Monitoring Nrf2/ARE Pathway Activity with a New Zebrafish Reporter System
Abstract
:1. Introduction
2. Results
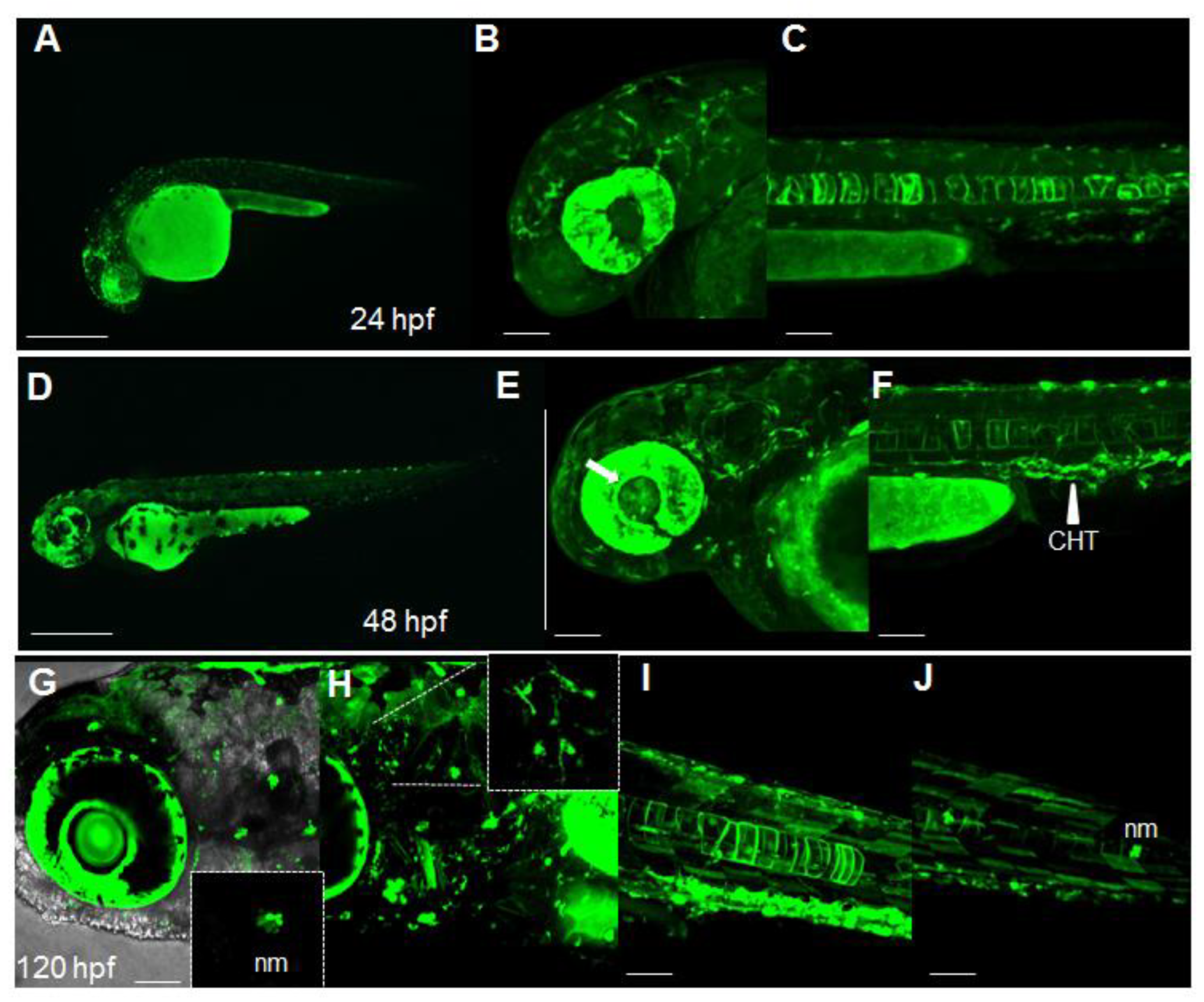
2.1. Dynamic Spatiotemporal Reporter Expression in the Transgenic Line Tg(8XAORE:EGFP)ia201
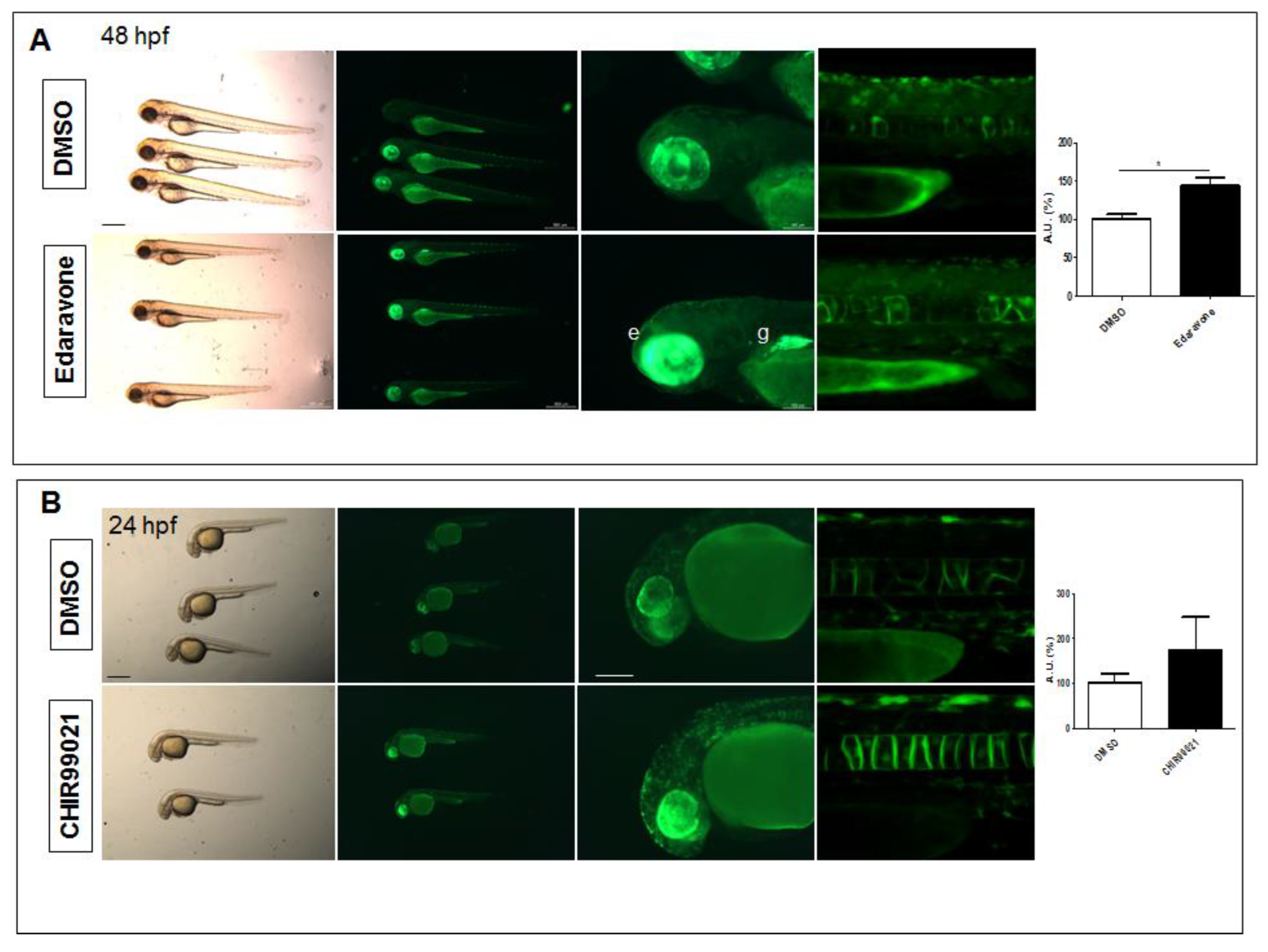
2.2. The Tg(8xAORE:EGFP)ia201 Line Is Responsive to Nrf2 Pathway Modulators
2.3. The Nrf2 Reporter Transgenics Serve as a Platform for the Identification of Nrf2 Pathway-Mediated Biological Activity of Small Molecules
2.4. Beta Glucocerebrosidase Knockdown Does Not Significantly Affect Nrf2 Reporter Activation but Induces the Upregulation of a Restricted Number of Nrf2 Target Genes
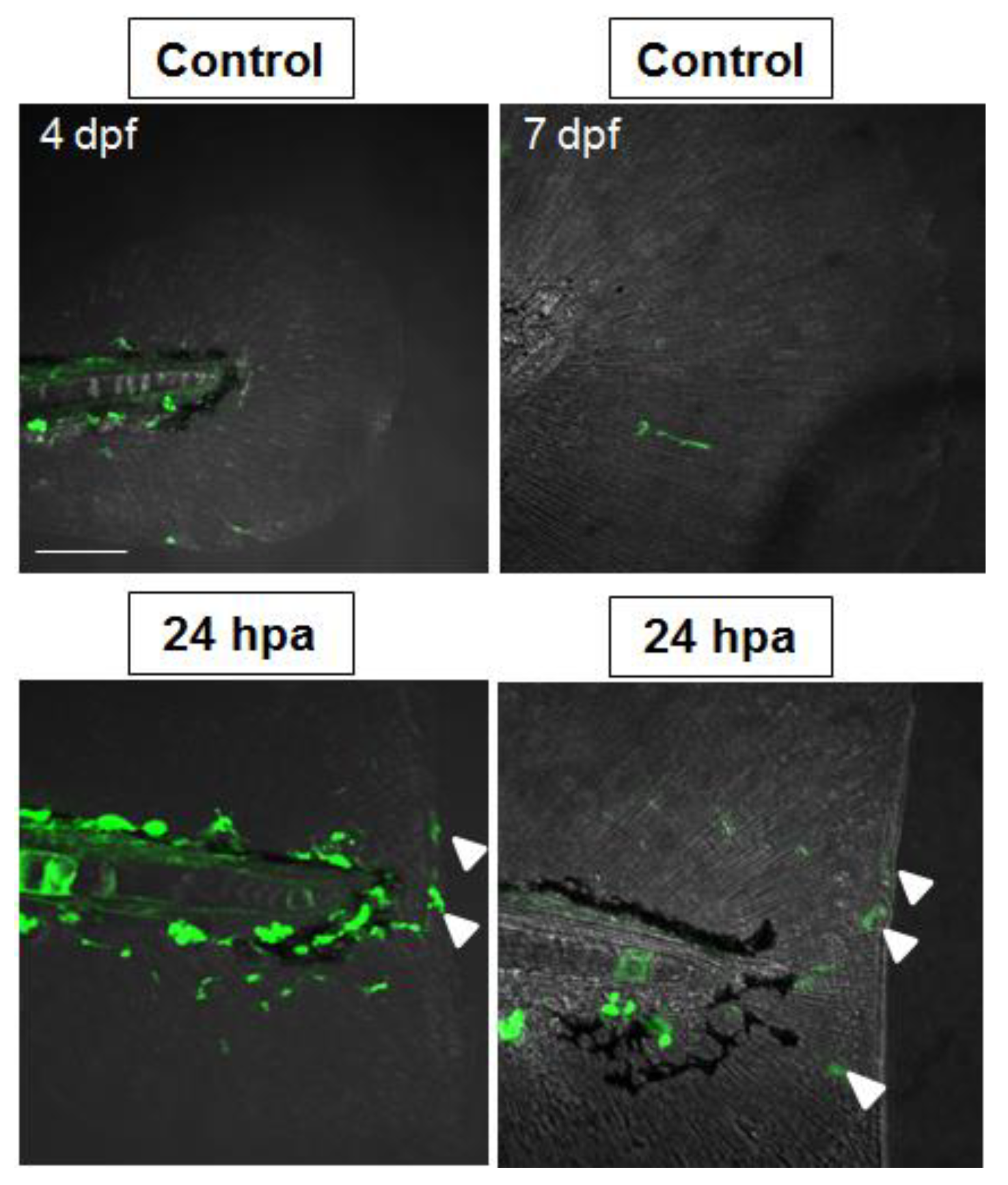
2.5. Nrf2/ARE Pathway Is Induced upon Fin Amputation
3. Discussion
4. Materials and Methods
4.1. Generation Tg(8XAORE:EGFP)ia201 Fish
4.2. Drug Treatments
4.3. Morpholino Injection, RNA Extraction and RQ-PCR
4.4. Tail Fin Amputation
4.5. Image Acquisition and Processing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Griffiths, H.R. ROS as signalling molecules in T cells--evidence for abnormal redox signaling in the autoimmune disease, rheumatoid arthritis. Redox Rep. 2005, 10, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-related factor 2 (NRF2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of NRF2. Mol. Cell. Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, A.; Sundarraj, K.; Nagarajan, R.; Arfuso, F.; Bian, J.; Kumar, A.P.; Sethi, G.; Perumal, E. Antioxidant response elements: Discovery, classes, regulation and potential applications. Redox Biol. 2018, 17, 297–314. [Google Scholar] [CrossRef]
