling Mechanisms. J. Biol. Chem. 2014, 289, 7812–7824. [Google Scholar] [CrossRef] [Green Version]
- Petkova, S.B.; Akilesh, S.; Sproule, T.J.; Christianson, G.J.; Al Khabbaz, H.; Brown, A.C.; Presta, L.G.; Meng, Y.G.; Roopenian, D.C. Enhanced Half-Life of Genetically Engineered Human IgG1 Antibodies in a Humanized FcRn Mouse Model: Potential Application in Humorally Mediated Autoimmune Disease. Int. Immunol. 2006, 18, 1759–1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monnet, C.; Jorieux, S.; Urbain, R.; Fournier, N.; Bouayadi, K.; De Romeuf, C.; Behrens, C.K.; Fontayne, A.; Mondon, P. Selection of IgG Variants with Increased FcRn Binding Using Random and Directed Mutagenesis: Impact on Effector Functions. Front. Immunol. 2015, 6, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Booth, B.J.; Ramakrishnan, B.; Narayan, K.; Wollacott, A.M.; Babcock, G.J.; Shriver, Z.; Viswanathan, K. Extending Human IgG Half-Life Using Structure-Guided Design. mAbs 2018, 10, 1098–1110. [Google Scholar] [CrossRef] [PubMed]
- Mackness, B.C.; Jaworski, J.A.; Boudanova, E.; Park, A.; Valente, D.; Mauriac, C.; Pasquier, O.; Schmidt, T.; Kabiri, M.; Kandira, A.; et al. Antibody Fc Engineering for Enhanced Neonatal Fc Receptor Binding and Prolonged Circulation Half-Life. mAbs 2019, 11, 1276–1288. [Google Scholar] [CrossRef] [Green Version]
- Pierce, B.; Weng, Z. ZRANK: Reranking Protein Docking Predictions with an Optimized Energy Function. Proteins Struct. Funct. Bioinform. 2007, 67, 1078–1086. [Google Scholar] [CrossRef]
- Kastritis, P.L.; Bonvin, A.M.J.J. Are Scoring Functions in Protein−Protein Docking Ready to Predict Interactomes? Clues from a Novel Binding Affinity Benchmark. J. Proteome Res. 2011, 10, 921–922. [Google Scholar] [CrossRef] [Green Version]
- Gromiha, M.M.; Yugandhar, K.; Jemimah, S. Protein–Protein Interactions: Scoring Schemes and Binding Affinity. Curr. Opin. Struct. Biol. 2017, 44, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Spassov, V.Z.; Yan, L. pH-Selective Mutagenesis of Protein-Protein Interfaces: In Silico Design of Therapeutic Antibodies with Prolonged Half-Life. Proteins: Struct. Funct. Bioinform. 2013, 81, 704–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Zheng, F.; Zhan, C.-G. Binding Structures and Energies of the Human Neonatal Fc Receptor with Human Fc and Its Mutants by Molecular Modeling and Dynamics Simulations. Mol. BioSyst. 2013, 9, 3047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igawa, T.; Maeda, A.; Haraya, K.; Tachibana, T.; Iwayanagi, Y.; Mimoto, F.; Higuchi, Y.; Ishii, S.; Tamba, S.; Hironiwa, N.; et al. Engineered Monoclonal Antibody with Novel Antigen-Sweeping Activity in Vivo. PLoS ONE 2013, 8, e63236. [Google Scholar] [CrossRef]
- Maas, B.M.; Cao, Y. A Minimal Physiologically Based Pharmacokinetic Model to Investigate FcRn-Mediated Monoclonal Antibody Salvage: Effects of Kon, Koff, Endosome Trafficking, and Animal Species. mAbs 2018, 10, 1322–1331. [Google Scholar] [CrossRef] [Green Version]
