Transcriptomic Study on Human Skin Samples: Identification of Two Subclasses of Actinic Keratoses
Abstract
1. Introduction
2. Results
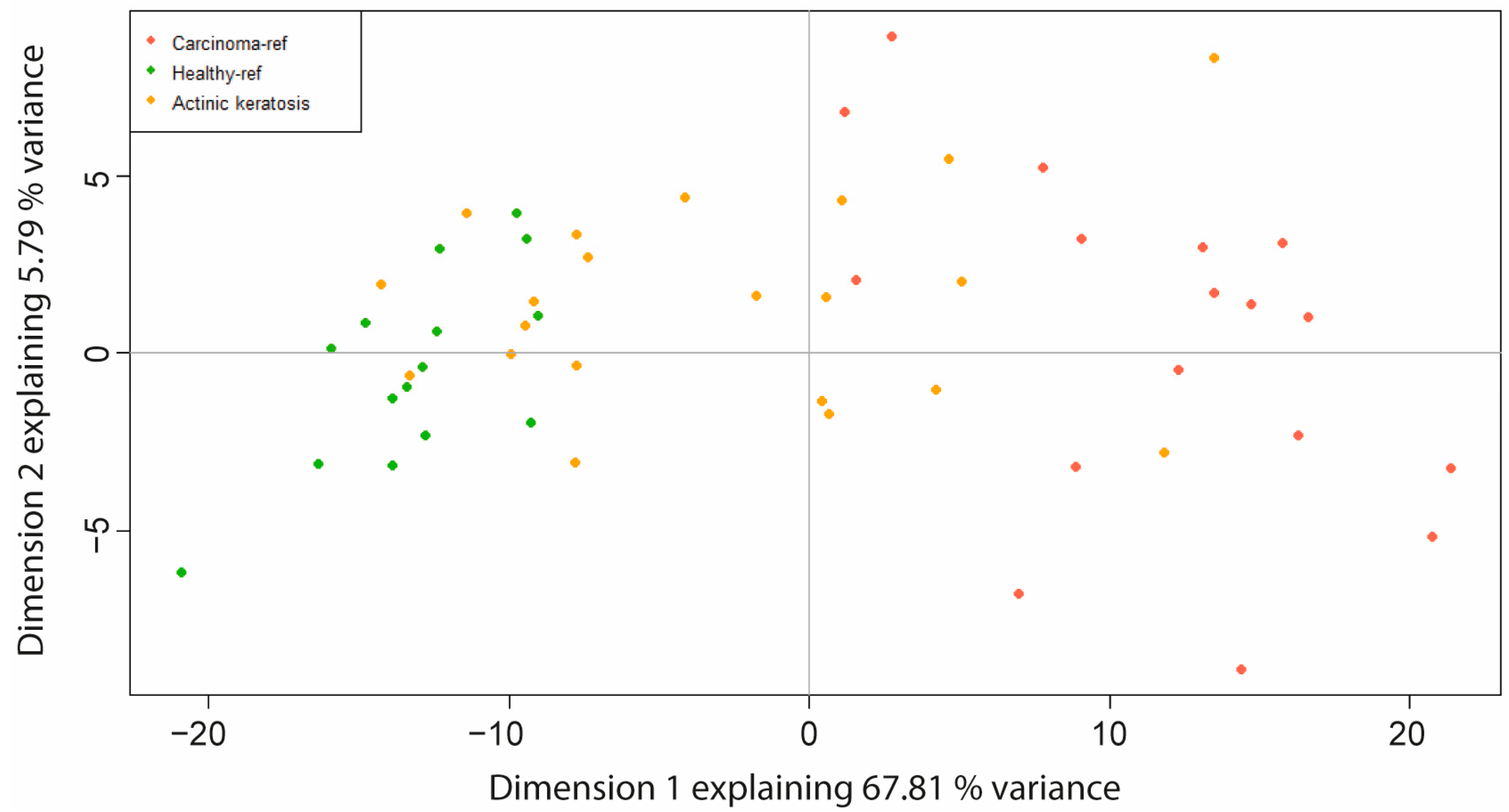
2.1. Principal Component Analysis (PCA) Based on Differentially Expressed Genes (DEGs)
2.2. Homemade Classification
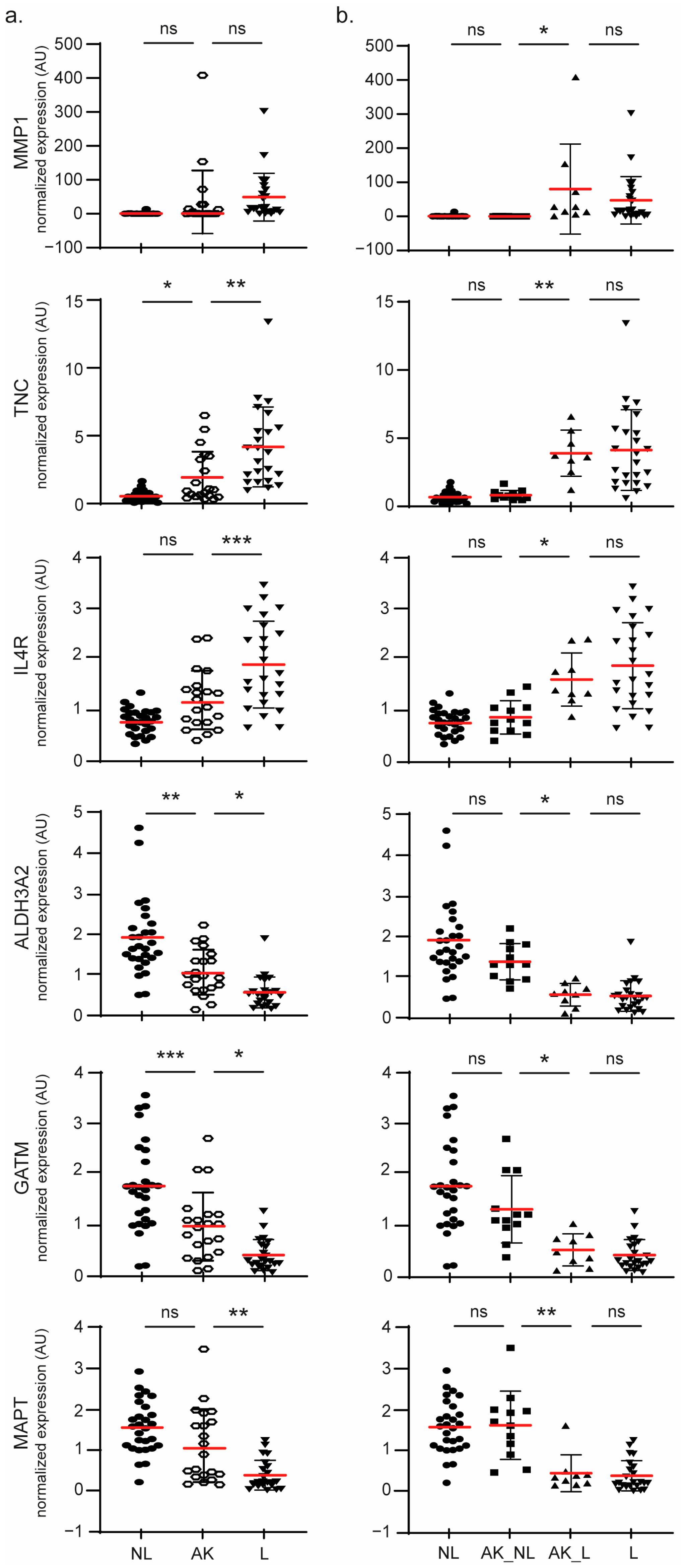
2.3. Molecular Profiling of SCC Lesions
2.4. Identification of Two AK Classes
2.5. Molecular Signature Involved in AK Progression
3. Discussion