- Wang, X.J.; Hayes, J.D.; Wolf, C.R. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of nrf2 by cancer chemotherapeutic agents. Cancer Res. 2006, 66, 10983–10994. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.A.; Andrews, G.K.; Xu, W.; Johnson, J.A. Activation of the antioxidant response element in primary cortical neuronal cultures derived from transgenic reporter mice. J. Neurochem. 2002, 81, 1233–1241. [Google Scholar] [CrossRef]
- Oikawa, D.; Akai, R.; Tokuda, M.; Iwawaki, T. A transgenic mouse model for monitoring oxidative stress. Sci. Rep. 2012, 2, 229. [Google Scholar] [CrossRef] [Green Version]
- Kraft, A.D.; Resch, J.M.; Johnson, D.A.; Johnson, J.A. Activation of the Nrf2/ARE pathway in muscle and spinal cord during ALS-like pathology in mice expressing mutant SOD1. Exp. Neurol. 2007, 207, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Mourabit, S.; Fitzgerald, J.A.; Ellis, R.P.; Takesono, A.; Porteus, C.S.; Trznadel, M.; Metz, J.; Winter, M.J.; Kudoh, T.; Tyler, C.R. New insights into organ-specific oxidative stress mechanisms using a novel biosensor zebrafish. Environ. Int. 2019, 133 Pt A, 105138. [Google Scholar] [CrossRef]
- Zancan, I.; Bellesso, S.; Costa, R.; Salvalaio, M.; Stroppiano, M.; Hammond, C.; Argenton, F.; Filocamo, M.; Moro, E. Glucocerebrosidase deficiency in zebrafish affects primary bone ossification through increased oxidative stress and reduced Wnt/β-catenin signaling. Hum. Mol. Genet. 2015, 24, 1280–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwan, K.M.; Fujimoto, E.; Grabher, C.; Mangum, B.D.; Hardy, M.E.; Campbell, D.S.; Parant, J.M.; Yost, H.J.; Kanki, J.P.; Chien, C.B. The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007, 236, 3088–3099. [Google Scholar] [CrossRef]
