Combined Inhibition of Hedgehog and HDAC6: In Vitro and In Vivo Studies Reveal a New Role for Lysosomal Stress in Reducing Glioblastoma Cell Viability
Abstract
1. Introduction
2. Results
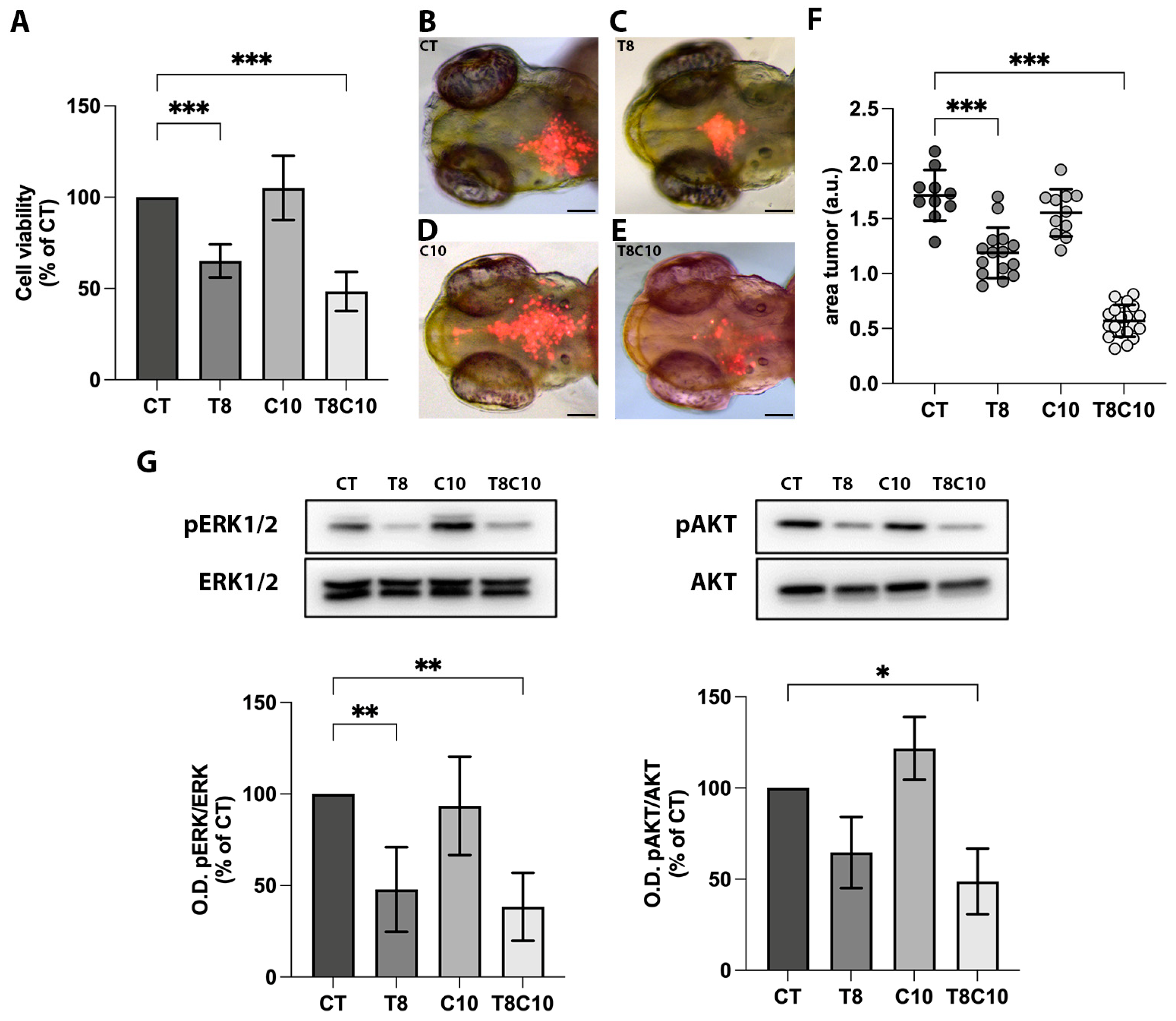
2.1. Combined Inhibition of HDAC6 and the Hh Pathway Synergistically Suppressed U87-MG Cell Viability In Vitro and In Vivo
2.2. Combined Inhibition of HDAC6 and the Hh Pathway Impaired Autophagy in U87-MG Cells and Zebrafish Embryos
2.3. Combined Inhibition of the HDAC6 and Hh Pathway Induced Lysosome Alteration in U87-MG Cells and Zebrafish Embryos
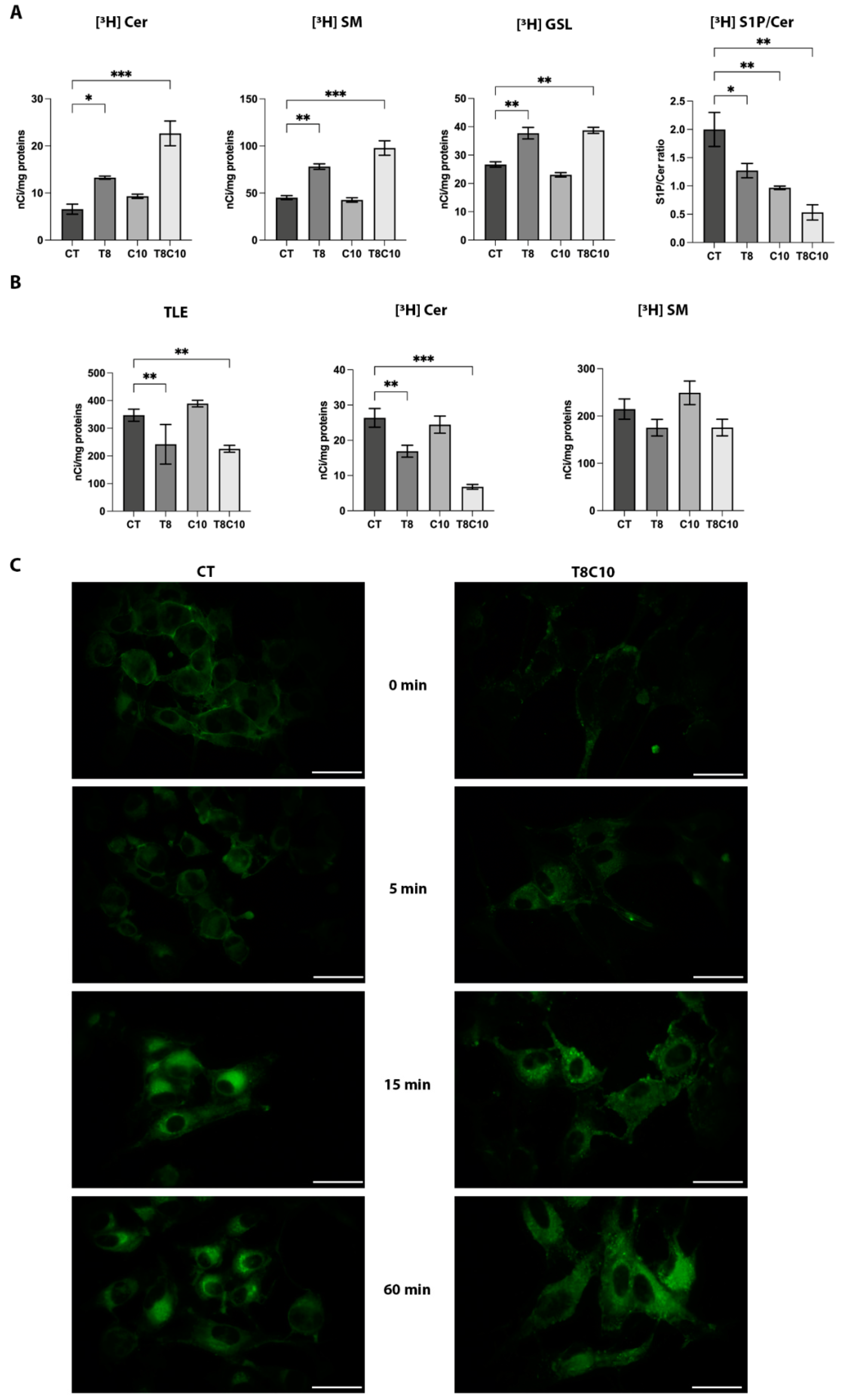
2.4. Combined Inhibition of HDAC6 and Hh Pathway Impaired the Lysosomal Catabolism of Complex Sphingolipids
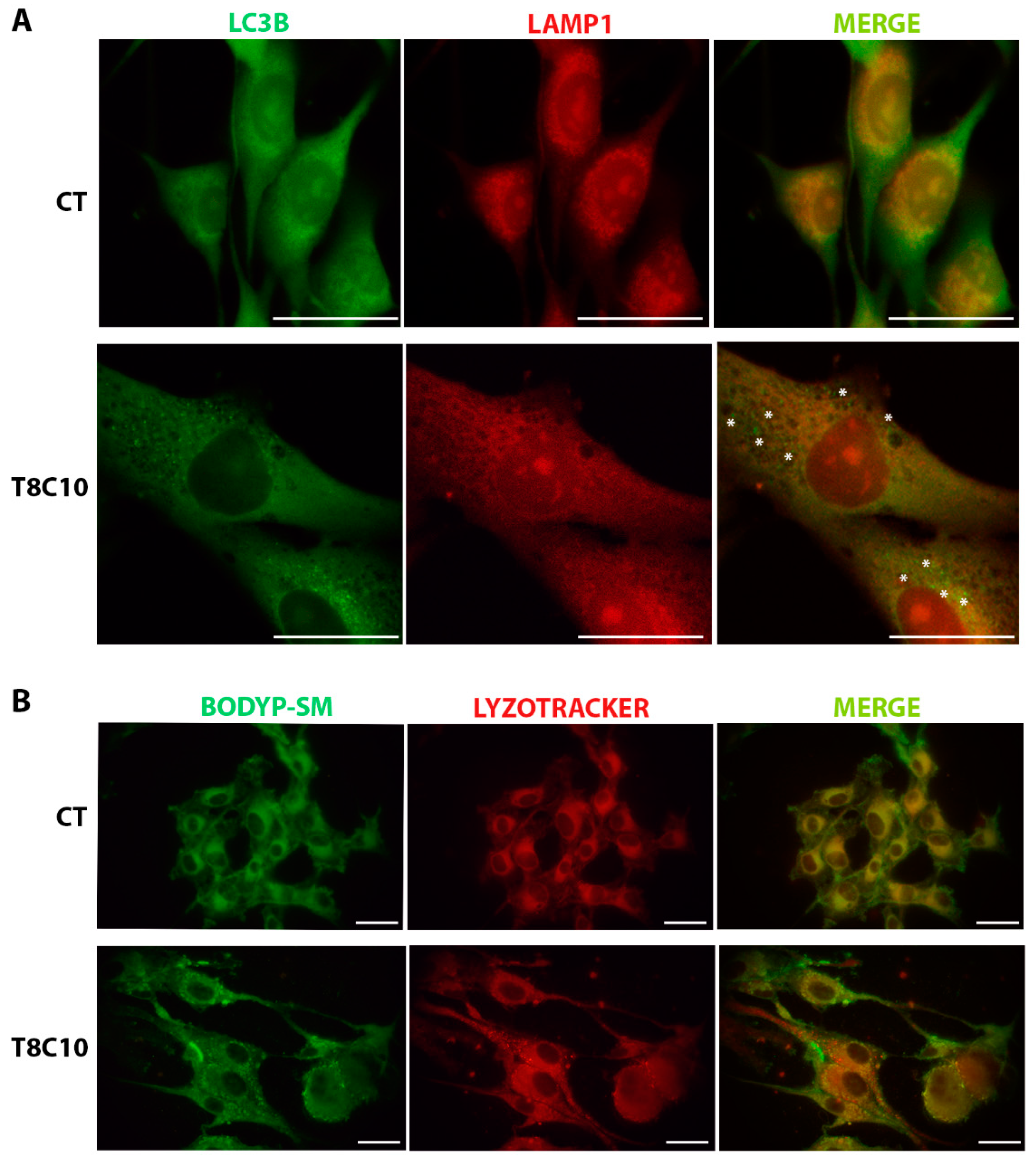
2.5. Tubastatin A and Cyclopamine Prevented Autophagosome and Endosome Fusion with Lysosomes
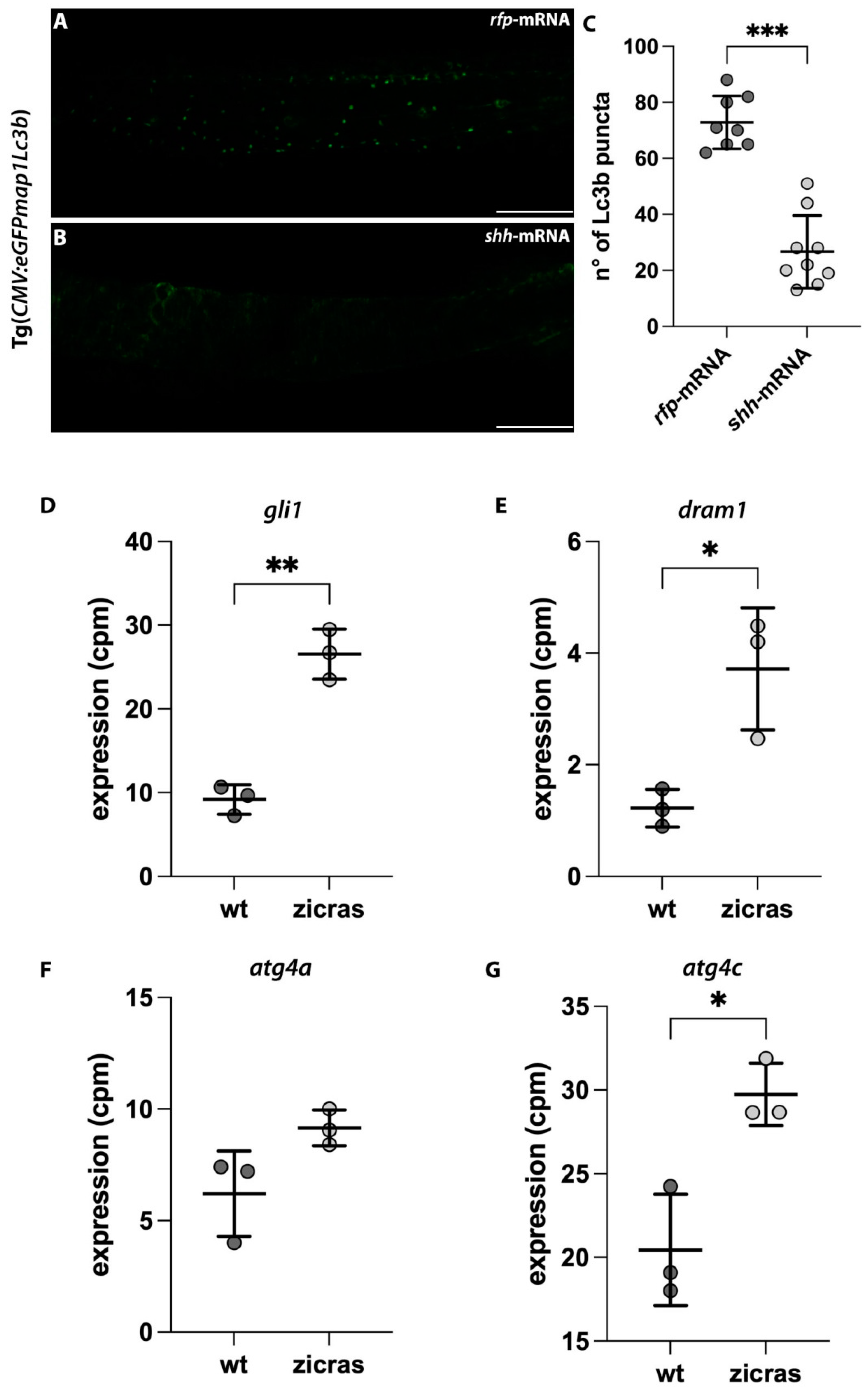
2.6. In Vivo Zebrafish Models of GBM Recapitulated the Alteration in the Autophagic Process Shown in U87-MG Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Animals
4.3. mRNA Microinjection
4.4. Pharmacological Treatments
4.5. Cell Viability Assays
4.6. Xenograft of U87-MG Treated Cells
4.7. Protein Quantification and Immunoblotting
4.8. Metabolic Labeling of U87-MG Sphingolipids with [1-3H] Sphingosine
4.9. [Sph-3H] Sphingomyelin Metabolism
4.10. Lysotracker Assays
4.11. Acridine Orange Staining
4.12. Uptake and Intracellular Distribution of BODIPY-C5-Sphingomyelin
4.13. Immunocytochemistry and Immunofluorescence
4.14. Image Processing and Statistical Analyses