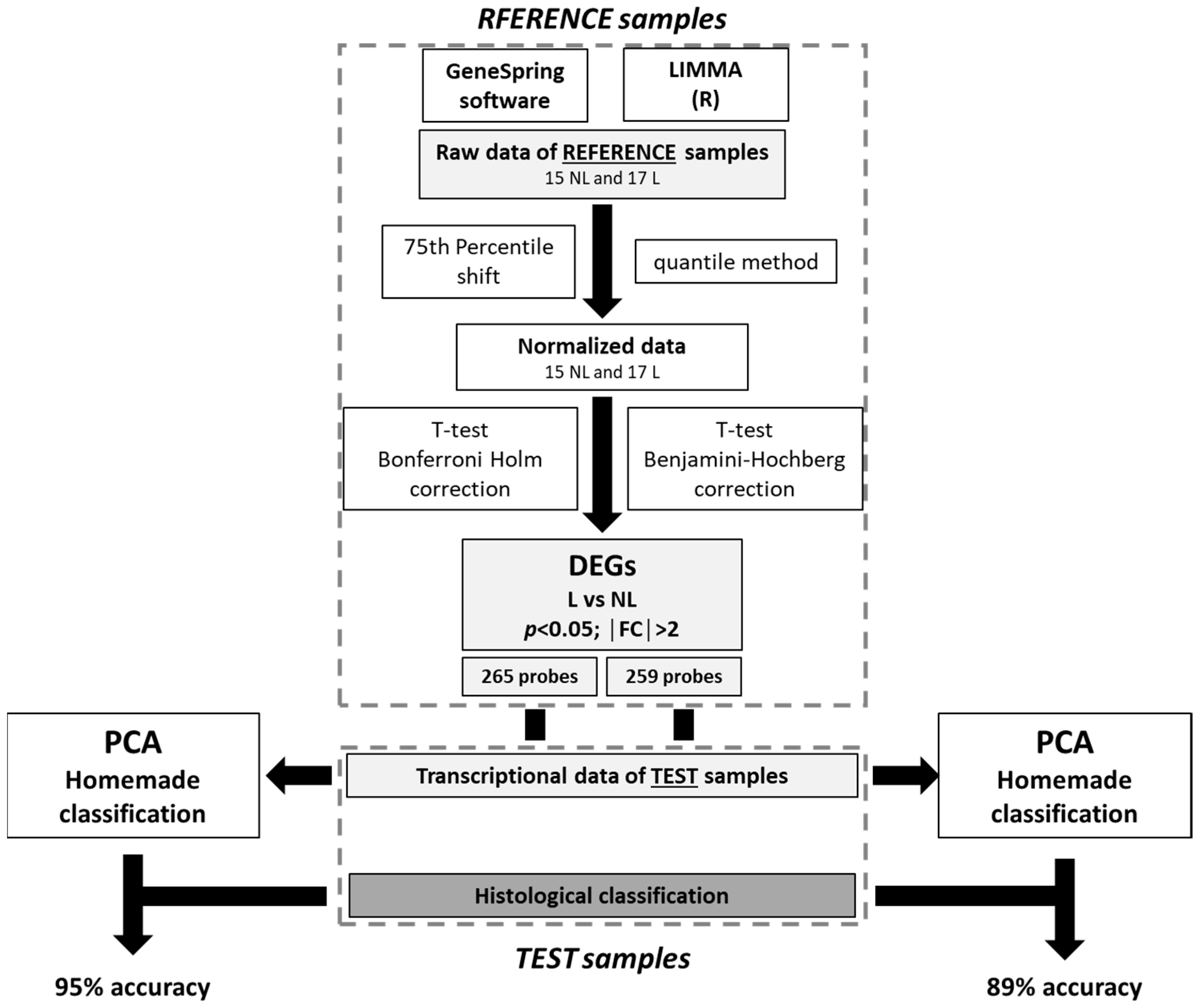
4. Materials and Methods
4.1. Human Tissue Samples
- The carcinoma (referred to in this study as the lesion (L) skin site) itself represented as a disk on the scheme; although SCCs may often display a round shape, their shape may vary and sometimes be random;
- The safety margins (referred to in this study as the peri-lesion (PL) skin site) are based on clinical and healthcare system recommendations. The margins are adapted to the cancer histological class (5 mm for an SCC, 3 mm for a nodular type of basal cell carcinoma, etc.) but also to the risk of recurrence of the cancer (e.g., higher for a trabecular basal cell carcinoma than for a nodular).
- The spindle tips (on each side of the carcinoma, referred to in this study as the non-lesion (NL) skin site) that exist in clinically considered healthy skin removed in order to allow for a regular suture from one side of the spindle to the other one.
