The Recent Advances in Molecular Diagnosis of Soft Tissue Tumors
Abstract
1. Introduction
2. Etiology
3. Classification of Sarcomas Based on Karyotypic Complexity
4. Molecular Tests
5. Immunohistochemistry
6. Practical Diagnostic Approach to Soft Tissue Tumors
7. Adipocytic Tumors
7.1. Spindle Cell Lipoma and Pleomorphic Lipoma
7.2. Atypical Spindle Cell/Pleomorphic Lipomatous Tumor
7.3. Atypical Lipomatous Tumor/Well-Differentiated Liposarcoma
7.4. Dedifferentiated Liposarcoma
7.5. Myxoid Liposarcoma
8. Fibroblastic and Myofibroblastic Tumors
8.1. Desmoid Fibromatosis
8.2. Solitary Fibrous Tumor
8.3. Inflammatory Myofibroblastic Tumor
8.4. Low-Grade Fibromyxoid Sarcoma
8.5. Sclerosing Epithelioid Fibrosarcoma
8.6. Infantile Fibrosarcoma
9. Vascular Tumors
9.1. Epithelioid Hemangioma
9.2. Pseudomyogenic Hemangioendothelioma
9.3. Epithelioid Hemangioendothelioma
10. Skeletal Muscle Tumors
10.1. Alveolar Rhabdomyosarcoma
10.2. Spindle Cell/Sclerosing Rhabdomyosarcoma
11. Gastrointestinal Stromal Tumor
12. Peripheral Nerve Sheath Tumors
12.1. Malignant Peripheral Nerve Sheath Tumor
12.2. Malignant Melanotic Nerve Sheath Tumor
13. Tumors of Uncertain Differentiation
13.1. Synovial Sarcoma
13.2. Epithelioid Sarcoma
13.3. Extrarenal Rhabdoid Tumor
13.4. Alveolar Soft Part Sarcoma
13.5. Desmoplastic Small Round Cell Tumor
13.6. Intimal Sarcoma
14. Undifferentiated Small Round Cell Sarcomas
14.1. Ewing Sarcoma
14.2. Round Cell Sarcoma with EWSR1-Non-ETS Fusions
14.3. CIC-Rearranged Sarcoma
14.4. Sarcoma with BCOR Genetic Alteration
15. Emerging Entities
15.1. EWSR1::SMAD3–Positive Fibroblastic Tumor
15.2. NTRK-Rearranged Spindle Cell Neoplasm
15.3. SWI/SNF Complex-Deficient Neoplasms
15.4. DICER1-Associated Sarcomas
16. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fletcher, C.D.M.; Baldini, E.H.; Blay, J.Y.; Gronchi, A.; Lazar, A.J.; Messiou, C.; Pollock, R.E.; Singer, S. Soft tissue tumours: Introduction. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 6–12. ISBN 978-92-832-4502-5. [Google Scholar]
- Sbaraglia, M.; Bellan, E.; Dei Tos, A.P. The 2020 WHO classification of soft tissue tumours: News and perspectives. Pathologica 2021, 113, 70–84. [Google Scholar] [CrossRef]
- Antonescu, C.R. The role of genetic testing in soft tissue sarcoma. Histopathology 2006, 48, 13–21. [Google Scholar] [CrossRef]
- Chan, J.K.; Ip, Y.T.; Cheuk, W. The utility of immunohistochemistry for providing genetic information on tumors. Int. J. Surg. Pathol. 2013, 21, 455–475. [Google Scholar] [CrossRef]
- Horvai, A. Bone, joints, and soft tissue tumors. In Robbins and Cotran Pathologic Basis of Disease, 10th ed.; Kumar, V., Abbas, A.K., Aster, J.C., Eds.; Elsevier: Philadelphia, PA, USA, 2021; pp. 1208–1209. [Google Scholar]
- Helman, L.J.; Meltzer, P. Mechanisms of sarcoma development. Nat. Rev. Cancer 2003, 3, 685–694. [Google Scholar] [CrossRef]
- Wang, W.L.; Lazar, A.J. Applications of molecular testing to differential diagnosis. In Practical Soft Tissue Pathology, 2nd ed.; Hornick, J.L., Ed.; Elsevier: Philadelphia, PA, USA, 2019; pp. 513–550. ISBN 978-0-323-49714-5. [Google Scholar]
- Gibault, L.; Pérot, G.; Chibon, F.; Bonnin, S.; Lagarde, P.; Terrier, P.; Coindre, J.M.; Aurias, A. New insights in sarcoma oncogenesis: A comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J. Pathol. 2011, 223, 64–71. [Google Scholar] [CrossRef]
- Golblum, J.R.; Folpe, A.L.; Weiss, S.E. (Eds.) Approach to the diagnosis of soft tissue tumors. In Enzinger & Weiss’s Soft Tissue Tumors, 7th ed.; Elsevier: Philadelphia, PA, USA, 2020; pp. 84–110. ISBN 978-0-323-61096-4. [Google Scholar]
- Tanas, M.R.; Rubin, B.P.; Tubbs, R.R.; Billings, S.D.; Downs-Kelly, E.; Goldblum, J.R. Utilization of fluorescence in situ hybridization in the diagnosis of 230 mesenchymal neoplasms: An institutional experience. Arch. Pathol. Lab. Med. 2010, 134, 1797–1803. [Google Scholar] [CrossRef]
- Tanas, M.R.; Goldblum, J.R. Fluorescence in situ hybridization in the diagnosis of soft tissue neoplasms: A review. Adv. Anat. Pathol. 2009, 16, 383–391. [Google Scholar] [CrossRef]
- Oda, Y.; Yamamoto, H.; Kohashi, K.; Yamada, Y.; Iura, K.; Ishii, T.; Maekawa, A.; Bekki, H. Soft tissue sarcomas: From a morphological to a molecular biological approach. Pathol. Int. 2017, 67, 435–446. [Google Scholar] [CrossRef]
- Groisberg, R.; Roszik, J.; Conley, A.; Patel, S.R.; Subbiah, V. The role of next-generation sequencing in sarcomas: Evolution from light microscope to molecular microscope. Curr. Oncol. Rep. 2017, 19, 78. [Google Scholar] [CrossRef]
- Cote, G.M.; He, J.; Choy, E. Next-generation sequencing for patients with sarcoma: A single center experience. Oncologist 2018, 23, 234–242. [Google Scholar] [CrossRef]
- Szurian, K.; Kashofer, K.; Liegl-Atzwanger, B. Role of next-generation sequencing as a diagnostic tool for the evaluation of bone and soft-tissue tumors. Pathobiology 2017, 84, 323–338. [Google Scholar] [CrossRef]
- Al-Zaid, T.; Wang, W.L.; Somaiah, N.; Lazar, A.J. Molecular profiling of sarcomas: New vistas for precision medicine. Virchows Arch. 2017, 471, 243–255. [Google Scholar] [CrossRef]
- ESMO/European Sarcoma Network Working Group. Soft tissue and visceral sarcomas: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25 (Suppl. S3), iii102–iii112. [Google Scholar] [CrossRef]
- Hornick, J.L. Novel uses of immunohistochemistry in the diagnosis and classification of soft tissue tumors. Mod. Pathol. 2014, 27 (Suppl. S1), S47–S63. [Google Scholar] [CrossRef]
- Fletcher, C.D.M. Tumors of soft tissue. In Diagnostic Histopathology of Tumors, 5th ed.; Fletcher, C.D.M., Ed.; Elsevier: Philadelphia, PA, USA, 2021; pp. 1919–1984. ISBN 978-0-323-42860-6. [Google Scholar]
- Billing, S.D.; Ud Din, N. Spindle cell lipoma and pleomorphic lipoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 29–30. ISBN 978-92-832-4502-5. [Google Scholar]
- Fletcher, C.D.; Akerman, M.; Dal Cin, P.; de Wever, I.; Mandahl, N.; Mertens, F.; Mitelman, F.; Rosai, J.; Rydholm, A.; Sciot, R.; et al. Correlation between clinicopathological features and karyotype in lipomatous tumors. A report of 178 cases from the Chromosomes and Morphology (CHAMP) Collaborative Study Group. Am. J. Pathol. 1996, 148, 623–630. [Google Scholar]
- Chen, B.J.; Mariño-Enríquez, A.; Fletcher, C.D.; Hornick, J.L. Loss of retinoblastoma protein expression in spindle cell/pleomorphic lipomas and cytogenetically related tumors: An immunohistochemical study with diagnostic implications. Am. J. Surg. Pathol. 2012, 36, 1119–1128. [Google Scholar] [CrossRef]
- Dahlén, A.; Debiec-Rychter, M.; Pedeutour, F.; Domanski, H.A.; Höglund, M.; Bauer, H.C.; Rydholm, A.; Sciot, R.; Mandahl, N.; Mertens, F. Clustering of deletions on chromosome 13 in benign and low-malignant lipomatous tumors. Int. J. Cancer 2003, 103, 616–623. [Google Scholar] [CrossRef]
- Magro, G.; Righi, A.; Casorzo, L.; Antonietta, T.; Salvatorelli, L.; Kacerovská, D.; Kazakov, D.; Michal, M. Mammary and vaginal myofibroblastomas are genetically related lesions: Fluorescence in situ hybridization analysis shows deletion of 13q14 region. Hum. Pathol. 2012, 43, 1887–1893. [Google Scholar] [CrossRef]
- Howitt, B.E.; Fletcher, C.D. Mammary-type myofibroblastoma: Clinicopathologic characterization in a series of 143 cases. Am. J. Surg. Pathol. 2016, 40, 361–367. [Google Scholar] [CrossRef]
- Flucke, U.; van Krieken, J.H.; Mentzel, T. Cellular angiofibroma: Analysis of 25 cases emphasizing its relationship to spindle cell lipoma and mammary-type myofibroblastoma. Mod. Pathol. 2011, 24, 82–89. [Google Scholar] [CrossRef]
- Creytens, D.; Marino-Enriquez, A. Atypical spindle cell/pleomorphic lipomatous tumour. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 34–35. ISBN 978-92-832-4502-5. [Google Scholar]
- Mariño-Enriquez, A.; Nascimento, A.F.; Ligon, A.H.; Liang, C.; Fletcher, C.D. Atypical spindle cell lipomatous tumor: Clinicopathologic characterization of 232 cases demonstrating a morphologic spectrum. Am. J. Surg. Pathol. 2017, 41, 234–244. [Google Scholar] [CrossRef]
- Creytens, D.; Mentzel, T.; Ferdinande, L.; Lecoutere, E.; van Gorp, J.; Atanesyan, L.; de Groot, K.; Savola, S.; Van Roy, N.; Van Dorpe, J.; et al. “Atypical” pleomorphic lipomatous tumor: A clinicopathologic, immunohistochemical and molecular study of 21 cases, emphasizing its relationship to atypical spindle cell lipomatous tumor and suggesting a morphologic spectrum (atypical spindle cell/pleomorphic lipomatous tumor). Am. J. Surg. Pathol. 2017, 41, 1443–1455. [Google Scholar] [CrossRef]
- Bahadır, B.; Behzatoğlu, K.; Hacıhasanoğlu, E.; Koca, S.B.; Sığırcı, B.B.; Tokat, F. Atypical spindle cell/pleomorphic lipomatous tumor: A clinicopathologic, immunohistochemical, and molecular study of 20 cases. Pathol. Int. 2018, 68, 550–556. [Google Scholar] [CrossRef]
- Mentzel, T.; Palmedo, G.; Kuhnen, C. Well-differentiated spindle cell liposarcoma (‘atypical spindle cell lipomatous tumor’) does not belong to the spectrum of atypical lipomatous tumor but has a close relationship to spindle cell lipoma: Clinicopathologic, immunohistochemical, and molecular analysis of six cases. Mod. Pathol. 2010, 23, 729–736. [Google Scholar] [CrossRef]
- Thway, K. What’s new in adipocytic neoplasia? Histopathology 2022, 80, 76–97. [Google Scholar] [CrossRef]
- Demicco, E.G. Molecular updates in adipocytic neoplasms. Semin. Diagn. Pathol. 2019, 36, 85–94. [Google Scholar] [CrossRef]
- Sbaraglia, M.; Dei Tos, A.P.; Pedeutour, F. Atypical lipomatous tumour/well-differentiated liposarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 36–38. ISBN 978-92-832-4502-5. [Google Scholar]
- Italiano, A.; Bianchini, L.; Gjernes, E.; Keslair, F.; Ranchere-Vince, D.; Dumollard, J.M.; Haudebourg, J.; Leroux, A.; Mainguené, C.; Terrier, P.; et al. Clinical and biological significance of CDK4 amplification in well-differentiated and dedifferentiated liposarcomas. Clin. Cancer Res. 2009, 15, 5696–5703. [Google Scholar] [CrossRef]
- Binh, M.B.; Sastre-Garau, X.; Guillou, L.; de Pinieux, G.; Terrier, P.; Lagacé, R.; Aurias, A.; Hostein, I.; Coindre, J.M. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: A comparative analysis of 559 soft tissue neoplasms with genetic data. Am. J. Surg. Pathol. 2005, 29, 1340–1347. [Google Scholar] [CrossRef]