4.15. GBM Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sasmita, A.O.; Wong, Y.; Ling, A.P.K. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia-Pac. J. Clin. Oncol. 2018, 14, 40–51. [Google Scholar] [CrossRef]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Stoyanov, G.S.; Dzhenkov, D.; Ghenev, P.; Iliev, B.; Enchev, Y.; Tonchev, A.B. Cell biology of glioblastoma multiforme: From basic science to diagnosis and treatment. Med. Oncol. 2018, 35, 27. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, S.K.; Brothers, S.; Wahlestedt, C. Emerging treatment strategies for glioblastoma multiforme. EMBO Mol Med 2014, 6, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Ou, A.; Yung, W.A.; Majd, N. Molecular mechanisms of treatment resistance in glioblastoma. Int. J. Mol. Sci. 2021, 22, 351. [Google Scholar] [CrossRef]
- Giussani, P.; Tringali, C.; Riboni, L.; Viani, P.; Venerando, B. Sphingolipids: Key regulators of apoptosis and pivotal players in cancer drug resistance. Int. J. Mol. Sci. 2014, 15, 4356–4392. [Google Scholar] [CrossRef]
- Giussani, P.; Bassi, R.; Anelli, V.; Brioschi, L.; De Zen, F.; Riccitelli, E.; Caroli, M.; Campanella, M.; Gaini, S.M.; Viani, P.; et al. Glucosylceramide synthase protects glioblastoma cells against autophagic and apoptotic death induced by temozolomide and paclitaxel. Cancer Invest. 2012, 30, 27–37. [Google Scholar] [CrossRef]
- Riboni, L.; Campanella, R.; Bassi, R.; Villani, R.; Gaini, S.M.; Boneschi, F.M.; Viani, P.; Tettamanti, G. Ceramide levels are inversely associated with malignant progression of human glial tumors. Glia 2002, 39, 105–113. [Google Scholar] [CrossRef]
- Cochrane, C.R.; Szczepny, A.; Watkins, D.; Cain, J.E. Hedgehog signaling in the maintenance of cancer stem cells. Cancers 2015, 7, 1554–1585. [Google Scholar] [CrossRef]
- Carballo, G.B.; Honorato, J.R.; De Lopes, G.P.F.; de Sampaio e Spohr, T.C.L. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. 2018, 16, 11. [Google Scholar] [CrossRef]