4.2. mRNA Extraction and Integrity
4.3. Microarray Analysis
4.4. Samples Grouping
4.5. Differentially Expressed Genes
4.6. Principal Components Analysis (PCA) and Home-Made Classification
4.7. Hierarchical Clustering
4.8. Functional Analysis
4.9. Gene Set Enrichment Analysis
4.10. Connectivity Map
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Green, A.C. Epidemiology of Actinic Keratoses. In Current Problems in Dermatology; Soyer, H.P., Prow, T.W., Jemec, G.B.E., Eds.; S. Karger AG: Berlin, Germany, 2015; Volume 46, pp. 1–7. [Google Scholar] [CrossRef]
- Warino, L.; Tusa, M.; Camacho, F.; Teuschler, H.; Fleischer, A.B.; Feldman, S.R. Frequency and cost of actinic keratosis treatment. Dermatol. Surg. 2006, 32, 1045–1049. [Google Scholar] [CrossRef]
- Skobowiat, C.; Dowdy, J.C.; Sayre, R.M.; Tuckey, R.C.; Slominski, A. Cutaneous hypothalamic-pituitary-adrenal axis homolog: Regulation by ultraviolet radiation. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E484–E493. [Google Scholar] [CrossRef]
- Holman, C.D.; Armstrong, B.K.; Evans, P.R.; Lumsden, G.J.; Dallimore, K.J.; Meehan, C.J.; Beagley, J.; Gibson, I.M. Relationship of solar keratosis and history of skin cancer to objective measures of actinic skin damage. Br. J. Dermatol. 1984, 110, 129–138. [Google Scholar] [CrossRef]
- Frost, C.A.; Green, A.C.; Williams, G.M. The prevalence and determinants of solar keratoses at a subtropical latitude (Queensland, Australia). Br. J. Dermatol. 1998, 139, 1033–1039. [Google Scholar] [CrossRef]
- Criscione, V.D.; Weinstock, M.A.; Naylor, M.F.; Luque, C.; Eide, M.J.; Bingham, S.F.; for the Department of Veteran Affairs Topical Tretinoin Chemoprevention Trial Group. Actinic keratoses: Natural history and risk of malignant transformation in the Veterans Affairs Topical Tretinoin Chemoprevention Trial. Cancer 2009, 115, 2523–2530. [Google Scholar] [CrossRef]
- Glogau, R.G. The risk of progression to invasive disease. J. Am. Acad. Dermatol. 2000, 42, 23–24. [Google Scholar] [CrossRef]
- Werner, R.N.; Stockfleth, E.; Connolly, S.M.; Correia, O.; Erdmann, R.; Foley, P.; Gupta, A.K.; Jacobs, A.; Kerl, H.; Lim, H.W.; et al. Evidence- and consensus-based (S3) Guidelines for the Treatment of Actinic Keratosis—International League of Dermatological Societies in cooperation with the European Dermatology Forum—Short version. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 2069–2079. [Google Scholar] [CrossRef]
- Caddick, J.; Green, L.; Stephenson, J.; Spyrou, G. The psycho-social impact of facial skin cancers. J. Plast. Reconstr. Aesthetic Surg. 2012, 65, e257–e259. [Google Scholar] [CrossRef]
- Röwert-Huber, J.; Patel, M.J.; Forschner, T.; Ulrich, C.; Eberle, J.; Kerl, H.; Sterry, W.; Stockfleth, E. Actinic keratosis is an early in situ squamous cell carcinoma: A proposal for reclassification. Br. J. Dermatol. 2007, 156 (Suppl. S3), 8–12. [Google Scholar] [CrossRef]
- Righi, V.; Reggiani, C.; Tarentini, E.; Mucci, A.; Paganelli, A.; Cesinaro, A.M.; Mataca, E.; Kaleci, S.; Ferrari, B.; Meleti, M.; et al. Metabolomic Analysis of Actinic Keratosis and SCC Suggests a Grade-Independent Model of Squamous Cancerization. Cancers 2021, 13, 5560. [Google Scholar] [CrossRef]