- Horton, N.; Lewis, M. Calculation of the Free Energy of Association for Protein Complexes. Protein Sci. 1992, 1, 169–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerois, R.; Nielsen, J.E.; Serrano, L. Predicting Changes in the Stability of Proteins and Protein Complexes: A Study of More than 1000 Mutations. J. Mol. Biol. 2002, 320, 369–387. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lai, L.; Wang, S. Further Development and Validation of Empirical Scoring Functions for Structure-Based Binding Affinity Prediction. J. Comput.-Aided Mol. Des. 2002, 16, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Varoquaux, G.; Buitinck, L.; Louppe, G.; Grisel, O.; Pedregosa, F.; Mueller, A. Scikit-Learn. GetMobile: Mob. Comput. Commun. 2015, 19, 29–33. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX Web Server: An Online Force Field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [Green Version]
- Borrok, M.J.; Wu, Y.; Beyaz, N.; Yu, X.-Q.; Oganesyan, V.; Dall’Acqua, W.F.; Tsui, P. pH-Dependent Binding Engineering Reveals an FcRn Affinity Threshold That Governs IgG Recycling. J. Biol. Chem. 2015, 290, 4282–4290. [Google Scholar] [CrossRef] [Green Version]
- Walters, B.T.; Jensen, P.F.; Larraillet, V.; Lin, K.; Patapoff, T.; Schlothauer, T.; Rand, K.D.; Zhang, J. Conformational Destabilization of Immunoglobulin G Increases the Low pH Binding Affinity with the Neonatal Fc Receptor. J. Biol. Chem. 2016, 291, 1817–1825. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Datasets | ||
| Name | Number of Variants | Selection Criteria |
| First learning set (FLS) | 1099 | Affinities measured by SPR at 25 °C, pH 7. |
| Second learning set (SLS) | 1323 | FLS variants + 224 variants with affinities only measured at pH 6. |
| Algorithms | ||
| Name | Description | |
| Support vector regressor (SVR) | The objective of support vector machines (SVMs) is to find the hyperplane separating at best the two categories of instances defined in a training sample. Support vector regression (SVR) uses the same principle, adding a constraint on the maximal distance between the instances and the hyperplane. | |
| Multi-linear regression (MLR) | Multiple linear regression optimizes a linear function of the parameters. | |
| Multi-layer perceptron (MLP) | An MLP is a class of feedforward artificial neural networks (ANNs) with at least three layers of nodes (input, hidden, and output) and the neurons of hidden and output layers using non-linear activation functions. | |
| Random forest regressor (RFR) | A random forest is a meta-estimator that fits a number of classifying decision trees on various sub-samples of the dataset and uses averaging to improve the predictive accuracy and control overfitting. | |
| Tocilizumab * | T8 * | T3 * | C7 | B5 | A3 | |
|---|---|---|---|---|---|---|
| Mutations | None | M252Y/N286E/T307Q/V308P/Q311A/N434Y/Y436V | M252Y/T307D/N434Y | T256E/N286H/K288E/V308P/L309D/N434Y/Y436K | T256Y/H285Q/N286D/V308A/N434Y | M252W/M428K/N434W |