- Bar, E.E.; Chaudhry, A.; Lin, A.; Fan, X.; Schreck, K.; Matsui, W.; Piccirillo, S.; Vescovi, A.L.; DiMeco, F.; Olivi, A.; et al. Cyclopamine-Mediated Hedgehog Pathway Inhibition Depletes Stem-Like Cancer Cells in Glioblastoma. Stem Cells 2007, 25, 2524–2533. [Google Scholar] [CrossRef]
- Morgenroth, A.; Vogg, A.; Ermert, K.; Zlatopolskiy, B.; Mottaghy, F.M. Hedgehog signaling sensitizes Glioma stem cells to endogenous nano-irradiation. Oncotarget 2014, 5, 5483–5493. [Google Scholar] [CrossRef]
- Shevde, L.A.; Samant, R.S. Nonclassical HEDGEHOG-GLI signaling and its clinical implications. Int. J. Cancer 2014, 135, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Aldape, K.; Zadeh, G.; Mansouri, S.; Reifenberger, G.; von Deimling, A. Glioblastoma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015, 129, 829–848. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hu, P.; Tang, F.; Lian, H.; Chen, X.; Zhang, Y.; He, X.; Liu, W.; Xie, C. HDAC6 promotes cell proliferation and confers resistance to temozolomide in glioblastoma. Cancer Lett. 2016, 379, 134–142. [Google Scholar] [CrossRef]
- Lee, S.Y. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016, 3, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Urdiciain, A.; Erausquin, E.; Meléndez, B.; Rey, J.; Idoate, M.; Castresana, J.S. Tubastatin A, an inhibitor of HDAC6, enhances temozolomide-induced apoptosis and reverses the malignant phenotype of glioblastoma cells. Int. J. Oncol. 2019, 54, 1797–1808. [Google Scholar] [CrossRef]
- Simpson, J.E.; Gammoh, N. The impact of autophagy during the development and survival of glioblastoma: Role of autophagy in glioblastoma. Open Biol. 2020, 10, 200184. [Google Scholar] [CrossRef]
- Xu, Y.; An, Y.; Wang, X.; Zha, W.; Li, X. Inhibition of the Hedgehog pathway induces autophagy in pancreatic ductal adenocarcinoma cells. Oncol. Rep. 2014, 31, 707–712. [Google Scholar] [CrossRef]