- Fernández-Figueras, M.T.; Carrato, C.; Sáenz, X.; Puig, L.; Musulen, E.; Ferrándiz, C.; Ariza, A. Actinic keratosis with atypical basal cells (AK I) is the most common lesion associated with invasive squamous cell carcinoma of the skin. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 991–997. [Google Scholar] [CrossRef]
- Schmitz, L.; Kahl, P.; Majores, M.; Bierhoff, E.; Stockfleth, E.; Dirschka, T. Actinic keratosis: Correlation between clinical and histological classification systems. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Hameetman, L.; Commandeur, S.; Bavinck, J.N.B.; Wisgerhof, H.C.; de Gruijl, F.R.; Willemze, R.; Mullenders, L.; Tensen, C.P.; Vrieling, H. Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer 2013, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Chitsazzadeh, V.; Coarfa, C.; Drummond, J.A.; Nguyen, T.; Joseph, A.; Chilukuri, S.; Charpiot, E.; Adelmann, C.H.; Ching, G.; Nguyen, T.N.; et al. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat. Commun. 2016, 7, 12601. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.R.; Mladkova, N.; Gulati, A.; Hamoudi, R.; Purdie, K.; Cerio, R.; Leigh, I.; Proby, C.; Harwood, C.A. Key differences identified between actinic keratosis and cutaneous squamous cell carcinoma by transcriptome profiling. Br. J. Cancer 2014, 110, 520–529. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Rajapakshe, K.; Nicholas, C.; Tordesillas, L.; Ehli, E.A.; Davis, C.M.; Coarfa, C.; Flores, E.R.; Dickinson, S.E.; Curiel-Lewandrowski, C.; et al. Integrative transcriptomic analysis for linking acute stress responses to squamous cell carcinoma development. Sci. Rep. 2020, 10, 17209. [Google Scholar] [CrossRef] [PubMed]
- Queen, D.; Shen, Y.; Trager, M.H.; Lopez, A.T.; Samie, F.H.; Lewin, J.M.; Niedt, G.W.; Geskin, L.J.; Liu, L. UV biomarker genes for classification and risk stratification of cutaneous actinic keratoses and squamous cell carcinoma subtypes. FASEB J. 2020, 34, 13022–13032. [Google Scholar] [CrossRef]
- Chen, H.; Yang, J.; Wu, W. Seven key hub genes identified by gene co-expression network in cutaneous squamous cell carcinoma. BMC Cancer 2021, 21, 852. [Google Scholar] [CrossRef]
- Das Mahapatra, K.; Pasquali, L.; Søndergaard, J.N.; Lapins, J.; Nemeth, I.B.; Baltás, E.; Kemény, L.; Homey, B.; Moldovan, L.-I.; Kjems, J.; et al. A comprehensive analysis of coding and non-coding transcriptomic changes in cutaneous squamous cell carcinoma. Sci. Rep. 2020, 10, 3637. [Google Scholar] [CrossRef]
- Hudson, L.G.; Gale, J.M.; Padilla, R.S.; Pickett, G.; Alexander, B.E.; Wang, J.; Kusewitt, D.F. Microarray analysis of cutaneous squamous cell carcinomas reveals enhanced expression of epidermal differentiation complex genes. Mol. Carcinog. 2010, 49, 619–629. [Google Scholar] [CrossRef]
- Zou, D.-D.; Xu, D.; Deng, Y.-Y.; Wu, W.-J.; Zhang, J.; Huang, L.; He, L. Identification of key genes in cutaneous squamous cell carcinoma: A transcriptome sequencing and bioinformatics profiling study. Ann. Transl. Med. 2021, 9, 1497. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Li, L.; Zhu, S.; Wu, Y.; Liu, Y.; Zhu, L.; Zhao, Z.; Wu, F.; Jia, N.; Liao, C.; et al. Single-cell transcriptomic analysis reveals the critical molecular pattern of UV-induced cutaneous squamous cell carcinoma. Cell Death Dis. 2021, 13, 23. [Google Scholar] [CrossRef] [PubMed]