- Makise, N.; Sekimizu, M.; Kubo, T.; Wakai, S.; Hiraoka, N.; Komiyama, M.; Fukayama, M.; Kawai, A.; Ichikawa, H.; Yoshida, A. Clarifying the distinction between malignant peripheral nerve sheath tumor and dedifferentiated liposarcoma: A critical reappraisal of the diagnostic utility of MDM2 and H3K27me3 Status. Am. J. Surg. Pathol. 2018, 42, 656–664. [Google Scholar] [CrossRef]
- Schoolmeester, J.K.; Sciallis, A.P.; Greipp, P.T.; Hodge, J.C.; Dal Cin, P.; Keeney, G.L.; Nucci, M.R. Analysis of MDM2 amplification in 43 endometrial stromal tumors: A potential diagnostic pitfall. Int. J. Gynecol. Pathol. 2015, 34, 576–583. [Google Scholar] [CrossRef]
- He, X.; Pang, Z.; Zhang, X.; Lan, T.; Chen, H.; Chen, M.; Yang, H.; Huang, J.; Chen, Y.; Zhang, Z.; et al. Consistent amplification of FRS2 and MDM2 in low-grade osteosarcoma: A genetic study of 22 cases with clinicopathologic analysis. Am. J. Surg. Pathol. 2018, 42, 1143–1155. [Google Scholar] [CrossRef]
- Wunder, J.S.; Eppert, K.; Burrow, S.R.; Gokgoz, N.; Bell, R.S.; Andrulis, I.L. Co-amplification and overexpression of CDK4, SAS and MDM2 occurs frequently in human parosteal osteosarcomas. Oncogene 1999, 18, 783–788. [Google Scholar] [CrossRef]
- Neuville, A.; Collin, F.; Bruneval, P.; Parrens, M.; Thivolet, F.; Gomez-Brouchet, A.; Terrier, P.; de Montpreville, V.T.; Le Gall, F.; Hostein, I.; et al. Intimal sarcoma is the most frequent primary cardiac sarcoma: Clinicopathologic and molecular retrospective analysis of 100 primary cardiac sarcomas. Am. J. Surg. Pathol. 2014, 38, 461–469. [Google Scholar] [CrossRef]
- Zhang, H.; Erickson-Johnson, M.; Wang, X.; Oliveira, J.L.; Nascimento, A.G.; Sim, F.H.; Wenger, D.E.; Zamolyi, R.Q.; Pannain, V.L.; Oliveira, A.M. Molecular testing for lipomatous tumors: Critical analysis and test recommendations based on the analysis of 405 extremity-based tumors. Am. J. Surg. Pathol. 2010, 34, 1304–1311. [Google Scholar] [CrossRef]
- Dei Tos, A.P.; Marino-Enriquez, A.; Pedeutour, F. Dedifferentiated liposarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 39–41. ISBN 978-92-832-4502-5. [Google Scholar]
- Mariño-Enríquez, A.; Fletcher, C.D.; Dal Cin, P.; Hornick, J.L. Dedifferentiated liposarcoma with “homologous” lipoblastic (pleomorphic liposarcoma-like) differentiation: Clinicopathologic and molecular analysis of a series suggesting revised diagnostic criteria. Am. J. Surg. Pathol. 2010, 34, 1122–1131. [Google Scholar] [CrossRef]
- Saâda-Bouzid, E.; Burel-Vandenbos, F.; Ranchère-Vince, D.; Birtwisle-Peyrottes, I.; Chetaille, B.; Bouvier, C.; Château, M.C.; Peoc’h, M.; Battistella, M.; Bazin, A.; et al. Prognostic value of MGA2, CDK4, and JUN amplification in well-differentiated and dedifferentiated liposarcomas. Mod. Pathol. 2015, 28, 1404–1414. [Google Scholar] [CrossRef]
- Thway, K.; Flora, R.; Shah, C.; Olmos, D.; Fisher, C. Diagnostic utility of p16, CDK4, and MDM2 as an immunohistochemical panel in distinguishing well-differentiated and dedifferentiated liposarcomas from other adipocytic tumors. Am. J. Surg. Pathol. 2012, 36, 462–469. [Google Scholar] [CrossRef]
- Sirvent, N.; Coindre, J.M.; Maire, G.; Hostein, I.; Keslair, F.; Guillou, L.; Ranchere-Vince, D.; Terrier, P.; Pedeutour, F. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: Utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. Am. J. Surg. Pathol. 2007, 31, 1476–1489. [Google Scholar] [CrossRef]
- Le Guellec, S.; Chibon, F.; Ouali, M.; Perot, G.; Decouvelaere, A.V.; Robin, Y.M.; Larousserie, F.; Terrier, P.; Coindre, J.M.; Neuville, A. Are peripheral purely undifferentiated pleomorphic sarcomas with MDM2 amplification dedifferentiated liposarcomas? Am. J. Surg. Pathol. 2014, 38, 293–304. [Google Scholar] [CrossRef]
- Thway, K.; Nielsen, T.O. Myxoid liposarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 42–44. ISBN 978-92-832-4502-5. [Google Scholar]
- Powers, M.P.; Wang, W.L.; Hernandez, V.S.; Patel, K.S.; Lev, D.C.; Lazar, A.J.; López-Terrada, D.H. Detection of myxoid liposarcoma-associated FUS-DDIT3 rearrangement variants including a newly identified breakpoint using an optimized RT-PCR assay. Mod. Pathol. 2010, 23, 1307–1315. [Google Scholar] [CrossRef]
- Han, J.; Murthy, R.; Wood, B.; Song, B.; Wang, S.; Sun, B.; Malhi, H.; Kaufman, R.J. ER stress signalling through eIF2α and CHOP, but not IRE1α, attenuates adipogenesis in mice. Diabetologia 2013, 56, 911–924. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S.; Bercovich, Z.; Feiler, Y.; Keshet, R.; Kahana, C. Dual regulatory role of polyamines in adipogenesis. J. Biol. Chem. 2015, 290, 27384–27392. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mancera, P.A.; Bermejo-Rodríguez, C.; Sánchez-Martín, M.; Abollo-Jiménez, F.; Pintado, B.; Sánchez-García, I. FUS-DDIT3 prevents the development of adipocytic precursors in liposarcoma by repressing PPARgamma and C/EBPalpha and activating eIF4E. PLoS ONE 2008, 3, e2569. [Google Scholar] [CrossRef] [PubMed]
- Koelsche, C.; Renner, M.; Hartmann, W.; Brandt, R.; Lehner, B.; Waldburger, N.; Alldinger, I.; Schmitt, T.; Egerer, G.; Penzel, R.; et al. TERT promoter hotspot mutations are recurrent in myxoid liposarcomas but rare in other soft tissue sarcoma entities. J. Exp. Clin. Cancer Res. 2014, 33, 33. [Google Scholar] [CrossRef] [PubMed]
- Trautmann, M.; Cyra, M.; Isfort, I.; Jeiler, B.; Krüger, A.; Grünewald, I.; Steinestel, K.; Altvater, B.; Rossig, C.; Hafner, S.; et al. Phosphatidylinositol-3-kinase (PI3K)/Akt signaling is functionally essential in myxoid liposarcoma. Mol. Cancer Ther. 2019, 18, 834–844. [Google Scholar] [CrossRef]
- Baranov, E.; Black, M.A.; Fletcher, C.D.M.; Charville, G.W.; Hornick, J.L. Nuclear expression of DDIT3 distinguishes high-grade myxoid liposarcoma from other round cell sarcomas. Mod. Pathol. 2021, 34, 1367–1372. [Google Scholar] [CrossRef]
- Italiano, A.; Di Mauro, I.; Rapp, J.; Pierron, G.; Auger, N.; Alberti, L.; Chibon, F.; Escande, F.; Voegeli, A.C.; Ghnassia, J.P.; et al. Clinical effect of molecular methods in sarcoma diagnosis (GENSARC): A prospective, multicentre, observational study. Lancet Oncol. 2016, 17, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Narendra, S.; Valente, A.; Tull, J.; Zhang, S. DDIT3 gene break-apart as a molecular marker for diagnosis of myxoid liposarcoma--assay validation and clinical experience. Diagn. Mol. Pathol. 2011, 20, 218–224. [Google Scholar] [CrossRef]
- Fritchie, K.J.; Crago, A.M.; van de Rijn, M. Desmoid fibromatosis. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 93–95. ISBN 978-92-832-4502-5. [Google Scholar]
- Amary, M.F.; Pauwels, P.; Meulemans, E.; Roemen, G.M.; Islam, L.; Idowu, B.; Bousdras, K.; Diss, T.C.; O’Donnell, P.; Flanagan, A.M. Detection of beta-catenin mutations in paraffin-embedded sporadic desmoid-type fibromatosis by mutation-specific restriction enzyme digestion (MSRED): An ancillary diagnostic tool. Am. J. Surg. Pathol. 2007, 31, 1299–1309. [Google Scholar] [CrossRef]
- Crago, A.M.; Chmielecki, J.; Rosenberg, M.; O’Connor, R.; Byrne, C.; Wilder, F.G.; Thorn, K.; Agius, P.; Kuk, D.; Socci, N.D.; et al. Near universal detection of alterations in CTNNB1 and Wnt pathway regulators in desmoid-type fibromatosis by whole-exome sequencing and genomic analysis. Genes Chromosomes Cancer 2015, 54, 606–615. [Google Scholar] [CrossRef]
- Giarola, M.; Stagi, L.; Presciuttini, S.; Mondini, P.; Radice, M.T.; Sala, P.; Pierotti, M.A.; Bertario, L.; Radice, P. Screening for mutations of the APC gene in 66 Italian familial adenomatous polyposis patients: Evidence for phenotypic differences in cases with and without identified mutation. Hum. Mutat. 1999, 13, 116–123. [Google Scholar] [CrossRef]
- Sturt, N.J.; Gallagher, M.C.; Bassett, P.; Philp, C.R.; Neale, K.F.; Tomlinson, I.P.; Silver, A.R.; Phillips, R.K. Evidence for genetic predisposition to desmoid tumours in familial adenomatous polyposis independent of the germline APC mutation. Gut 2004, 53, 1832–1836. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.L.; Nero, C.; Pappo, A.; Lev, D.; Lazar, A.J.; López-Terrada, D. CTNNB1 genotyping and APC screening in pediatric desmoid tumors: A proposed algorithm. Pediatr. Dev. Pathol. 2012, 15, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Barker, N. The canonical Wnt/beta-catenin signalling pathway. Methods Mol. Biol. 2008, 468, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, B.; Dilworth, H.P.; Iacobuzio-Donahue, C.; Ricci, F.; Weber, K.; Furlong, M.A.; Fisher, C.; Montgomery, E. Nuclear beta-catenin expression distinguishes deep fibromatosis from other benign and malignant fibroblastic and myofibroblastic lesions. Am. J. Surg. Pathol. 2005, 29, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Carlson, J.W.; Fletcher, C.D. Immunohistochemistry for beta-catenin in the differential diagnosis of spindle cell lesions: Analysis of a series and review of the literature. Histopathology 2007, 51, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.L.; Gown, A.M.; Barry, T.S.; Cheang, M.C.; Chan, A.K.; Turbin, D.A.; Hsu, F.D.; West, R.B.; Nielsen, T.O. Nuclear beta-catenin in mesenchymal tumors. Mod. Pathol. 2005, 18, 68–74. [Google Scholar] [CrossRef]
- Colombo, C.; Bolshakov, S.; Hajibashi, S.; Lopez-Terrada, L.; Wang, W.L.; Rao, P.; Benjamin, R.S.; Lazar, A.J.; Lev, D. ‘Difficult to diagnose’ desmoid tumours: A potential role for CTNNB1 mutational analysis. Histopathology 2011, 59, 336–340. [Google Scholar] [CrossRef]
- Le Guellec, S.; Soubeyran, I.; Rochaix, P.; Filleron, T.; Neuville, A.; Hostein, I.; Coindre, J.M. CTNNB1 mutation analysis is a useful tool for the diagnosis of desmoid tumors: A study of 260 desmoid tumors and 191 potential morphologic mimics. Mod. Pathol. 2012, 25, 1551–1558. [Google Scholar] [CrossRef]
- Colombo, C.; Miceli, R.; Lazar, A.J.; Perrone, F.; Pollock, R.E.; Le Cesne, A.; Hartgrink, H.H.; Cleton-Jansen, A.M.; Domont, J.; Bovée, J.V.; et al. CTNNB1 45F mutation is a molecular prognosticator of increased postoperative primary desmoid tumor recurrence: An independent, multicenter validation study. Cancer 2013, 119, 3696–3702. [Google Scholar] [CrossRef]
- Demicco, E.G.; Fritchie, K.J.; Han, A. Solitary fibrous tumour. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 104–106. ISBN 978-92-832-4502-5. [Google Scholar]