- Chang, P.; Li, H.; Hu, H.; Li, Y.; Wang, T. The Role of HDAC6 in Autophagy and NLRP3 Inflammasome. Front. Immunol. 2021, 12, 763831. [Google Scholar] [CrossRef] [PubMed]
- Pezzotta, A.; Gentile, I.; Genovese, D.; Totaro, M.G.; Battaglia, C.; Leung, A.Y.-H.; Fumagalli, M.; Parma, M.; Cazzaniga, G.; Fazio, G.; et al. HDAC6 inhibition decreases leukemic stem cell expansion driven by Hedgehog hyperactivation by restoring primary ciliogenesis. Pharmacol. Res. 2022, 183, 106378. [Google Scholar] [CrossRef] [PubMed]
- Mizgirev, I.; Revskoy, S. Generation of clonal zebrafish lines and transplantable hepatic tumors. Nat. Protoc. 2010, 5, 383–394. [Google Scholar] [CrossRef]
- Zhao, S.; Huang, J.; Ye, J. A fresh look at zebrafish from the perspective of cancer research. J. Exp. Clin. Cancer Res. 2015, 34, 80. [Google Scholar] [CrossRef] [PubMed]
- Marques, I.J.; Weiss, F.U.; Vlecken, D.H.; Nitsche, C.; Bakkers, J.; Lagendijk, A.K.; Partecke, L.I.; Heidecke, C.-D.; Lerch, M.M.; Bagowski, C.P. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer 2009, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Kitambi, S.S.; Toledo, E.M.; Usoskin, D.; Wee, S.; Harisankar, A.; Svensson, R.; Sigmundsson, K.; Kalderén, C.; Niklasson, M.; Kundu, S.; et al. Vulnerability of glioblastoma cells to catastrophic vacuolization and death induced by a small molecule. Cell 2014, 157, 313–328. [Google Scholar] [CrossRef]
- Vittori, M.; Motaln, H.; Turnšek, T.L. The Study of Glioma by Xenotransplantation in Zebrafish Early Life Stages. J. Histochem. Cytochem. 2015, 63, 749–761. [Google Scholar] [CrossRef]