- Campione, E.; Di Prete, M.; Di Raimondo, C.; Costanza, G.; Palumbo, V.; Garofalo, V.; Mazzilli, S.; Franceschini, C.; Dika, E.; Bianchi, L.; et al. Topical Treatment of Actinic Keratosis and Metalloproteinase Expression: A Clinico-Pathological Retrospective Study. IJMS 2022, 23, 11351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Huang, X.; Huang, R.; Zhu, H.; Ye, P.; Lin, X.; Zhang, S.; Wu, M.; Jiang, F. MMP1 Overexpression Promotes Cancer Progression and Associates with Poor Outcome in Head and Neck Carcinoma. Comput. Math. Methods Med. 2022, 2022, 3058342. [Google Scholar] [CrossRef]
- Mitsui, H.; Suárez-Fariñas, M.; Gulati, N.; Shah, K.R.; Cannizzaro, M.V.; Coats, I.; Felsen, D.; Krueger, J.G.; Carucci, J.A. Gene Expression Profiling of the Leading Edge of Cutaneous Squamous Cell Carcinoma: IL-24-Driven MMP-7. J. Investig. Dermatol. 2014, 134, 1418–1427. [Google Scholar] [CrossRef]
- Nindl, I.; Dang, C.; Forschner, T.; Kuban, R.J.; Meyer, T.; Sterry, W.; Stockfleth, E. Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Mol. Cancer 2006, 5, 30. [Google Scholar] [CrossRef]
- Zheng, L.-Q.; Wang, R.; Chi, S.-M.; Li, C.-X. Matrix metalloproteinase 1: A better biomarker for squamous cell carcinoma by multiple microarray analyses. G. Ital. Dermatol. Venereol. 2019, 154, 327–337. [Google Scholar] [CrossRef]
- Dang, C.; Gottschling, M.; Roewert, J.; Forschner, T.; Stockfleth, E.; Nindl, I. Tenascin-C patterns and splice variants in actinic keratosis and cutaneous squamous cell carcinoma. Br. J. Dermatol. 2006, 155, 763–770. [Google Scholar] [CrossRef]
- Azin, M.; Demehri, S. Innate Lymphoid Cells: New Targets for Cutaneous Squamous Cell Carcinoma Immunotherapy. J. Investig. Dermatol. 2021, 141, 2320–2322. [Google Scholar] [CrossRef]
- Steeb, T.; Petzold, A.; Hornung, A.; Wessely, A.; Berking, C.; Heppt, M.V. Spontaneous regression rates of actinic keratosis: A systematic review and pooled analysis of randomized controlled trials. Sci. Rep. 2022, 12, 5884. [Google Scholar] [CrossRef]
- Lessard, J.C.; Piña-Paz, S.; Rotty, J.D.; Hickerson, R.P.; Kaspar, R.L.; Balmain, A.; Coulombe, P.A. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc. Natl. Acad. Sci. USA 2013, 110, 19537–19542. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Loercher, A.; Lee, T.L.; Ricker, J.L.; Howard, A.; Geoghegen, J.; Chen, Z.; Sunwoo, J.B.; Sitcheran, R.; Chuang, E.Y.; Mitchell, J.B.; et al. Nuclear factor-kappaB is an important modulator of the altered gene expression profile and malignant phenotype in squamous cell carcinoma. Cancer Res. 2004, 64, 6511–6523. [Google Scholar] [CrossRef]
- Poligone, B.; Hayden, M.S.; Chen, L.; Pentland, A.P.; Jimi, E.; Ghosh, S. A Role for NF-κB Activity in Skin Hyperplasia and the Development of Keratoacanthomata in Mice. PLoS ONE 2013, 8, e71887. [Google Scholar] [CrossRef] [PubMed]
- Rini, B.I.; Pal, S.K.; Escudier, B.J.; Atkins, M.B.; Hutson, T.E.; Porta, C.; Verzoni, E.; Needle, M.N.; McDermott, D.F. Tivozanib versus sorafenib in patients with advanced renal cell carcinoma (TIVO-3): A phase 3, multicentre, randomised, controlled, open-label study. Lancet Oncol. 2020, 21, 95–104. [Google Scholar] [CrossRef]
- Chang, E.; Weinstock, C.; Zhang, L.; Fiero, M.H.; Zhao, M.; Zahalka, E.; Ricks, T.K.; Fourie Zirkelbach, J.; Qiu, J.; Yu, J.; et al. FDA Approval Summary: Tivozanib for Relapsed or Refractory Renal Cell Carcinoma. Clin. Cancer Res. 2022, 28, 441–445. [Google Scholar] [CrossRef]