- Chmielecki, J.; Crago, A.M.; Rosenberg, M.; O’Connor, R.; Walker, S.R.; Ambrogio, L.; Auclair, D.; McKenna, A.; Heinrich, M.C.; Frank, D.A.; et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat. Genet. 2013, 45, 131–132. [Google Scholar] [CrossRef] [PubMed]
- Mohajeri, A.; Tayebwa, J.; Collin, A.; Nilsson, J.; Magnusson, L.; von Steyern, F.V.; Brosjö, O.; Domanski, H.A.; Larsson, O.; Sciot, R.; et al. Comprehensive genetic analysis identifies a pathognomonic NAB2/STAT6 fusion gene, nonrandom secondary genomic imbalances, and a characteristic gene expression profile in solitary fibrous tumor. Genes Chromosomes Cancer 2013, 52, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.R.; Wu, Y.M.; Kalyana-Sundaram, S.; Cao, X.; Lonigro, R.; Sung, Y.S.; Chen, C.L.; Zhang, L.; Wang, R.; Su, F.; et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat. Genet. 2013, 45, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, M.; Singer, S.; Maki, R.G.; Schwartz, G.K.; Keohan, M.L.; Antonescu, C.R. IGF2 over-expression in solitary fibrous tumours is independent of anatomical location and is related to loss of imprinting. J. Pathol. 2010, 221, 300–307. [Google Scholar] [CrossRef]
- Bertucci, F.; Bouvier-Labit, C.; Finetti, P.; Metellus, P.; Adelaide, J.; Mokhtari, K.; Figarella-Branger, D.; Decouvelaere, A.V.; Miquel, C.; Coindre, J.M.; et al. Gene expression profiling of solitary fibrous tumors. PLoS ONE 2013, 8, e64497. [Google Scholar] [CrossRef]
- Demicco, E.G.; Wani, K.; Ingram, D.; Wagner, M.; Maki, R.G.; Rizzo, A.; Meeker, A.; Lazar, A.J.; Wang, W.L. TERT promoter mutations in solitary fibrous tumour. Histopathology 2018, 73, 843–851. [Google Scholar] [CrossRef]
- Kurisaki-Arakawa, A.; Akaike, K.; Hara, K.; Arakawa, A.; Takahashi, M.; Mitani, K.; Yao, T.; Saito, T. A case of dedifferentiated solitary fibrous tumor in the pelvis with TP53 mutation. Virchows Arch. 2014, 465, 615–621. [Google Scholar] [CrossRef]
- Doyle, L.A.; Vivero, M.; Fletcher, C.D.; Mertens, F.; Hornick, J.L. Nuclear expression of STAT6 distinguishes solitary fibrous tumor from histologic mimics. Mod. Pathol. 2014, 27, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.A.; Tao, D.; Mariño-Enríquez, A. STAT6 is amplified in a subset of dedifferentiated liposarcoma. Mod. Pathol. 2014, 27, 1231–1237. [Google Scholar] [CrossRef]
- Schneider, N.; Hallin, M.; Thway, K. STAT6 loss in dedifferentiated solitary fibrous tumor. Int. J. Surg. Pathol. 2017, 25, 58–60. [Google Scholar] [CrossRef] [PubMed]
- Demicco, E.G.; Wagner, M.J.; Maki, R.G.; Gupta, V.; Iofin, I.; Lazar, A.J.; Wang, W.L. Risk assessment in solitary fibrous tumors: Validation and refinement of a risk stratification model. Mod. Pathol. 2017, 30, 1433–1442. [Google Scholar] [CrossRef]
- Yamamoto, H. Inflammatory myofibroblastic tumour. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 109–111. ISBN 978-92-832-4502-5. [Google Scholar]
- Bridge, J.A.; Kanamori, M.; Ma, Z.; Pickering, D.; Hill, D.A.; Lydiatt, W.; Lui, M.Y.; Colleoni, G.W.; Antonescu, C.R.; Ladanyi, M.; et al. Fusion of the ALK gene to the clathrin heavy chain gene, CLTC, in inflammatory myofibroblastic tumor. Am. J. Pathol. 2001, 159, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, B.; Perez-Atayde, A.; Hibbard, M.K.; Rubin, B.P.; Dal Cin, P.; Pinkus, J.L.; Pinkus, G.S.; Xiao, S.; Yi, E.S.; Fletcher, C.D.; et al. TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am. J. Pathol. 2000, 157, 377–384. [Google Scholar] [CrossRef]
- Hornick, J.L.; Sholl, L.M.; Dal Cin, P.; Childress, M.A.; Lovely, C.M. Expression of ROS1 predicts ROS1 gene rearrangement in inflammatory myofibroblastic tumors. Mod. Pathol. 2015, 28, 732–739. [Google Scholar] [CrossRef]
- Alassiri, A.H.; Ali, R.H.; Shen, Y.; Lum, A.; Strahlendorf, C.; Deyell, R.; Rassekh, R.; Sorensen, P.H.; Laskin, J.; Marra, M.; et al. ETV6-NTRK3 is expressed in a subset of ALK-negative inflammatory myofibroblastic tumors. Am. J. Surg. Pathol. 2016, 40, 1051–1061. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Suurmeijer, A.J.; Zhang, L.; Sung, Y.S.; Jungbluth, A.A.; Travis, W.D.; Al-Ahmadie, H.; Fletcher, C.D.; Alaggio, R. Molecular characterization of inflammatory myofibroblastic tumors with frequent ALK and ROS1 gene fusions and rare novel RET rearrangement. Am. J. Surg. Pathol. 2015, 39, 957–967. [Google Scholar] [CrossRef]
- Mariño-Enríquez, A.; Wang, W.L.; Roy, A.; Lopez-Terrada, D.; Lazar, A.J.; Fletcher, C.D.; Coffin, C.M.; Hornick, J.L. Epithelioid inflammatory myofibroblastic sarcoma: An aggressive intra-abdominal variant of inflammatory myofibroblastic tumor with nuclear membrane or perinuclear ALK. Am. J. Surg. Pathol. 2011, 35, 135–144. [Google Scholar] [CrossRef]
- Lee, J.C.; Li, C.F.; Huang, H.Y.; Zhu, M.J.; Mariño-Enríquez, A.; Lee, C.T.; Ou, W.B.; Hornick, J.L.; Fletcher, J.A. ALK oncoproteins in atypical inflammatory myofibroblastic tumours: Novel RRBP1-ALK fusions in epithelioid inflammatory myofibroblastic sarcoma. J. Pathol. 2017, 241, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Yoshida, A.; Taguchi, K.; Kohashi, K.; Hatanaka, Y.; Yamashita, A.; Mori, D.; Oda, Y. ALK, ROS1 and NTRK3 gene rearrangements in inflammatory myofibroblastic tumours. Histopathology 2016, 69, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.A.; Mertens, F. Low-grade fibromyxoid sarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 127–129. ISBN 978-92-832-4502-5. [Google Scholar]
- Mertens, F.; Fletcher, C.D.; Antonescu, C.R.; Coindre, J.M.; Colecchia, M.; Domanski, H.A.; Downs-Kelly, E.; Fisher, C.; Goldblum, J.R.; Guillou, L.; et al. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS/CREB3L1 fusion gene. Lab. Investig. 2005, 85, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, A.; Hisaoka, M.; Shimajiri, S.; Hayashi, T.; Imamura, T.; Ishida, T.; Fukunaga, M.; Fukuhara, T.; Minato, H.; Nakajima, T.; et al. Molecular detection of FUS-CREB3L2 fusion transcripts in low-grade fibromyxoid sarcoma using formalin-fixed, paraffin-embedded tissue specimens. Am. J. Surg. Pathol. 2006, 30, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Guillou, L.; Benhattar, J.; Gengler, C.; Gallagher, G.; Ranchère-Vince, D.; Collin, F.; Terrier, P.; Terrier-Lacombe, M.J.; Leroux, A.; Marquès, B.; et al. Translocation-positive low-grade fibromyxoid sarcoma: Clinicopathologic and molecular analysis of a series expanding the morphologic spectrum and suggesting potential relationship to sclerosing epithelioid fibrosarcoma: A study from the French Sarcoma Group. Am. J. Surg. Pathol. 2007, 31, 1387–1402. [Google Scholar] [CrossRef] [PubMed]
- Möller, E.; Hornick, J.L.; Magnusson, L.; Veerla, S.; Domanski, H.A.; Mertens, F. FUS-CREB3L2/L1-positive sarcomas show a specific gene expression profile with upregulation of CD24 and FOXL1. Clin. Cancer Res. 2011, 17, 2646–2656. [Google Scholar] [CrossRef]
- Lau, P.P.; Lui, P.C.; Lau, G.T.; Yau, D.T.; Cheung, E.T.; Chan, J.K. EWSR1-CREB3L1 gene fusion: A novel alternative molecular aberration of low-grade fibromyxoid sarcoma. Am. J. Surg. Pathol. 2013, 37, 734–738. [Google Scholar] [CrossRef]
- Cowan, M.L.; Thompson, L.D.; Leon, M.E.; Bishop, J.A. Low-grade fibromyxoid sarcoma of the head and neck: A clinicopathologic series and review of the literature. Head Neck Pathol. 2016, 10, 161–166. [Google Scholar] [CrossRef]
- Doyle, L.A.; Möller, E.; Dal Cin, P.; Fletcher, C.D.; Mertens, F.; Hornick, J.L. MUC4 is a highly sensitive and specific marker for low-grade fibromyxoid sarcoma. Am. J. Surg. Pathol. 2011, 35, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.A.; Mertens, F. Sclerosing epithelioid fibrosarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 130–132. ISBN 978-92-832-4502-5. [Google Scholar]
- Arbajian, E.; Puls, F.; Magnusson, L.; Thway, K.; Fisher, C.; Sumathi, V.P.; Tayebwa, J.; Nord, K.H.; Kindblom, L.G.; Mertens, F. Recurrent EWSR1-CREB3L1 gene fusions in sclerosing epithelioid fibrosarcoma. Am. J. Surg. Pathol. 2014, 38, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Granada, C.; Zhang, L.; Chen, H.W.; Sung, Y.S.; Agaram, N.P.; Jungbluth, A.A.; Antonescu, C.R. A genetic dichotomy between pure sclerosing epithelioid fibrosarcoma (SEF) and hybrid SEF/low-grade fibromyxoid sarcoma: A pathologic and molecular study of 18 cases. Genes Chromosomes Cancer 2015, 54, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.A.; Hornick, J.L. EWSR1 rearrangements in sclerosing epithelioid fibrosarcoma. Am. J. Surg. Pathol. 2013, 37, 1630–1631. [Google Scholar] [CrossRef]
- Dewaele, B.; Libbrecht, L.; Levy, G.; Brichard, B.; Vanspauwen, V.; Sciot, R.; Debiec-Rychter, M. A novel EWS-CREB3L3 gene fusion in a mesenteric sclerosing epithelioid fibrosarcoma. Genes Chromosomes Cancer 2017, 56, 695–699. [Google Scholar] [CrossRef]
- Arbajian, E.; Puls, F.; Antonescu, C.R.; Amary, F.; Sciot, R.; Debiec-Rychter, M.; Sumathi, V.P.; Järås, M.; Magnusson, L.; Nilsson, J.; et al. In-depth genetic analysis of sclerosing epithelioid fibrosarcoma reveals recurrent genomic alterations and potential treatment targets. Clin. Cancer Res. 2017, 23, 7426–7434. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.A.; Wang, W.L.; Dal Cin, P.; Lopez-Terrada, D.; Mertens, F.; Lazar, A.J.; Fletcher, C.D.; Hornick, J.L. MUC4 is a sensitive and extremely useful marker for sclerosing epithelioid fibrosarcoma: Association with FUS gene rearrangement. Am. J. Surg. Pathol. 2012, 36, 1444–1451. [Google Scholar] [CrossRef]
- Kao, Y.C.; Lee, J.C.; Zhang, L.; Sung, Y.S.; Swanson, D.; Hsieh, T.H.; Liu, Y.R.; Agaram, N.P.; Huang, H.Y.; Dickson, B.C.; et al. Recurrent YAP1 and KMT2A gene rearrangements in a subset of MUC4-negative sclerosing epithelioid fibrosarcoma. Am. J. Surg. Pathol. 2020, 44, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Warmke, L.M.; Meis, J.M. Sclerosing epithelioid fibrosarcoma: A distinct sarcoma with aggressive features. Am. J. Surg. Pathol. 2021, 45, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.L.; Antonescu, C.R.; Bahrami, A. Infantile fibrosarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 119–121. ISBN 978-92-832-4502-5. [Google Scholar]