- Auzmendi-Iriarte, J.; Saenz-Antoñanzas, A.; Mikelez-Alonso, I.; Carrasco-Garcia, E.; Tellaetxe-Abete, M.; Lawrie, C.H.; Sampron, N.; Cortajarena, A.L.; Matheu, A. Characterization of a new small-molecule inhibitor of HDAC6 in glioblastoma. Cell Death Dis. 2020, 11, 417. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Runwal, G.; Stamatakou, E.; Siddiqi, F.; Puri, C.; Zhu, Y.; Rubinsztein, D.C. LC3-positive structures are prominent in autophagy-deficient cells. Sci. Rep. 2019, 9, 10147. [Google Scholar] [CrossRef]
- He, C.; Bartholomew, C.; Zhou, W.; Klionsky, D.J. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy 2009, 5, 520–526. [Google Scholar] [CrossRef]
- Mayrhofer, M.; Gourain, V.; Reischl, M.; Affaticati, P.; Jenett, A.; Joly, J.-S.; Benelli, M.; Demichelis, F.; Poliani, P.L.; Sieger, D.; et al. A novel brain tumour model in zebrafish reveals the role of YAP activation in MAPK- and PI3K-induced malignant growth. DMM Dis. Model. Mech. 2017, 10, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Galavotti, S.; Bartesaghi, S.; Faccenda, D.; Shaked-Rabi, M.; Sanzone, S.; McEvoy, A.; Dinsdale, D.; Condorelli, F.; Brandner, S.; Campanella, M.; et al. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene 2013, 32, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.; Chen, W.; Orr, A.B.; Spitsbergen, J.M.; Jia, S.; Eden, C.J.; Henson, E.H.; Taylor, M.R. Oncogenic KRAS promotes malignant brain tumors in zebrafish. Mol. Cancer 2015, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, D.; Qian, Z.; Cui, D.; Gao, L.; Lou, M. Hedgehog/Gli1 signaling pathway regulates MGMT expression and chemoresistance to temozolomide in human glioblastoma. Cancer Cell Int. 2017, 17, 117. [Google Scholar] [CrossRef]
- Kim, G.W.; Lee, D.H.; Yeon, S.-K.; Jeon, Y.H.; Yoo, J.; Lee, S.W.; Kwon, S.H. Temozolomide-resistant glioblastoma depends on HDAC6 activity through regulation of DNA mismatch repair. Anticancer Res. 2019, 39, 6731–6741. [Google Scholar] [CrossRef]