- Probst, B.L.; Trevino, I.; McCauley, L.; Bumeister, R.; Dulubova, I.; Wigley, W.C.; Ferguson, D.A. RTA-408, A Novel Synthetic Triterpenoid with Broad Anticancer and Anti-Inflammatory Activity. PLoS ONE 2015, 10, e0122942. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, P.S.; Ellison, T.; Waqas, B.; Sultan, D.; Abdou, S.; David, J.A.; Cohen, J.M.; Gomez-Viso, A.; Lam, G.; Kim, C.; et al. Targeted Nrf2 activation therapy with RTA-408 enhances regenerative capacity of diabetic wounds. Diabetes Res. Clin. Pract. 2018, 139, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Villeneuve, N.F.; Jiang, T.; Wu, T.; Lau, A.; Toppin, H.A.; Zhang, D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. USA 2011, 108, 1433–1438. [Google Scholar] [CrossRef] [Green Version]
- Linker, R.A.; Lee, D.H.; Ryan, S.; van Dam, A.M.; Conrad, R.; Bista, P.; Zeng, W.; Hronowsky, X.; Buko, A.; Chollate, S.; et al. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain 2011, 134 Pt 3, 678–692. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Venkannagari, S.; Oh, K.H.; Zhang, Y.Q.; Rohde, J.M.; Liu, L.; Nimmagadda, S.; Sudini, K.; Brimacombe, K.R.; Gajghate, S.; et al. Small molecule inhibitor of Nrf2 selectively intervenes therapeutic resistance in Keap1-deficient NSCLC tumors. ACS Chem. Biol. 2016, 11, 3214–3225. [Google Scholar] [CrossRef] [Green Version]
- Rothstein, J.D. Edaravone: A new drug approved for ALS. Cell 2017, 171, 725. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, Y.; Zhang, G.; Lin, Z.; Du, S. Protective effect of edaravone on blood-brain barrier by affecting NRF-2/HO-1 signaling pathway. Exp. Ther. Med. 2019, 18, 2437–2442. [Google Scholar] [CrossRef] [Green Version]