- Brauchle, M.; Funk, J.O.; Kind, P.; Werner, S. Ultraviolet B and H2O2 Are Potent Inducers of Vascular Endothelial Growth Factor Expression in Cultured Keratinocytes. J. Biol. Chem. 1996, 271, 21793–21797. [Google Scholar] [CrossRef]
- Alitalo, A.K.; Proulx, S.T.; Karaman, S.; Aebischer, D.; Martino, S.; Jost, M.; Schneider, N.; Bry, M.; Detmar, M. VEGF-C and VEGF-D Blockade Inhibits Inflammatory Skin Carcinogenesis. Cancer Res. 2013, 73, 4212–4221. [Google Scholar] [CrossRef]
- Bottomley, M.J.; Thomson, J.; Harwood, C.; Leigh, I. The Role of the Immune System in Cutaneous Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019, 20, 2009. [Google Scholar] [CrossRef]
- Berriz, G.F.; Beaver, J.E.; Cenik, C.; Tasan, M.; Roth, F.P. Next generation software for functional trend analysis. Bioinformatics 2009, 25, 3043–3044. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]


| Patient Inclusion Number | Sample Name | Histopathological Diagnostic (Grade) | Transcriptomic Profile | ||
|---|---|---|---|---|---|
| DistToNL | DistToL | AK Classification | |||
| 1 | 001_C_AK | AK_I | 6.58 | 19.58 | AK_NL |
| 6 | 006_A_AK | AK_II | 25.09 | 3.19 | AK_L |
| 16 | 016_B_AK | AK_I | 6.60 | 19.09 | AK_NL |
| 17 | 017_A_AK | AK_I | 2.62 | 25.89 | AK_NL |
| 28 | 028_F_AK | AK_II | 28.06 | 8.21 | AK_L |
| 29 | 029_C_AK | AK_II | 3.22 | 21.54 | AK_NL |
| 35 = 41 | 041_A_AK | AK_III | 4.73 | 23.30 | AK_NL |
| 48 | 048_F_AK | AK_I | 5.37 | 19.38 | AK_NL |
| 56 | 056_B_AK | AK_I | 17.39 | 7.48 | AK_L |
| 75 | 075_U_AK | AK_I | 3.87 | 21.06 | AK_NL |
| 77 | 077_B_AK_1 | AK_I | 4.37 | 20.82 | AK_NL |
| 077_B_AK_2 | AK_I | 11.55 | 13.42 | AK_NL | |
| 82 | 082_al_AK | AK_III | 18.73 | 8.62 | AK_L |
| 082_B_AK_1 | AK_I | 13.85 | 11.14 | AK_L | |
| 082_B_AK_2 | AK_I | 13.87 | 11.08 | AK_L | |
| 90 | 090_A_AK | AK_I | 5.98 | 19.69 | AK_NL |
| 109 | 109_C’_AK_1 | AK_I | 0.26 | 24.91 | AK_NL |
| 109_C’_AK_2 | AK_II | 13.58 | 11.31 | AK_L | |
| 115 | 115_F_AK | AK_II | 18.38 | 6.72 | AK_L |
| 119 | 119_C_AK_1 | AK_I | 14.99 | 11.23 | AK_L |
| 119_C_AK_2 | AK_I | 10.21 | 16.26 | AK_NL | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubois-Pot-Schneider, H.; Khairallah, G.; Brzenczek, C.; Plénat, F.; Marchal, F.; Amouroux, M. Transcriptomic Study on Human Skin Samples: Identification of Two Subclasses of Actinic Keratoses. Int. J. Mol. Sci. 2023, 24, 5937. https://doi.org/10.3390/ijms24065937
Dubois-Pot-Schneider H, Khairallah G, Brzenczek C, Plénat F, Marchal F, Amouroux M. Transcriptomic Study on Human Skin Samples: Identification of Two Subclasses of Actinic Keratoses. International Journal of Molecular Sciences. 2023; 24(6):5937. https://doi.org/10.3390/ijms24065937
Chicago/Turabian StyleDubois-Pot-Schneider, Hélène, Grégoire Khairallah, Cyril Brzenczek, François Plénat, Frédéric Marchal, and Marine Amouroux. 2023. "Transcriptomic Study on Human Skin Samples: Identification of Two Subclasses of Actinic Keratoses" International Journal of Molecular Sciences 24, no. 6: 5937. https://doi.org/10.3390/ijms24065937
APA StyleDubois-Pot-Schneider, H., Khairallah, G., Brzenczek, C., Plénat, F., Marchal, F., & Amouroux, M. (2023). Transcriptomic Study on Human Skin Samples: Identification of Two Subclasses of Actinic Keratoses. International Journal of Molecular Sciences, 24(6), 5937. https://doi.org/10.3390/ijms24065937