- Tognon, C.; Garnett, M.; Kenward, E.; Kay, R.; Morrison, K.; Sorensen, P.H. The chimeric protein tyrosine kinase ETV6-NTRK3 requires both Ras-Erk1/2 and PI3-kinase-Akt signaling for fibroblast transformation. Cancer Res. 2001, 61, 8909–8916. [Google Scholar] [PubMed]
- Church, A.J.; Calicchio, M.L.; Nardi, V.; Skalova, A.; Pinto, A.; Dillon, D.A.; Gomez-Fernandez, C.R.; Manoj, N.; Haimes, J.D.; Stahl, J.A.; et al. Recurrent EML4-NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod. Pathol. 2018, 31, 463–473. [Google Scholar] [CrossRef]
- Wegert, J.; Vokuhl, C.; Collord, G.; Del Castillo Velasco-Herrera, M.; Farndon, S.J.; Guzzo, C.; Jorgensen, M.; Anderson, J.; Slater, O.; Duncan, C.; et al. Recurrent intragenic rearrangements of EGFR and BRAF in soft tissue tumors of infants. Nat. Commun. 2018, 9, 2378. [Google Scholar] [CrossRef]
- Flucke, U.; van Noesel, M.M.; Wijnen, M.; Zhang, L.; Chen, C.L.; Sung, Y.S.; Antonescu, C.R. TFG-MET fusion in an infantile spindle cell sarcoma with neural features. Genes Chromosomes Cancer 2017, 56, 663–667. [Google Scholar] [CrossRef]
- Hung, Y.P.; Fletcher, C.D.M.; Hornick, J.L. Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumour and histological mimics. Histopathology 2018, 73, 634–644. [Google Scholar] [CrossRef]
- Del Castillo, M.; Chibon, F.; Arnould, L.; Croce, S.; Ribeiro, A.; Perot, G.; Hostein, I.; Geha, S.; Bozon, C.; Garnier, A.; et al. Secretory breast carcinoma: A histopathologic and genomic spectrum characterized by a joint specific ETV6-NTRK3 gene fusion. Am. J. Surg. Pathol. 2015, 39, 1458–1467. [Google Scholar] [CrossRef]
- Skalova, A.; Michal, M.; Simpson, R.H. Newly described salivary gland tumors. Mod. Pathol. 2017, 30 (Suppl. S1), S27–S43. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Folpe, A.L. Adult fibrosarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 122–123. ISBN 978-92-832-4502-5. [Google Scholar]
- Bovée, J.V.M.G.; Huang, S.C.; Wang, J. Epithelioid haemangioma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 152–153. ISBN 978-92-832-4502-5. [Google Scholar]
- Huang, S.C.; Zhang, L.; Sung, Y.S.; Chen, C.L.; Krausz, T.; Dickson, B.C.; Kao, Y.C.; Agaram, N.P.; Fletcher, C.D.; Antonescu, C.R. Frequent FOS gene rearrangements in epithelioid hemangioma: A molecular study of 58 cases with morphologic reappraisal. Am. J. Surg. Pathol. 2015, 39, 1313–1321. [Google Scholar] [CrossRef]
- van IJzendoorn, D.G.; de Jong, D.; Romagosa, C.; Picci, P.; Benassi, M.S.; Gambarotti, M.; Daugaard, S.; van de Sande, M.; Szuhai, K.; Bovée, J.V. Fusion events lead to truncation of FOS in epithelioid hemangioma of bone. Genes Chromosomes Cancer 2015, 54, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Agaram, N.P.; Zhang, L.; Cotzia, P.; Antonescu, C.R. Expanding the spectrum of genetic alterations in pseudomyogenic hemangioendothelioma with recurrent novel ACTB-FOSB gene fusions. Am. J. Surg. Pathol. 2018, 42, 1653–1661. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Chen, H.W.; Zhang, L.; Sung, Y.S.; Panicek, D.; Agaram, N.P.; Dickson, B.C.; Krausz, T.; Fletcher, C.D. ZFP36-FOSB fusion defines a subset of epithelioid hemangioma with atypical features. Genes Chromosomes Cancer 2014, 53, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.P.; Fletcher, C.D.; Hornick, J.L. FOSB is a useful diagnostic marker for pseudomyogenic hemangioendothelioma. Am. J. Surg. Pathol. 2017, 41, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Hornick, J.L.; Agaram, N.P.; Bovée, J.V.M.G. Pseudomyogenic haemangioendothelioma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 169–171. ISBN 978-92-832-4502-5. [Google Scholar]
- Trombetta, D.; Magnusson, L.; von Steyern, F.V.; Hornick, J.L.; Fletcher, C.D.; Mertens, F. Translocation t(7;19)(q22;q13)−a recurrent chromosome aberration in pseudomyogenic hemangioendothelioma? Cancer Genet. 2011, 204, 211–215. [Google Scholar] [CrossRef]
- Walther, C.; Tayebwa, J.; Lilljebjörn, H.; Magnusson, L.; Nilsson, J.; von Steyern, F.V.; Øra, I.; Domanski, H.A.; Fioretos, T.; Nord, K.H.; et al. A novel SERPINE1-FOSB fusion gene results in transcriptional up-regulation of FOSB in pseudomyogenic haemangioendothelioma. J. Pathol. 2014, 232, 534–540. [Google Scholar] [CrossRef]
- Sugita, S.; Hirano, H.; Kikuchi, N.; Kubo, T.; Asanuma, H.; Aoyama, T.; Emori, M.; Hasegawa, T. Diagnostic utility of FOSB immunohistochemistry in pseudomyogenic hemangioendothelioma and its histological mimics. Diagn. Pathol. 2016, 11, 75. [Google Scholar] [CrossRef]
- Mirra, J.M.; Kessler, S.; Bhuta, S.; Eckardt, J. The fibroma-like variant of epithelioid sarcoma. A fibrohistiocytic/myoid cell lesion often confused with benign and malignant spindle cell tumors. Cancer 1992, 69, 1382–1395. [Google Scholar] [CrossRef]
- Hornick, J.L.; Fletcher, C.D. Pseudomyogenic hemangioendothelioma: A distinctive, often multicentric tumor with indolent behavior. Am. J. Surg. Pathol. 2011, 35, 190–201. [Google Scholar] [CrossRef]
- Panagopoulos, I.; Lobmaier, I.; Gorunova, L.; Heim, S. Fusion of the genes WWTR1 and FOSB in pseudomyogenic hemangioendothelioma. Cancer Genomics Proteomics 2019, 16, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Bridge, J.A.; Sumegi, J.; Royce, T.; Baker, M.; Linos, K. A novel CLTC-FOSB gene fusion in pseudomyogenic hemangioendothelioma of bone. Genes Chromosomes Cancer 2021, 60, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.P.; Deyrup, A.T.; Doyle, L.A. Epithelioid haemangioendothelioma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 172–175. ISBN 978-92-832-4502-5. [Google Scholar]
- Errani, C.; Zhang, L.; Sung, Y.S.; Hajdu, M.; Singer, S.; Maki, R.G.; Healey, J.H.; Antonescu, C.R. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer 2011, 50, 644–653. [Google Scholar] [CrossRef]
- Mendlick, M.R.; Nelson, M.; Pickering, D.; Johansson, S.L.; Seemayer, T.A.; Neff, J.R.; Vergara, G.; Rosenthal, H.; Bridge, J.A. Translocation t(1;3)(p36.3;q25) is a nonrandom aberration in epithelioid hemangioendothelioma. Am. J. Surg. Pathol. 2001, 25, 684–687. [Google Scholar] [CrossRef] [PubMed]
- Tanas, M.R.; Sboner, A.; Oliveira, A.M.; Erickson-Johnson, M.R.; Hespelt, J.; Hanwright, P.J.; Flanagan, J.; Luo, Y.; Fenwick, K.; Natrajan, R.; et al. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci. Transl. Med. 2011, 3, 98ra82. [Google Scholar] [CrossRef]
- Tanas, M.R.; Ma, S.; Jadaan, F.O.; Ng, C.K.; Weigelt, B.; Reis-Filho, J.S.; Rubin, B.P. Mechanism of action of a WWTR1(TAZ)-CAMTA1 fusion oncoprotein. Oncogene 2016, 35, 929–938. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Le Loarer, F.; Mosquera, J.M.; Sboner, A.; Zhang, L.; Chen, C.L.; Chen, H.W.; Pathan, N.; Krausz, T.; Dickson, B.C.; et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer 2013, 52, 775–784. [Google Scholar] [CrossRef]
- Doyle, L.A.; Fletcher, C.D.; Hornick, J.L. Nuclear expression of CAMTA1 distinguishes epithelioid hemangioendothelioma from histologic mimics. Am. J. Surg. Pathol. 2016, 40, 94–102. [Google Scholar] [CrossRef]
- Lee, S.J.; Yang, W.I.; Chung, W.S.; Kim, S.K. Epithelioid hemangioendotheliomas with TFE3 gene translocations are compossible with CAMTA1 gene rearrangements. Oncotarget 2016, 7, 7480–7488. [Google Scholar] [CrossRef]
- Kohashi, K.; Bode-Lesniewska, B. Alveolar rhabdomyosarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 205–208. ISBN 978-92-832-4502-5. [Google Scholar]
- Barr, F.G.; Galili, N.; Holick, J.; Biegel, J.A.; Rovera, G.; Emanuel, B.S. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 1993, 3, 113–117. [Google Scholar] [CrossRef]
- Davis, R.J.; D’Cruz, C.M.; Lovell, M.A.; Biegel, J.A.; Barr, F.G. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994, 54, 2869–2872. [Google Scholar]
- Buckingham, M.; Relaix, F. PAX3 and PAX7 as upstream regulators of myogenesis. Semin. Cell Dev. Biol. 2015, 44, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Yu, Y.; Bilke, S.; Walker, R.L.; Mayeenuddin, L.H.; Azorsa, D.O.; Yang, F.; Pineda, M.; Helman, L.J.; Meltzer, P.S. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010, 70, 6497–6508. [Google Scholar] [CrossRef] [PubMed]
- Gryder, B.E.; Yohe, M.E.; Chou, H.C.; Zhang, X.; Marques, J.; Wachtel, M.; Schaefer, B.; Sen, N.; Song, Y.; Gualtieri, A.; et al. PAX3-FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov. 2017, 7, 884–899. [Google Scholar] [CrossRef] [PubMed]
- Skapek, S.X.; Ferrari, A.; Gupta, A.A.; Lupo, P.J.; Butler, E.; Shipley, J.; Barr, F.G.; Hawkins, D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Primers 2019, 5, 1. [Google Scholar] [CrossRef]
- Hostein, I.; Andraud-Fregeville, M.; Guillou, L.; Terrier-Lacombe, M.J.; Deminière, C.; Ranchère, D.; Lussan, C.; Longavenne, E.; Bui, N.B.; Delattre, O.; et al. Rhabdomyosarcoma: Value of myogenin expression analysis and molecular testing in diagnosing the alveolar subtype: An analysis of 109 paraffin-embedded specimens. Cancer 2004, 101, 2817–2824. [Google Scholar] [CrossRef]
- Rekhi, B.; Gupta, C.; Chinnaswamy, G.; Qureshi, S.; Vora, T.; Khanna, N.; Laskar, S. Clinicopathologic features of 300 rhabdomyosarcomas with emphasis upon differential expression of skeletal muscle specific markers in the various subtypes: A single institutional experience. Ann. Diagn. Pathol. 2018, 36, 50–60. [Google Scholar] [CrossRef]
- Lindberg, M.R. (Ed.) Alveolar rhabdomyosarcoma. In Diagnostic Pathology. Soft Tissue Tumors, 3rd ed.; Elsevier: Philadelphia, PA, USA, 2019; pp. 385–391. ISBN 978-0-323-66110-2. [Google Scholar]
- Williamson, D.; Missiaglia, E.; de Reyniès, A.; Pierron, G.; Thuille, B.; Palenzuela, G.; Thway, K.; Orbach, D.; Laé, M.; Fréneaux, P.; et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J. Clin. Oncol. 2010, 28, 2151–2158. [Google Scholar] [CrossRef]
- Agaram, N.P.; Szuhai, K. Spindle cell/sclerosing rhabdomyosarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 211–213. ISBN 978-92-832-4502-5. [Google Scholar]
- Mosquera, J.M.; Sboner, A.; Zhang, L.; Kitabayashi, N.; Chen, C.L.; Sung, Y.S.; Wexler, L.H.; LaQuaglia, M.P.; Edelman, M.; Sreekantaiah, C.; et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer 2013, 52, 538–550. [Google Scholar] [CrossRef]
- Alaggio, R.; Zhang, L.; Sung, Y.S.; Huang, S.C.; Chen, C.L.; Bisogno, G.; Zin, A.; Agaram, N.P.; LaQuaglia, M.P.; Wexler, L.H.; et al. A molecular study of pediatric spindle and sclerosing rhabdomyosarcoma: Identification of novel and recurrent VGLL2-related fusions in infantile cases. Am. J. Surg. Pathol. 2016, 40, 224–235. [Google Scholar] [CrossRef]