- Paredes, L.C.; Luz, R.B.D.S.; Tozzi, O.N.; de Carvalho, L.Â.S.J.; Calado, S.L.M.; Padovani, B.N.; Fénero, C.I.M.; do Amaral, M.A.; de Assis, H.C.D.S.; Câmara, N.O.S.; et al. Distinct macrophage phenotypes and redox environment during the fin fold regenerative process in zebrafish. Scand. J. Immunol. 2021, 94, e13026. [Google Scholar] [CrossRef]
- Soares, M.A.; Cohen, O.D.; Low, Y.C.; Sartor, R.A.; Ellison, T.; Anil, U.; Anzai, L.; Chang, J.B.; Saadeh, P.B.; Rabbani, P.S.; et al. Restoration of Nrf2 Signaling Normalizes the Regenerative Niche. Diabetes 2016, 65, 633–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, J.D.; Dinkova-Kostova, A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef] [PubMed]
- Mozaheb, N.; Arefian, E.; Amoozegar, M.A. Designing a whole cell bioreporter to show antioxidant activities of agents that work by promotion of the KEAP1-NRF2 signaling pathway. Sci. Rep. 2019, 9, 3248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moro, E.; Vettori, A.; Porazzi, P.; Schiavone, M.; Rampazzo, E.; Casari, A.; Ek, O.; Facchinello, N.; Astone, M.; Zancan, I.; et al. Generation and application of signaling pathway reporter lines in zebrafish. Mol. Genet. Genom. 2013, 288, 231–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, T.Y.; Choi, T.I.; Lee, Y.R.; Choe, S.K.; Kim, C.H. Zebrafish as an animal model for biomedical research. Exp. Mol. Med. 2021, 53, 310–317. [Google Scholar] [CrossRef]