- Yang, W.; Liu, Y.; Gao, R.; Yu, H.; Sun, T. HDAC6 inhibition induces glioma stem cells differentiation and enhances cellular radiation sensitivity through the SHH/Gli1 signaling pathway. Cancer Lett. 2018, 415, 164–176. [Google Scholar] [CrossRef]
- Urdiciain, A.; Erausquin, E.; Zelaya, M.; Zazpe, I.; Lanciego, J.; Meléndez, B.; Rey, J.; Idoate, M.; Galdo, N.R.-D.; Castresana, J. Silencing of histone deacetylase 6 decreases cellular malignancy and contributes to primary cilium restoration, epithelial-to-mesenchymal transition reversion, and autophagy inhibition in glioblastoma cell lines. Biology 2021, 10, 467. [Google Scholar] [CrossRef]
- Traver, D.; Herbomel, P.; Patton, E.; Murphey, R.D.; Yoder, J.A.; Litman, G.W.; Catic, A.; Amemiya, C.T.; Zon, L.I.; Trede, N.S. The Zebrafish as a Model Organism to Study Development of the Immune System. Adv. Immunol. 2003, 81, 253–330. [Google Scholar] [CrossRef]
- Xiao, J.; Glasgow, E.; Agarwal, S. Zebrafish Xenografts for Drug Discovery and Personalized Medicine. Trends Cancer 2020, 6, 569–579. [Google Scholar] [CrossRef]
- Ke, X.X.; Pang, Y.; Chen, K.; Zhang, D.; Wang, F.; Zhu, S.; Mao, J.; Hu, X.; Zhang, G.; Cui, H. Knockdown of arsenic resistance protein 2 inhibits human glioblastoma cell proliferation through the MAPK/ERK pathway. Oncol. Rep. 2018, 40, 3313–3322. [Google Scholar] [CrossRef]
- Li, X.; Wu, C.; Chen, N.; Gu, H.; Yen, A.; Cao, L.; Wang, E.; Wang, L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 2016, 7, 33440. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Ju, D. Hedgehog signaling pathway and autophagy in cancer. Int. J. Mol. Sci. 2018, 19, 2279. [Google Scholar] [CrossRef]
- Ito, H.; Daido, S.; Kanzawa, T.; Kondo, S.; Kondo, Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int. J. Oncol. 2005, 26, 1401–1410. [Google Scholar] [CrossRef] [PubMed]