- Agaram, N.P.; Chen, C.L.; Zhang, L.; LaQuaglia, M.P.; Wexler, L.; Antonescu, C.R. Recurrent MYOD1 mutations in pediatric and adult sclerosing and spindle cell rhabdomyosarcomas: Evidence for a common pathogenesis. Genes Chromosomes Cancer 2014, 53, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Agaram, N.P.; LaQuaglia, M.P.; Alaggio, R.; Zhang, L.; Fujisawa, Y.; Ladanyi, M.; Wexler, L.H.; Antonescu, C.R. MYOD1-mutant spindle cell and sclerosing rhabdomyosarcoma: An aggressive subtype irrespective of age. A reappraisal for molecular classification and risk stratification. Mod. Pathol. 2019, 32, 27–36. [Google Scholar] [CrossRef]
- Dashti, N.K.; Wehrs, R.N.; Thomas, B.C.; Nair, A.; Davila, J.; Buckner, J.C.; Martinez, A.P.; Sukov, W.R.; Halling, K.C.; Howe, B.M.; et al. Spindle cell rhabdomyosarcoma of bone with FUS-TFCP2 fusion: Confirmation of a very recently described rhabdomyosarcoma subtype. Histopathology 2018, 73, 514–520. [Google Scholar] [CrossRef]
- Rekhi, B.; Upadhyay, P.; Ramteke, M.P.; Dutt, A. MYOD1 (L122R) mutations are associated with spindle cell and sclerosing rhabdomyosarcomas with aggressive clinical outcomes. Mod. Pathol. 2016, 29, 1532–1540. [Google Scholar] [CrossRef] [PubMed]
- Dei Tos, A.P.; Hornick, J.L.; Miettinen, M.; Wanless, I.R.; Wardelmann, E. Gastrointestinal stromal tumour. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 216–221. ISBN 978-92-832-4502-5. [Google Scholar]
- Emile, J.F.; Brahimi, S.; Coindre, J.M.; Bringuier, P.P.; Monges, G.; Samb, P.; Doucet, L.; Hostein, I.; Landi, B.; Buisine, M.P.; et al. Frequencies of KIT and PDGFRA mutations in the MolecGIST prospective population-based study differ from those of advanced GISTs. Med. Oncol. 2012, 29, 1765–1772. [Google Scholar] [CrossRef]
- Joensuu, H.; Hohenberger, P.; Corless, C.L. Gastrointestinal stromal tumour. Lancet 2013, 382, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.C.; Corless, C.L.; Duensing, A.; McGreevey, L.; Chen, C.J.; Joseph, N.; Singer, S.; Griffith, D.J.; Haley, A.; Town, A.; et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science 2003, 299, 708–710. [Google Scholar] [CrossRef]
- Corless, C.L.; Schroeder, A.; Griffith, D.; Town, A.; McGreevey, L.; Harrell, P.; Shiraga, S.; Bainbridge, T.; Morich, J.; Heinrich, M.C. PDGFRA mutations in gastrointestinal stromal tumors: Frequency, spectrum and in vitro sensitivity to imatinib. J. Clin. Oncol. 2005, 23, 5357–5364. [Google Scholar] [CrossRef] [PubMed]
- Corless, C.L.; Barnett, C.M.; Heinrich, M.C. Gastrointestinal stromal tumours: Origin and molecular oncology. Nat. Rev. Cancer 2011, 11, 865–878. [Google Scholar] [CrossRef]
- Boikos, S.A.; Pappo, A.S.; Killian, J.K.; LaQuaglia, M.P.; Weldon, C.B.; George, S.; Trent, J.C.; von Mehren, M.; Wright, J.A.; Schiffman, J.D.; et al. Molecular subtypes of KIT/PDGFRA wild-type gastrointestinal stromal tumors: A report from the National Institutes of Health Gastrointestinal Stromal Tumor Clinic. JAMA Oncol. 2016, 2, 922–928. [Google Scholar] [CrossRef]
- Janeway, K.A.; Kim, S.Y.; Lodish, M.; Nosé, V.; Rustin, P.; Gaal, J.; Dahia, P.L.; Liegl, B.; Ball, E.R.; Raygada, M.; et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc. Natl. Acad. Sci. USA 2011, 108, 314–318. [Google Scholar] [CrossRef]
- Miettinen, M.; Wang, Z.F.; Sarlomo-Rikala, M.; Osuch, C.; Rutkowski, P.; Lasota, J. Succinate dehydrogenase-deficient GISTs: A clinicopathologic, immunohistochemical, and molecular genetic study of 66 gastric GISTs with predilection to young age. Am. J. Surg. Pathol. 2011, 35, 1712–1721. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.J.; Remillard, S.P.; Zhang, Y.X.; Doyle, L.A.; George, S.; Hornick, J.L. Loss of expression of SDHA predicts SDHA mutations in gastrointestinal stromal tumors. Mod. Pathol. 2013, 26, 289–294. [Google Scholar] [CrossRef]
- Espinosa, I.; Lee, C.H.; Kim, M.K.; Rouse, B.T.; Subramanian, S.; Montgomery, K.; Varma, S.; Corless, C.L.; Heinrich, M.C.; Smith, K.S.; et al. A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am. J. Surg. Pathol. 2008, 32, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Joensuu, H.; Rutkowski, P.; Nishida, T.; Steigen, S.E.; Brabec, P.; Plank, L.; Nilsson, B.; Braconi, C.; Bordoni, A.; Magnusson, M.K.; et al. KIT and PDGFRA mutations and the risk of GI stromal tumor recurrence. J. Clin. Oncol. 2015, 33, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.C.; Jones, R.L.; von Mehren, M.; Schöffski, P.; Serrano, C.; Kang, Y.K.; Cassier, P.A.; Mir, O.; Eskens, F.; Tap, W.D.; et al. Avapritinib in advanced PDGFRA D842V-mutant gastrointestinal stromal tumour (NAVIGATOR): A multicentre, open-label, phase 1 trial. Lancet Oncol. 2020, 21, 935–946. [Google Scholar] [CrossRef]
- Brčić, I.; Argyropoulos, A.; Liegl-Atzwanger, B. Update on molecular genetics of gastrointestinal stromal tumors. Diagnostics 2021, 11, 194. [Google Scholar] [CrossRef]
- Nielsen, G.P.; Chi, P. Malignant peripheral nerve sheath tumour. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 254–257. ISBN 978-92-832-4502-5. [Google Scholar]
- Lee, W.; Teckie, S.; Wiesner, T.; Ran, L.; Prieto Granada, C.N.; Lin, M.; Zhu, S.; Cao, Z.; Liang, Y.; Sboner, A.; et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet. 2014, 46, 1227–1232. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Y.; Jones, S.; Sausen, M.; McMahon, K.; Sharma, R.; Wang, Q.; Belzberg, A.J.; Chaichana, K.; Gallia, G.L.; et al. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat. Genet. 2014, 46, 1170–1172. [Google Scholar] [CrossRef]
- Pemov, A.; Hansen, N.F.; Sindiri, S.; Patidar, R.; Higham, C.S.; Dombi, E.; Miettinen, M.M.; Fetsch, P.; Brems, H.; Chandrasekharappa, S.C.; et al. Low mutation burden and frequent loss of CDKN2A/B and SMARCA2, but not PRC2, define premalignant neurofibromatosis type 1-associated atypical neurofibromas. Neuro-Oncol. 2019, 21, 981–992. [Google Scholar] [CrossRef]
- Prieto-Granada, C.N.; Wiesner, T.; Messina, J.L.; Jungbluth, A.A.; Chi, P.; Antonescu, C.R. Loss of H3K27me3 expression is a highly sensitive marker for sporadic and radiation-induced MPNST. Am. J. Surg. Pathol. 2016, 40, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, I.M.; Fletcher, C.D.; Hornick, J.L. Loss of H3K27 trimethylation distinguishes malignant peripheral nerve sheath tumors from histologic mimics. Mod. Pathol. 2016, 29, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Pekmezci, M.; Cuevas-Ocampo, A.K.; Perry, A.; Horvai, A.E. Significance of H3K27me3 loss in the diagnosis of malignant peripheral nerve sheath tumors. Mod. Pathol. 2017, 30, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, I.M.; Dong, F.; Garcia, E.P.; Fletcher, C.D.M.; Jo, V.Y. Recurrent SMARCB1 inactivation in epithelioid malignant peripheral nerve sheath tumors. Am. J. Surg. Pathol. 2019, 43, 835–843. [Google Scholar] [CrossRef]
- Hornick, J.L.; Nielsen, G.P. Beyond “Triton”: Malignant peripheral nerve sheath tumors with complete heterologous rhabdomyoblastic differentiation mimicking spindle cell rhabdomyosarcoma. Am. J. Surg. Pathol. 2019, 43, 1323–1330. [Google Scholar] [CrossRef]
- Folpe, A.L.; Hameed, M. Malignant melanotic nerve sheath tumour. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 258–260. ISBN 978-92-832-4502-5. [Google Scholar]
- Wang, L.; Zehir, A.; Sadowska, J.; Zhou, N.; Rosenblum, M.; Busam, K.; Agaram, N.; Travis, W.; Arcila, M.; Dogan, S.; et al. Consistent copy number changes and recurrent PRKAR1A mutations distinguish Melanotic Schwannomas from Melanomas: SNP-array and next generation sequencing analysis. Genes Chromosomes Cancer 2015, 54, 463–471. [Google Scholar] [CrossRef]
- Torres-Mora, J.; Dry, S.; Li, X.; Binder, S.; Amin, M.; Folpe, A.L. Malignant melanotic schwannian tumor: A clinicopathologic, immunohistochemical, and gene expression profiling study of 40 cases, with a proposal for the reclassification of “melanotic schwannoma”. Am. J. Surg. Pathol. 2014, 38, 94–105. [Google Scholar] [CrossRef]
- Vallat-Decouvelaere, A.V.; Wassef, M.; Lot, G.; Catala, M.; Moussalam, M.; Caruel, N.; Mikol, J. Spinal melanotic schwannoma: A tumour with poor prognosis. Histopathology 1999, 35, 558–566. [Google Scholar] [CrossRef]
- Khoo, M.; Pressney, I.; Hargunani, R.; Tirabosco, R. Melanotic schwannoma: An 11-year case series. Skelet. Radiol. 2016, 45, 29–34. [Google Scholar] [CrossRef]
- Suumeijer, A.J.H.; Ladanyi, M.; Nielsen, T.O. Synovial sarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 290–293. ISBN 978-92-832-4502-5. [Google Scholar]
- Ladanyi, M.; Antonescu, C.R.; Leung, D.H.; Woodruff, J.M.; Kawai, A.; Healey, J.H.; Brennan, M.F.; Bridge, J.A.; Neff, J.R.; Barr, F.G.; et al. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: A multi-institutional retrospective study of 243 patients. Cancer Res. 2002, 62, 135–140. [Google Scholar] [PubMed]
- dos Santos, N.R.; de Bruijn, D.R.; van Kessel, A.G. Molecular mechanisms underlying human synovial sarcoma development. Genes Chromosomes Cancer 2001, 30, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Amary, M.F.; Berisha, F.; Bernardi Fdel, C.; Herbert, A.; James, M.; Reis-Filho, J.S.; Fisher, C.; Nicholson, A.G.; Tirabosco, R.; Diss, T.C.; et al. Detection of SS18-SSX fusion ranscripts in formalin-fixed paraffin-embedded neoplasms: Analysis of conventional RT-PCR, qRT-PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod. Pathol. 2007, 20, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.B.; Barrott, J.J.; Xie, M.; Haldar, M.; Jin, H.; Zhu, J.F.; Monument, M.J.; Mosbruger, T.L.; Langer, E.M.; Randall, R.L.; et al. The impact of chromosomal translocation locus and fusion oncogene coding sequence in synovial sarcomagenesis. Oncogene 2016, 35, 5021–5032. [Google Scholar] [CrossRef]
- Barrott, J.J.; Illum, B.E.; Jin, H.; Hedberg, M.L.; Wang, Y.; Grossmann, A.; Haldar, M.; Capecchi, M.R.; Jones, K.B. Paracrine osteoprotegerin and β-catenin stabilization support synovial sarcomagenesis in periosteal cells. J. Clin. Investig. 2018, 128, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Baranov, E.; McBride, M.J.; Bellizzi, A.M.; Ligon, A.H.; Fletcher, C.D.M.; Kadoch, C.; Hornick, J.L. A novel SS18-SSX fusion-specific antibody for the diagnosis of synovial sarcoma. Am. J. Surg. Pathol. 2020, 44, 922–933. [Google Scholar] [CrossRef]