| KD at pH7 (patent) | 8.8 × 10−5 | 4.4 × 10−9 | 2.1 × 10−7 | |||
| KD at pH7 (this work) | NB | 7.8 × 10−9 | 3.8 × 10−7 | 1.6 × 10−7 | 6.2 × 10−7 | 5.7 × 10−7 |
| KD at pH6 (this work) | 3.8 × 10−7 | 1.3 × 10−9 | 1.3 × 10−8 | 3.4 × 10−8 | 4.5 × 10−8 | 1.1 × 10−8 |
| Prediction setting | ||||||
| SVR/FLS | 6.91 × 10−7 | 7.29 × 10−9 | 2.54 × 10−8 | 1.90 × 10−7 | 1.90 × 10−7 | 1.40 × 10−7 |
| SVR/SLS | 6.70 × 10−6 | 1.30 × 10−8 | 9.50 × 10−8 | 2.30 × 10−7 | 4.20 × 10−7 | 4.20 × 10−7 |
| MLR/FLD | 6.20 × 10−7 | 1.30 × 10−8 | 8.00 × 10−8 | 1.80 × 10−8 | 4.40 × 10−8 | 1.30 × 10−7 |
| MLR/SLS | 1.00 × 10−4 | 5.40 × 10−8 | 6.50 × 10−8 | 7.40 × 10−8 | 1.40 × 10−7 | 2.70 × 10−7 |
| MLP/FLS | 8.00 × 10−7 | 1.30 × 10−8 | 8.00 × 10−8 | 8.70 × 10−8 | 1.30 × 10−7 | 7.70 × 10−8 |
| MLP/SLS | 1.90 × 10−5 | 4.40 × 10−8 | 6.00 × 10−8 | 6.80 × 10−7 | 7.10 × 10−6 | 6.90 × 10−7 |
| RFR/FLS | 1.20 × 10−6 | 3.80 × 10−9 | 2.47 × 10−7 | 6.00 × 10−8 | 1.60 × 10−7 | 3.30 × 10−7 |
| RFR/SLS | 3.40 × 10−6 | 4.10 × 10−9 | 1.50 × 10−7 | 4.90 × 10−8 | 2.10 × 10−7 | 3.20 × 10−7 |
| SVR/FLS | SVR/SLS | MLR/FLS | MLR/SLS | MLP/FLS | MLP/SLS | RFR/FLS | RFR/SLS | |
|---|---|---|---|---|---|---|---|---|
| Log KD MAE (6 Abs) | 0.64 | 0.19 | 0.81 | 0.11 | 0.63 | 0.26 | 0.52 | 0.47 |
| Pearson correlation coefficient (6 Abs) | 0.88 | 0.98 | 0.91 | 0.89 | 0.98 | 0.84 | 0.91 | 0.97 |
| Log KD Maximum error (6 Abs) | 2.11 | 1.12 | 2.15 | 1.09 | 2.04 | 1.06 | 1.87 | 1.41 |
| Log KD MAE (A, B, C Abs) | 0.35 | 0.05 | 0.91 | 0.44 | 0.60 | 0.59 | 0.42 | 0.41 |
| Pearson correlation coefficient (A, B, C Abs) | −0.45 | 0.99 | 0.81 | 0.83 | 0.35 | 0.55 | 0.88 | 0.96 |
| Log KD Maximum error (A, B, C Abs) | 0.61 | 0.17 | 1.15 | 0.65 | 0.87 | 1.06 | 0.59 | 0.51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dumet, C.; Pugnière, M.; Henriquet, C.; Gouilleux-Gruart, V.; Poupon, A.; Watier, H. Harnessing Fc/FcRn Affinity Data from Patents with Different Machine Learning Methods. Int. J. Mol. Sci. 2023, 24, 5724. https://doi.org/10.3390/ijms24065724
Dumet C, Pugnière M, Henriquet C, Gouilleux-Gruart V, Poupon A, Watier H. Harnessing Fc/FcRn Affinity Data from Patents with Different Machine Learning Methods. International Journal of Molecular Sciences. 2023; 24(6):5724. https://doi.org/10.3390/ijms24065724
Chicago/Turabian StyleDumet, Christophe, Martine Pugnière, Corinne Henriquet, Valérie Gouilleux-Gruart, Anne Poupon, and Hervé Watier. 2023. "Harnessing Fc/FcRn Affinity Data from Patents with Different Machine Learning Methods" International Journal of Molecular Sciences 24, no. 6: 5724. https://doi.org/10.3390/ijms24065724
APA StyleDumet, C., Pugnière, M., Henriquet, C., Gouilleux-Gruart, V., Poupon, A., & Watier, H. (2023). Harnessing Fc/FcRn Affinity Data from Patents with Different Machine Learning Methods. International Journal of Molecular Sciences, 24(6), 5724. https://doi.org/10.3390/ijms24065724