- Moro, E.; Ozhan-Kizil, G.; Mongera, A.; Beis, D.; Wierzbicki, C.; Young, R.M.; Bournele, D.; Domenichini, A.; Valdivia, L.E.; Lum, L.; et al. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev. Biol. 2012, 366, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, H.; Nakajima-Takagi, Y.; Tsujita, T.; Akiyama, S.; Wakasa, T.; Mukaigasa, K.; Kaneko, H.; Tamaru, Y.; Yamamoto, M.; Kobayashi, M. Tissue-restricted expression of Nrf2 and its target genes in zebrafish with gene-specific variations in the induction profiles. PLoS ONE 2011, 6, e26884. [Google Scholar] [CrossRef] [Green Version]
- Murakami, S.; Motohashi, H. Roles of Nrf2 in cell proliferation and differentiation. Free Radic. Biol. Med. 2015, 88 Pt B, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, M.; Morita, R.; Mizoguchi, T.; Matsuo, H.; Isoda, M.; Ishitani, T.; Chitnis, A.B.; Matsumoto, K.; Crump, J.G.; Hozumi, K.; et al. Mib-Jag1-Notch signalling regulates patterning and structural roles of the notochord by controlling cell-fate decisions. Development 2010, 137, 2527–2537. [Google Scholar] [CrossRef] [Green Version]
- Wakabayashi, N.; Shin, S.; Slocum, S.L.; Agoston, E.S.; Wakabayashi, J.; Kwak, M.K.; Misra, V.; Biswal, S.; Yamamoto, M.; Kensler, T.W. Regulation of notch1 signaling by nrf2: Implications for tissue regeneration. Sci. Signal. 2010, 3, ra52. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Sun, W.; Zhang, Z.; Zheng, Y. The role of Nrf2-mediated pathway in cardiac remodeling and heart failure. Oxid. Med. Cell. Longev. 2014, 2014, 260429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unni, S.; Deshmukh, P.; Krishnappa, G.; Kommu, P.; Padmanabhan, B. Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1. FEBS J. 2021, 288, 1599–1613. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Goedert, M. GSK3 inhibitors: Development and therapeutic potential. Nat. Rev. Drug Discov. 