- Escamilla-Ramírez, A.; Castillo-Rodríguez, R.A.; Zavala-Vega, S.; Jimenez-Farfan, D.; Anaya-Rubio, I.; Briseño, E.; Palencia, G.; Guevara, P.; Cruz-Salgado, A.; Sotelo, J.; et al. Autophagy as a potential therapy for malignant glioma. Pharmaceuticals 2020, 13, 156. [Google Scholar] [CrossRef]
- Jimenez-Sanchez, M.; Menzies, F.; Chang, Y.; Simecek, N.; Neufeld, T.; Rubinsztein, D.C. The Hedgehog signalling pathway regulates autophagy. Nat. Commun. 2012, 3, 1200. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, L.; Lotfi, P.; Pal, R.; di Ronza, A.; Sharma, J.; Sardiello, M. Lysosome biogenesis in health and disease. J. Neurochem. 2019, 148, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, Z.; Zhu, J.; Zhan, X. Roles of cortactin, an actin polymerization mediator, in cell endocytosis. Acta Biochim. Biophys. Sin. 2006, 38, 95–103. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, Z.; Zhang, Y.; Yong, S.; Salas-Burgos, A.; Koomen, J.; Olashaw, N.; Parsons, J.T.; Yang, X.-J.; Dent, S.R.; et al. HDAC6 Modulates Cell Motility by Altering the Acetylation Level of Cortactin. Mol. Cell 2007, 27, 197–213. [Google Scholar] [CrossRef]
- Dowdy, T.; Zhang, L.; Celiku, O.; Movva, S.; Lita, A.; Ruiz-Rodado, V.; Gilbert, M.R.; Larion, M. Sphingolipid pathway as a source of vulnerability in IDH1mut glioma. Cancers 2020, 12, 2910. [Google Scholar] [CrossRef]
- Tea, M.N.; Poonnoose, S.I.; Pitson, S.M. Targeting the sphingolipid system as a therapeutic direction for glioblastoma. Cancers 2020, 12, 111. [Google Scholar] [CrossRef]