- Perret, R.; Velasco, V.; Le Guellec, S.; Coindre, J.M.; Le Loarer, F. The SS18-SSX antibody has perfect specificity for the SS18-SSX fusion protein: A validation study of 609 neoplasms including 2 unclassified tumors with SS18-Non-SSX fusions. Am. J. Surg. Pathol. 2021, 45, 582–584. [Google Scholar] [CrossRef]
- Foo, W.C.; Cruise, M.W.; Wick, M.R.; Hornick, J.L. Immunohistochemical staining for TLE1 distinguishes synovial sarcoma from histologic mimics. Am. J. Clin. Pathol. 2011, 135, 839–844. [Google Scholar] [CrossRef]
- Yoshida, A.; Arai, Y.; Satomi, K.; Kubo, T.; Ryo, E.; Matsushita, Y.; Hama, N.; Sudo, K.; Komiyama, M.; Yatabe, Y.; et al. Identification of novel SSX1 fusions in synovial sarcoma. Mod. Pathol. 2022, 35, 228–239. [Google Scholar] [CrossRef]
- Oda, Y.; Dal Cin, P.; Le Loarer, F.; Nielsen, T.O. Epithelioid sarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 294–296. ISBN 978-92-832-4502-5. [Google Scholar]
- Hornick, J.L.; Dal Cin, P.; Fletcher, C.D. Loss of INI1 expression is characteristic of both conventional and proximal-type epithelioid sarcoma. Am. J. Surg. Pathol. 2009, 33, 542–550. [Google Scholar] [CrossRef]
- Kohashi, K.; Izumi, T.; Oda, Y.; Yamamoto, H.; Tamiya, S.; Taguchi, T.; Iwamoto, Y.; Hasegawa, T.; Tsuneyoshi, M. Infrequent SMARCB1/INI1 gene alteration in epithelioid sarcoma: A useful tool in distinguishing epithelioid sarcoma from malignant rhabdoid tumor. Hum. Pathol. 2009, 40, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Sakharpe, A.; Lahat, G.; Gulamhusein, T.; Liu, P.; Bolshakov, S.; Nguyen, T.; Zhang, P.; Belousov, R.; Young, E.; Xie, X.; et al. Epithelioid sarcoma and unclassified sarcoma with epithelioid features: Clinicopathological variables, molecular markers, and a new experimental model. Oncologist 2011, 16, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.; Wang, Z.; Sarlomo-Rikala, M.; Abdullaev, Z.; Pack, S.D.; Fetsch, J.F. ERG expression in epithelioid sarcoma: A diagnostic pitfall. Am. J. Surg. Pathol. 2013, 37, 1580–1585. [Google Scholar] [CrossRef] [PubMed]
- Stockman, D.L.; Hornick, J.L.; Deavers, M.T.; Lev, D.C.; Lazar, A.J.; Wang, W.L. ERG and FLI1 protein expression in epithelioid sarcoma. Mod. Pathol. 2014, 27, 496–501. [Google Scholar] [CrossRef]
- Kohashi, K.; Yamada, Y.; Hotokebuchi, Y.; Yamamoto, H.; Taguchi, T.; Iwamoto, Y.; Oda, Y. ERG and SALL4 expressions in SMARCB1/INI1-deficient tumors: A useful tool for distinguishing epithelioid sarcoma from malignant rhabdoid tumor. Hum. Pathol. 2015, 46, 225–230. [Google Scholar] [CrossRef]
- Kohashi, K.; Yamamoto, H.; Yamada, Y.; Kinoshita, I.; Taguchi, T.; Iwamoto, Y.; Oda, Y. SWI/SNF chromatin-remodeling complex status in SMARCB1/INI1-preserved epithelioid sarcoma. Am. J. Surg. Pathol. 2018, 42, 312–318. [Google Scholar] [CrossRef]
- Oda, Y.; Biegel, J.A.; Pfister, S.M. Extrarenal rhabdoid tumour. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 309–311. ISBN 978-92-832-4502-5. [Google Scholar]
- Schneppenheim, R.; Frühwald, M.C.; Gesk, S.; Hasselblatt, M.; Jeibmann, A.; Kordes, U.; Kreuz, M.; Leuschner, I.; Martin Subero, J.I.; Obser, T.; et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am. J. Hum. Genet. 2010, 86, 279–284. [Google Scholar] [CrossRef]
- Hoot, A.C.; Russo, P.; Judkins, A.R.; Perlman, E.J.; Biegel, J.A. Immunohistochemical analysis of hSNF5/INI1 distinguishes renal and extra-renal malignant rhabdoid tumors from other pediatric soft tissue tumors. Am. J. Surg. Pathol. 2004, 28, 1485–1491. [Google Scholar] [CrossRef]
- Hollmann, T.J.; Hornick, J.L. INI1-deficient tumors: Diagnostic features and molecular genetics. Am. J. Surg. Pathol. 2011, 35, e47–e63. [Google Scholar] [CrossRef]
- Yoshida, A.; Asano, N.; Kawai, A.; Kawamoto, H.; Nakazawa, A.; Kishimoto, H.; Kushima, R. Differential SALL4 immunoexpression in malignant rhabdoid tumours and epithelioid sarcomas. Histopathology 2015, 66, 252–261. [Google Scholar] [CrossRef]
- Kohashi, K.; Nakatsura, T.; Kinoshita, Y.; Yamamoto, H.; Yamada, Y.; Tajiri, T.; Taguchi, T.; Iwamoto, Y.; Oda, Y. Glypican 3 expression in tumors with loss of SMARCB1/INI1 protein expression. Hum. Pathol. 2013, 44, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Versteege, I.; Sévenet, N.; Lange, J.; Rousseau-Merck, M.F.; Ambros, P.; Handgretinger, R.; Aurias, A.; Delattre, O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998, 394, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Eaton, K.W.; Tooke, L.S.; Wainwright, L.M.; Judkins, A.R.; Biegel, J.A. Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr. Blood Cancer 2011, 56, 7–15. [Google Scholar] [CrossRef]
- Jambhekar, N.A.; Landanyi, M. Alveolar soft part sarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 297–299. ISBN 978-92-832-4502-5. [Google Scholar]
- Tsuda, M.; Davis, I.J.; Argani, P.; Shukla, N.; McGill, G.G.; Nagai, M.; Saito, T.; Laé, M.; Fisher, D.E.; Ladanyi, M. TFE3 fusions activate MET signaling by transcriptional up-regulation, defining another class of tumors as candidates for therapeutic MET inhibition. Cancer Res. 2007, 67, 919–929. [Google Scholar] [CrossRef]
- Kobos, R.; Nagai, M.; Tsuda, M.; Merl, M.Y.; Saito, T.; Laé, M.; Mo, Q.; Olshen, A.; Lianoglou, S.; Leslie, C.; et al. Combining integrated genomics and functional genomics to dissect the biology of a cancer-associated, aberrant transcription factor, the ASPSCR1-TFE3 fusion oncoprotein. J. Pathol. 2013, 229, 743–754. [Google Scholar] [CrossRef]
- Mukaihara, K.; Tanabe, Y.; Kubota, D.; Akaike, K.; Hayashi, T.; Mogushi, K.; Hosoya, M.; Sato, S.; Kobayashi, E.; Okubo, T.; et al. Cabozantinib and dastinib exert anti-tumor activity in alveolar soft part sarcoma. PLoS ONE 2017, 12, e0185321. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.J.; Chang, B.; Zou, H.; Qi, Y.; Jiang, J.F.; Li, H.A.; Hu, W.H.; Chen, Y.Z.; Liu, C.X.; Zhang, W.J.; et al. Alveolar soft part sarcoma: A bimarker diagnostic strategy using TFE3 immunoassay and ASPL-TFE3 fusion transcripts in paraffin-embedded tumor tissues. Diagn. Mol. Pathol. 2008, 17, 245–252. [Google Scholar] [CrossRef]
- Williams, A.; Bartle, G.; Sumathi, V.P.; Meis, J.M.; Mangham, D.C.; Grimer, R.J.; Kindblom, L.G. Detection of ASPL/TFE3 fusion transcripts and the TFE3 antigen in formalin-fixed, paraffin-embedded tissue in a series of 18 cases of alveolar soft part sarcoma: Useful diagnostic tools in cases with unusual histological features. Virchows Arch. 2011, 458, 291–300. [Google Scholar] [CrossRef]
- Martignoni, G.; Gobbo, S.; Camparo, P.; Brunelli, M.; Munari, E.; Segala, D.; Pea, M.; Bonetti, F.; Illei, P.B.; Netto, G.J.; et al. Differential expression of cathepsin K in neoplasms harboring TFE3 gene fusions. Mod. Pathol. 2011, 24, 1313–1319. [Google Scholar] [CrossRef]
- Chamberlain, B.K.; McClain, C.M.; Gonzalez, R.S.; Coffin, C.M.; Cates, J.M. Alveolar soft part sarcoma and granular cell tumor: An immunohistochemical comparison study. Hum. Pathol. 2014, 45, 1039–1044. [Google Scholar] [CrossRef]
- Folpe, A.L.; Deyrup, A.T. Alveolar soft-part sarcoma: A review and update. J. Clin. Pathol. 2006, 59, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Antonescu, C.R.; Illei, P.B.; Lui, M.Y.; Timmons, C.F.; Newbury, R.; Reuter, V.E.; Garvin, A.J.; Perez-Atayde, A.R.; Fletcher, J.A.; et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: A distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am. J. Pathol. 2001, 159, 179–192. [Google Scholar] [CrossRef]
- Schöffski, P.; Wozniak, A.; Kasper, B.; Aamdal, S.; Leahy, M.G.; Rutkowski, P.; Bauer, S.; Gelderblom, H.; Italiano, A.; Lindner, L.H.; et al. Activity and safety of crizotinib in patients with alveolar soft part sarcoma with rearrangement of TFE3: European Organization for Research and Treatment of Cancer (EORTC) phase II trial 90101 ‘CREATE’. Ann. Oncol. 2018, 29, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Agaram, N.P.; Antonescu, C.R.; Ladanyi, M. Desmoplastic small round cell tumour. In WHO Classification of Tumours. Soft tissue and bone tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 306–308. ISBN 978-92-832-4502-5. [Google Scholar]
- Sawyer, J.R.; Tryka, A.F.; Lewis, J.M. A novel reciprocal chromosome translocation t(11;22)(p13;q12) in an intraabdominal desmoplastic small round-cell tumor. Am. J. Surg. Pathol. 1992, 16, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Biegel, J.A.; Conard, K.; Brooks, J.J. Translocation (11;22)(p13;q12): Primary change in intra-abdominal desmoplastic small round cell tumor. Genes Chromosomes Cancer 1993, 7, 119–121. [Google Scholar] [CrossRef]
- Ladanyi, M.; Gerald, W. Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res. 1994, 54, 2837–2840. [Google Scholar]
- Gerald, W.L.; Rosai, J.; Ladanyi, M. Characterization of the genomic breakpoint and chimeric transcripts in the EWS-WT1 gene fusion of desmoplastic small round cell tumor. Proc. Natl. Acad. Sci. USA 1995, 92, 1028–1032. [Google Scholar] [CrossRef]
- Gerald, W.L.; Haber, D.A. The EWS-WT1 gene fusion in desmoplastic small round cell tumor. Semin. Cancer Biol. 2005, 15, 197–205. [Google Scholar] [CrossRef]
- Kang, H.J.; Park, J.H.; Chen, W.; Kang, S.I.; Moroz, K.; Ladanyi, M.; Lee, S.B. EWS-WT1 oncoprotein activates neuronal reprogramming factor ASCL1 and promotes neural differentiation. Cancer Res. 2014, 74, 4526–4535. [Google Scholar] [CrossRef]
- Barnoud, R.; Sabourin, J.C.; Pasquier, D.; Ranchère, D.; Bailly, C.; Terrier-Lacombe, M.J.; Pasquier, B. Immunohistochemical expression of WT1 by desmoplastic small round cell tumor: A comparative study with other small round cell tumors. Am. J. Surg. Pathol. 2000, 24, 830–836. [Google Scholar] [CrossRef]
- Schoolmeester, J.K.; Folpe, A.L.; Nair, A.A.; Halling, K.; Sutton, B.C.; Landers, E.; Karnezis, A.N.; Dickson, B.C.; Nucci, M.R.; Kolin, D.L. EWSR1-WT1 gene fusions in neoplasms other than desmoplastic small round cell tumor: A report of three unusual tumors involving the female genital tract and review of the literature. Mod. Pathol. 2021, 34, 1912–1920. [Google Scholar] [CrossRef] [PubMed]