2004, 3, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Rada, P.; Rojo, A.I.; Chowdhry, S.; McMahon, M.; Hayes, J.D.; Cuadrado, A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011, 31, 1121–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, S.J.; Kim, K. Effects of the Edaravone, a Drug Approved for the Treatment of Amyotrophic Lateral Sclerosis, on Mitochondrial Function and Neuroprotection. Antioxidants 2022, 11, 195. [Google Scholar] [CrossRef]
- Suzuki, T.; Kelly, V.P.; Motohashi, H.; Nakajima, O.; Takahashi, S.; Nishimura, S.; Yamamoto, M. Deletion of the selenocysteine tRNA gene in macrophages and liver results in compensatory gene induction of cytoprotective enzymes by Nrf2. J. Biol Chem. 2008, 283, 2021–2030. [Google Scholar] [CrossRef] [Green Version]
- Renshaw, S.A.; Loynes, C.A.; Trushell, D.M.; Elworthy, S.; Ingham, P.W.; Whyte, M.K. A transgenic zebrafish model of neutrophilic inflammation. Blood 2006, 108, 3976–3978. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badenetti, L.; Manzoli, R.; Rubin, M.; Cozza, G.; Moro, E. Monitoring Nrf2/ARE Pathway Activity with a New Zebrafish Reporter System. Int. J. Mol. Sci. 2023, 24, 6804. https://doi.org/10.3390/ijms24076804
Badenetti L, Manzoli R, Rubin M, Cozza G, Moro E. Monitoring Nrf2/ARE Pathway Activity with a New Zebrafish Reporter System. International Journal of Molecular Sciences. 2023; 24(7):6804. https://doi.org/10.3390/ijms24076804
Chicago/Turabian StyleBadenetti, Lorenzo, Rosa Manzoli, Michela Rubin, Giorgio Cozza, and Enrico Moro. 2023. "Monitoring Nrf2/ARE Pathway Activity with a New Zebrafish Reporter System" International Journal of Molecular Sciences 24, no. 7: 6804. https://doi.org/10.3390/ijms24076804