- Gaudenzi, G.; Vitale, G. Transplantable zebrafish models of neuroendocrine tumors. Ann. Endocrinol. 2019, 80, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Riboni, L.; Viani, P.; Bassi, R.; Giussani, P.; Tettamanti, G. Cultured granule cells and astrocytes from cerebellum differ in metabolizing sphingosine. J. Neurochem. 2000, 75, 503–510. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pezzotta, A.; Brioschi, L.; Carbone, S.; Mazzoleni, B.; Bontempi, V.; Monastra, F.; Mauri, L.; Marozzi, A.; Mione, M.; Pistocchi, A.; et al. Combined Inhibition of Hedgehog and HDAC6: In Vitro and In Vivo Studies Reveal a New Role for Lysosomal Stress in Reducing Glioblastoma Cell Viability. Int. J. Mol. Sci. 2023, 24, 5771. https://doi.org/10.3390/ijms24065771
Pezzotta A, Brioschi L, Carbone S, Mazzoleni B, Bontempi V, Monastra F, Mauri L, Marozzi A, Mione M, Pistocchi A, et al. Combined Inhibition of Hedgehog and HDAC6: In Vitro and In Vivo Studies Reveal a New Role for Lysosomal Stress in Reducing Glioblastoma Cell Viability. International Journal of Molecular Sciences. 2023; 24(6):5771. https://doi.org/10.3390/ijms24065771
Chicago/Turabian StylePezzotta, Alex, Loredana Brioschi, Sabrina Carbone, Beatrice Mazzoleni, Vittorio Bontempi, Federica Monastra, Laura Mauri, Anna Marozzi, Marina Mione, Anna Pistocchi, and et al. 2023. "Combined Inhibition of Hedgehog and HDAC6: In Vitro and In Vivo Studies Reveal a New Role for Lysosomal Stress in Reducing Glioblastoma Cell Viability" International Journal of Molecular Sciences 24, no. 6: 5771. https://doi.org/10.3390/ijms24065771
APA StylePezzotta, A., Brioschi, L., Carbone, S., Mazzoleni, B., Bontempi, V., Monastra, F., Mauri, L., Marozzi, A., Mione, M., Pistocchi, A., & Viani, P. (2023). Combined Inhibition of Hedgehog and HDAC6: In Vitro and In Vivo Studies Reveal a New Role for Lysosomal Stress in Reducing Glioblastoma Cell Viability. International Journal of Molecular Sciences, 24(6), 5771. https://doi.org/10.3390/ijms24065771