- Gerald, W.L.; Ladanyi, M.; de Alava, E.; Cuatrecasas, M.; Kushner, B.H.; LaQuaglia, M.P.; Rosai, J. Clinical, pathologic, and molecular spectrum of tumors associated with t(11;22)(p13;q12): Desmoplastic small round-cell tumor and its variants. J. Clin. Oncol. 1998, 16, 3028–3036. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Lamhamedi-Cherradi, S.E.; Cuglievan, B.; Menegaz, B.A.; Camacho, P.; Huh, W.; Ramamoorthy, V.; Anderson, P.M.; Pollock, R.E.; Lev, D.C.; et al. Multimodality treatment of desmoplastic small round cell tumor: Chemotherapy and complete cytoreductive surgery improve patient survival. Clin. Cancer Res. 2018, 24, 4865–4873. [Google Scholar] [CrossRef] [PubMed]
- Mello, C.A.; Campos, F.A.B.; Santos, T.G.; Silva, M.L.G.; Torrezan, G.T.; Costa, F.D.; Formiga, M.N.; Nicolau, U.; Nascimento, A.G.; Silva, C.; et al. Desmoplastic small round cell tumor: A review of main molecular abnormalities and emerging therapy. Cancers 2021, 13, 498. [Google Scholar] [CrossRef] [PubMed]
- Bode-Lesniewska, B.; Debiec-Rychter, M.; Tavora, F. Intimal sarcoma. In WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 315–317. ISBN 978-92-832-4502-5. [Google Scholar]
- Bode-Lesniewska, B.; Zhao, J.; Speel, E.J.; Biraima, A.M.; Turina, M.; Komminoth, P.; Heitz, P.U. Gains of 12q13-14 and overexpression of mdm2 are frequent findings inintimal sarcomas of the pulmonary artery. Virchows Arch. 2001, 438, 57–65. [Google Scholar] [CrossRef]
- Zhang, H.; Macdonald, W.D.; Erickson-Johnson, M.; Wang, X.; Jenkins, R.B.; Oliveira, A.M. Cytogenetic and molecular cytogenetic findings of intimal sarcoma. Cancer Genet. Cytogenet. 2007, 179, 146–149. [Google Scholar] [CrossRef]
- Zhao, J.; Roth, J.; Bode-Lesniewska, B.; Pfaltz, M.; Heitz, P.U.; Komminoth, P. Combined comparative genomic hybridization and genomic microarray for detection of gene amplifications in pulmonary artery intimal sarcomas and adrenocortical tumors. Genes Chromosomes Cancer 2002, 34, 48–57. [Google Scholar] [CrossRef]
- Dewaele, B.; Floris, G.; Finalet-Ferreiro, J.; Fletcher, C.D.; Coindre, J.M.; Guillou, L.; Hogendoorn, P.C.; Wozniak, A.; Vanspauwen, V.; Schöffski, P.; et al. Coactivated platelet-derived growth factor receptor α and epidermal growth factor receptor are potential therapeutic targets in intimal sarcoma. Cancer Res. 2010, 70, 7304–7314. [Google Scholar] [CrossRef]
- Koelsche, C.; Benhamida, J.K.; Kommoss, F.K.F.; Stichel, D.; Jones, D.T.W.; Pfister, S.M.; Heilig, C.E.; Fröhling, S.; Stenzinger, A.; Buslei, R.; et al. Intimal sarcomas and undifferentiated cardiac sarcomas carry mutually exclusive MDM2, MDM4, and CDK6 amplifications and share a common DNA methylation signature. Mod. Pathol. 2021, 34, 2122–2129. [Google Scholar] [CrossRef]
- Burke, A.P. Cardiac undifferentiated pleomorphic sarcoma. In WHO Classification of Tumours Thoracic Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 262–264. ISBN 978-92-832-4502-5. [Google Scholar]
- de Álava, E.; Lessnick, S.L.; Stamenkovic, I. Ewing sarcoma. In WHO Classification of Tumours Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 323–325. ISBN 978-92-832-4502-5. [Google Scholar]
- Brohl, A.S.; Solomon, D.A.; Chang, W.; Wang, J.; Song, Y.; Sindiri, S.; Patidar, R.; Hurd, L.; Chen, L.; Shern, J.F.; et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014, 10, e1004475. [Google Scholar] [CrossRef]
- Crompton, B.D.; Stewart, C.; Taylor-Weiner, A.; Alexe, G.; Kurek, K.C.; Calicchio, M.L.; Kiezun, A.; Carter, S.L.; Shukla, S.A.; Mehta, S.S.; et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014, 4, 1326–1341. [Google Scholar] [CrossRef]
- Tirode, F.; Surdez, D.; Ma, X.; Parker, M.; Le Deley, M.C.; Bahrami, A.; Zhang, Z.; Lapouble, E.; Grossetête-Lalami, S.; Rusch, M.; et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014, 4, 1342–1353. [Google Scholar] [CrossRef] [PubMed]
- Machado, I.; Yoshida, A.; Morales, M.G.N.; Abrahão-Machado, L.F.; Navarro, S.; Cruz, J.; Lavernia, J.; Parafioriti, A.; Picci, P.; Llombart-Bosch, A. Review with novel markers facilitates precise categorization of 41 cases of diagnostically challenging, “undifferentiated small round cell tumors”. A clinicopathologic, immunophenotypic and molecular analysis. Ann. Diagn. Pathol. 2018, 34, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.L.; Patel, N.R.; Caragea, M.; Hogendoorn, P.C.; López-Terrada, D.; Hornick, J.L.; Lazar, A.J. Expression of ERG, an Ets family transcription factor, identifies ERG-rearranged Ewing sarcoma. Mod. Pathol. 2012, 25, 1378–1383. [Google Scholar] [CrossRef] [PubMed]
- Folpe, A.L.; Goldblum, J.R.; Rubin, B.P.; Shehata, B.M.; Liu, W.; Dei Tos, A.P.; Weiss, S.W. Morphologic and immunophenotypic diversity in Ewing family tumors: A study of 66 genetically confirmed cases. Am. J. Surg. Pathol. 2005, 29, 1025–1033. [Google Scholar] [CrossRef]
- Hornick, J.L. Soft tissues. In Mills and Sternberg’s Diagnostic Surgical Pathology, 7th ed.; Longacre, T.A., Greenson, J.K., Hornick, J.L., Reuter, V.E., Eds.; Wolters Kluwer: Philadelphia, PA, USA, 2022; pp. 220–223. ISBN 978-1-975150-72-3. [Google Scholar]
- Chen, S.; Deniz, K.; Sung, Y.S.; Zhang, L.; Dry, S.; Antonescu, C.R. Ewing sarcoma with ERG gene rearrangements: A molecular study focusing on the prevalence of FUS-ERG and common pitfalls in detecting EWSR1-ERG fusions by FISH. Genes Chromosomes Cancer 2016, 55, 340–349. [Google Scholar] [CrossRef]
- Newby, R.; Rowe, D.; Paterson, L.; Farquharson, M.A.; MacDuff, E.; Coupe, A.; Hale, J.; Dildey, P.; Bown, N. Cryptic EWSR1-FLI1 fusions in Ewing sarcoma: Potential pitfalls in the diagnostic use of fluorescence in situ hybridization probes. Cancer Genet. Cytogenet. 2010, 200, 60–64. [Google Scholar] [CrossRef]
- Le Loarer, F.; Szuhai, K.; Tirode, F. Round cell sarcoma with EWSR1–non-ETS fusions. In WHO Classification of Tumours Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 326–329. ISBN 978-92-832-4502-5. [Google Scholar]
- Diaz-Perez, J.A.; Nielsen, G.P.; Antonescu, C.; Taylor, M.S.; Lozano-Calderon, S.A.; Rosenberg, A.E. EWSR1/FUS-NFATc2 rearranged round cell sarcoma: Clinicopathological series of 4 cases and literature review. Hum. Pathol. 2019, 90, 45–53. [Google Scholar] [CrossRef]
- Chougule, A.; Taylor, M.S.; Nardi, V.; Chebib, I.; Cote, G.M.; Choy, E.; Nielsen, G.P.; Deshpande, V. Spindle and round cell sarcoma with EWSR1-PATZ1 gene fusion: A sarcoma with polyphenotypic differentiation. Am. J. Surg. Pathol. 2019, 43, 220–228. [Google Scholar] [CrossRef]
- Toki, S.; Wakai, S.; Sekimizu, M.; Mori, T.; Ichikawa, H.; Kawai, A.; Yoshida, A. PAX7 immunohistochemical evaluation of Ewing sarcoma and other small round cell tumours. Histopathology 2018, 73, 645–652. [Google Scholar] [CrossRef]
- Charville, G.W.; Wang, W.L.; Ingram, D.R.; Roy, A.; Thomas, D.; Patel, R.M.; Hornick, J.L.; van de Rijn, M.; Lazar, A.J. EWSR1 fusion proteins mediate PAX7 expression in Ewing sarcoma. Mod. Pathol. 2017, 30, 1312–1320. [Google Scholar] [CrossRef] [PubMed]
- Perret, R.; Escuriol, J.; Velasco, V.; Mayeur, L.; Soubeyran, I.; Delfour, C.; Aubert, S.; Polivka, M.; Karanian, M.; Meurgey, A.; et al. NFATc2-rearranged sarcomas: Clinicopathologic, molecular, and cytogenetic study of 7 cases with evidence of AGGRECAN as a novel diagnostic marker. Mod. Pathol. 2020, 33, 1930–1944. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.I.; Machado, I.; Motoi, T.; Parafioriti, A.; Lacambra, M.; Ichikawa, H.; Kawai, A.; Antonescu, C.R.; Yoshida, A. NKX3-1 is a useful immunohistochemical marker of EWSR1-NFATC2 sarcoma and mesenchymal chondrosarcoma. Am. J. Surg. Pathol. 2020, 44, 719–728. [Google Scholar] [CrossRef]
- Yoshida, A.; Hashimoto, T.; Ryo, E.; Yoshida, K.I.; Motoi, T.; Yatabe, Y.; Mori, T. Confirmation of NKX3-1 expression in EWSR1-NFATC2 sarcoma and mesenchymal chondrosarcoma using monoclonal antibody immunohistochemistry, RT-PCR, and RNA in situ hybridization. Am. J. Surg. Pathol. 2021, 45, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.; Thomas, D.G.; Davis, J.L.; Ng, T.; Patel, R.M.; Harms, P.W.; Betz, B.L.; Schuetze, S.M.; McHugh, J.B.; Horvai, A.E.; et al. EWSR1-NFATC2 translocation-associated sarcoma clinicopathologic findings in a rare aggressive primary bone or soft tissue tumor. Am. J. Surg. Pathol. 2019, 43, 1112–1122. [Google Scholar] [CrossRef]
- Bode-Lesniewska, B.; Fritz, C.; Exner, G.U.; Wagner, U.; Fuchs, B. EWSR1-NFATC2 and FUS-NFATC2 gene fusion-associated mesenchymal tumors: Clinicopathologic correlation and literature review. Sarcoma 2019, 2019, 9386390. [Google Scholar] [CrossRef]
- Bridge, J.A.; Sumegi, J.; Druta, M.; Bui, M.M.; Henderson-Jackson, E.; Linos, K.; Baker, M.; Walko, C.M.; Millis, S.; Brohl, A.S. Clinical, pathological, and genomic features of EWSR1-PATZ1 fusion sarcoma. Mod. Pathol. 2019, 32, 1593–1604. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Yoshida, A. CIC-rearranged sarcoma. In WHO Classification of Tumours Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 330–332. ISBN 978-92-832-4502-5. [Google Scholar]
- Kawamura-Saito, M.; Yamazaki, Y.; Kaneko, K.; Kawaguchi, N.; Kanda, H.; Mukai, H.; Gotoh, T.; Motoi, T.; Fukayama, M.; Aburatani, H.; et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 2006, 15, 2125–2137. [Google Scholar] [CrossRef]
- Italiano, A.; Sung, Y.S.; Zhang, L.; Singer, S.; Maki, R.G.; Coindre, J.M.; Antonescu, C.R. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer 2012, 51, 207–218. [Google Scholar] [CrossRef]
- Le Loarer, F.; Pissaloux, D.; Watson, S.; Godfraind, C.; Galmiche-Rolland, L.; Silva, K.; Mayeur, L.; Italiano, A.; Michot, A.; Pierron, G.; et al. Clinicopathologic features of CIC-NUTM1 sarcomas, a new molecular variant of the family of CIC-fused sarcomas. Am. J. Surg. Pathol. 2019, 43, 268–276. [Google Scholar] [CrossRef]
- Sugita, S.; Arai, Y.; Tonooka, A.; Hama, N.; Totoki, Y.; Fujii, T.; Aoyama, T.; Asanuma, H.; Tsukahara, T.; Kaya, M.; et al. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: A genetically distinct variant of Ewing-like sarcoma. Am. J. Surg. Pathol. 2014, 38, 1571–1576. [Google Scholar] [CrossRef]
- Huang, S.C.; Zhang, L.; Sung, Y.S.; Chen, C.L.; Kao, Y.C.; Agaram, N.P.; Singer, S.; Tap, W.D.; D’Angelo, S.; Antonescu, C.R. Recurrent CIC gene abnormalities in angiosarcomas: A molecular study of 120 cases with concurrent investigation of PLCG1, KDR, MYC, and FLT4 Gene Alterations. Am. J. Surg. Pathol. 2016, 40, 645–655. [Google Scholar] [CrossRef]
- Specht, K.; Sung, Y.S.; Zhang, L.; Richter, G.H.; Fletcher, C.D.; Antonescu, C.R. Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas: Further evidence toward distinct pathologic entities. Genes Chromosomes Cancer 2014, 53, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.C.; Buehler, D.; Choi, E.Y.; McHugh, J.B.; Rubin, B.P.; Billings, S.D.; Balzer, B.; Thomas, D.G.; Lucas, D.R.; Goldblum, J.R.; et al. CIC-DUX sarcomas demonstrate frequent MYC amplification and ETS-family transcription factor expression. Mod. Pathol. 2015, 28, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Siegele, B.; Roberts, J.; Black, J.O.; Rudzinski, E.; Vargas, S.O.; Galambos, C. DUX4 immunohistochemistry is a highly sensitive and specific marker for CIC-DUX4 fusion-positive round cell tumor. Am. J. Surg. Pathol. 2017, 41, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.P.; Fletcher, C.D.; Hornick, J.L. Evaluation of ETV4 and WT1 expression in CIC-rearranged sarcomas and histologic mimics. Mod. Pathol. 2016, 29, 1324–1334. [Google Scholar] [CrossRef]
- Le Guellec, S.; Velasco, V.; Pérot, G.; Watson, S.; Tirode, F.; Coindre, J.M. ETV4 is a useful marker for the diagnosis of CIC-rearranged undifferentiated round-cell sarcomas: A study of 127 cases including mimicking lesions. Mod. Pathol. 2016, 29, 1523–1531. [Google Scholar] [CrossRef]
- Yoshida, A.; Goto, K.; Kodaira, M.; Kobayashi, E.; Kawamoto, H.; Mori, T.; Yoshimoto, S.; Endo, O.; Kodama, N.; Kushima, R.; et al. CIC-rearranged sarcomas: A study of 20 cases and comparisons with Ewing sarcomas. Am. J. Surg. Pathol. 2016, 40, 313–323. [Google Scholar] [CrossRef]
- Sturm, D.; Orr, B.A.; Toprak, U.H.; Hovestadt, V.; Jones, D.T.W.; Capper, D.; Sill, M.; Buchhalter, I.; Northcott, P.A.; Leis, I.; et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 2016, 164, 1060–1072. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Owosho, A.A.; Zhang, L.; Chen, S.; Deniz, K.; Huryn, J.M.; Kao, Y.C.; Huang, S.C.; Singer, S.; Tap, W.; et al. Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: A clinicopathologic and molecular study of 115 cases. Am. J. Surg. Pathol. 2017, 41, 941–949. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Puls, F.; Tirode, F. Sarcoma with BCOR genetic alterations. In WHO Classification of Tumours Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 333–335. ISBN 978-92-832-4502-5. [Google Scholar]
- Kao, Y.C.; Sung, Y.S.; Zhang, L.; Huang, S.C.; Argani, P.; Chung, C.T.; Graf, N.S.; Wright, D.C.; Kellie, S.J.; Agaram, N.P.; et al. Recurrent BCOR internal tandem duplication and YWHAE-NUTM2B fusions in soft tissue undifferentiated round cell sarcoma of infancy: Overlapping genetic features with clear cell sarcoma of kidney. Am. J. Surg. Pathol. 2016, 40, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kumar, V.; Zorman, B.; Fang, E.; Haines, K.M.; Doddapaneni, H.; Hampton, O.A.; White, S.; Bavle, A.A.; Patel, N.R.; et al. Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat. Commun. 2015, 6, 8891. [Google Scholar] [CrossRef] [PubMed]
- Specht, K.; Zhang, L.; Sung, Y.S.; Nucci, M.; Dry, S.; Vaiyapuri, S.; Richter, G.H.; Fletcher, C.D.; Antonescu, C.R. Novel BCOR-MAML3 and ZC3H7B-BCOR gene fusions in undifferentiated small blue round cell sarcomas. Am. J. Surg. Pathol. 2016, 40, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.C.; Sung, Y.S.; Zhang, L.; Jungbluth, A.A.; Huang, S.C.; Argani, P.; Agaram, N.P.; Zin, A.; Alaggio, R.; Antonescu, C.R. BCOR overexpression is a highly sensitive marker in round cell sarcomas with BCOR genetic abnormalities. Am. J. Surg. Pathol. 2016, 40, 1670–1678. [Google Scholar] [CrossRef]
- Kao, Y.C.; Owosho, A.A.; Sung, Y.S.; Zhang, L.; Fujisawa, Y.; Lee, J.C.; Wexler, L.; Argani, P.; Swanson, D.; Dickson, B.C.; et al. BCOR-CCNB3 fusion positive sarcomas: A clinicopathologic and molecular analysis of 36 cases with comparison to morphologic spectrum and clinical behavior of other round cell sarcomas. Am. J. Surg. Pathol. 2018, 42, 604–615. [Google Scholar] [CrossRef]
- Matsuyama, A.; Shiba, E.; Umekita, Y.; Nosaka, K.; Kamio, T.; Yanai, H.; Miyasaka, C.; Watanabe, R.; Ito, I.; Tamaki, T.; et al. Clinicopathologic diversity of undifferentiated sarcoma with bcor-ccnb3 fusion: Analysis of 11 cases with a reappraisal of the utility of immunohistochemistry for BCOR and CCNB3. Am. J. Surg. Pathol. 2017, 41, 1713–1721. [Google Scholar] [CrossRef]
- Shibayama, T.; Okamoto, T.; Nakashima, Y.; Kato, T.; Sakurai, T.; Minamiguchi, S.; Kataoka, T.R.; Shibuya, S.; Yoshizawa, A.; Toguchida, J.; et al. Screening of BCOR-CCNB3 sarcoma using immunohistochemistry for CCNB3: A clinicopathological report of three pediatric cases. Pathol. Int. 2015, 65, 410–414. [Google Scholar] [CrossRef]
- Salgado, C.M.; Zin, A.; Garrido, M.; Kletskaya, I.; DeVito, R.; Reyes-Múgica, M.; Bisogno, G.; Donofrio, V.; Alaggio, R. Pediatric soft tissue tumors with BCOR ITD express EGFR but not OLIG2. Pediatr. Dev. Pathol. 2020, 23, 424–430. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Suurmeijer, A.J.H. EWSR1-SMAD3–positive fibroblastic tumour (emerging). In WHO Classification of Tumours Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 76–77. ISBN 978-92-832-4502-5. [Google Scholar]
- Michal, M.; Berry, R.S.; Rubin, B.P.; Kilpatrick, S.E.; Agaimy, A.; Kazakov, D.V.; Steiner, P.; Ptakova, N.; Martinek, P.; Hadravsky, L.; et al. EWSR1-SMAD3–rearranged fibroblastic tumor: An emerging entity in an increasingly more complex group of fibroblastic/myofibroblastic neoplasms. Am. J. Surg. Pathol. 2018, 42, 1325–1333. [Google Scholar] [CrossRef]
- Kao, Y.C.; Flucke, U.; Eijkelenboom, A.; Zhang, L.; Sung, Y.S.; Suurmeijer, A.J.H.; Antonescu, C.R. Novel EWSR1-SMAD3 gene fusions in a group of acral fibroblastic spindle cell neoplasms. Am. J. Surg. Pathol. 2018, 42, 522–528. [Google Scholar] [CrossRef]
- Armstrong, S.M.; Demicco, E.G. What’s new in fibroblastic tumors? Virchows Arch. 2020, 476, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.W.; Silva, T.M.; Bovée, J.V.M.G. New molecular entities of soft tissue and bone tumors. Curr. Opin. Oncol. 2022, 34, 354–361. [Google Scholar] [CrossRef]
- Suurmeijer, A.J.H.; Antonescu, C.R. NTRK-rearranged spindle cell neoplasm (emerging). In WHO Classification of Tumours Soft Tissue and Bone Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 287–289. ISBN 978-92-832-4502-5. [Google Scholar]
- Agaram, N.P.; Zhang, L.; Sung, Y.S.; Chen, C.L.; Chung, C.T.; Antonescu, C.R.; Fletcher, C.D. Recurrent NTRK1 gene fusions define a novel subset of locally aggressive lipofibromatosis-like neural tumors. Am. J. Surg. Pathol. 2016, 40, 1407–1416. [Google Scholar] [CrossRef]
- Suurmeijer, A.J.H.; Dickson, B.C.; Swanson, D.; Zhang, L.; Sung, Y.S.; Cotzia, P.; Fletcher, C.D.M.; Antonescu, C.R. A novel group of spindle cell tumors defined by S100 and CD34 co-expression shows recurrent fusions involving RAF1, BRAF, and NTRK1/2 genes. Genes Chromosomes Cancer 2018, 57, 611–621. [Google Scholar] [CrossRef]
- Haller, F.; Knopf, J.; Ackermann, A.; Bieg, M.; Kleinheinz, K.; Schlesner, M.; Moskalev, E.A.; Will, R.; Satir, A.A.; Abdelmagid, I.E.; et al. Paediatric and adult soft tissue sarcomas with NTRK1 gene fusions: A subset of spindle cell sarcomas unified by a prominent myopericytic/haemangiopericytic pattern. J. Pathol. 2016, 238, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, F.; Nakatani, F.; Asano, N.; Wakai, S.; Sekimizu, M.; Mitani, S.; Kubo, T.; Kawai, A.; Ichikawa, H.; Yoshida, A. Novel NTRK3 fusions in fibrosarcomas of adults. Am. J. Surg. Pathol. 2019, 43, 523–530. [Google Scholar] [CrossRef]
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef] [PubMed]
- Hechtman, J.F.; Benayed, R.; Hyman, D.M.; Drilon, A.; Zehir, A.; Frosina, D.; Arcila, M.E.; Dogan, S.; Klimstra, D.S.; Ladanyi, M.; et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am. J. Surg. Pathol. 2017, 41, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Rudzinski, E.R.; Lockwood, C.M.; Stohr, B.A.; Vargas, S.O.; Sheridan, R.; Black, J.O.; Rajaram, V.; Laetsch, T.W.; Davis, J.L. Pan-Trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. Am. J. Surg. Pathol. 2018, 42, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Antonescu, C.R. Emerging soft tissue tumors with kinase fusions: An overview of the recent literature with an emphasis on diagnostic criteria. Genes Chromosomes Cancer 2020, 59, 437–444. [Google Scholar] [CrossRef]
- Lam, S.W.; Briaire-de Bruijn, I.H.; van Wezel, T.; Cleven, A.H.G.; Hogendoorn, P.C.W.; Cleton-Jansen, A.M.; Bovée, J.V.M.G. NTRK fusions are extremely rare in bone tumours. Histopathology 2021, 79, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.L.; Lockwood, C.M.; Stohr, B.; Boecking, C.; Al-Ibraheemi, A.; DuBois, S.G.; Vargas, S.O.; Black, J.O.; Cox, M.C.; Luquette, M.; et al. Expanding the spectrum of pediatric NTRK-rearranged mesenchymal tumors. Am. J. Surg. Pathol. 2019, 43, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Tauziède-Espariat, A.; Duchesne, M.; Baud, J.; Le Quang, M.; Bochaton, D.; Azmani, R.; Croce, S.; Hostein, I.; Kesrouani, C.; Guillemot, D.; et al. NTRK-rearranged spindle cell neoplasms are ubiquitous tumors of myofibroblastic lineage with a distinct methylation class. Histopathology 2023, 82, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Masliah-Planchon, J.; Bièche, I.; Guinebretière, J.M.; Bourdeaut, F.; Delattre, O. SWI/SNF chromatin remodeling and human malignancies. Annu. Rev. Pathol. 2015, 10, 145–171. [Google Scholar] [CrossRef]
- Agaimy, A. Moving from “single gene” concept to “functionally homologous multigene complex”: The SWI/SNF paradigm. Semin. Diagn. Pathol. 2021, 38, 165–166. [Google Scholar] [CrossRef]
- Schaefer, I.M.; Hornick, J.L. SWI/SNF complex-deficient soft tissue neoplasms: An update. Semin. Diagn. Pathol. 2021, 38, 222–231. [Google Scholar] [CrossRef]
- Nambirajan, A.; Jain, D. Recent updates in thoracic SMARCA4-deficient undifferentiated tumor. Semin. Diagn. Pathol. 2021, 38, 83–89. [Google Scholar] [CrossRef]
- Yoshida, A.; Boland, J.M.; Jain, D.; Le Loarer, F.; Rekhtman, N. Thoracic SMARCA4-deficient undifferentiated tumor. In WHO Classification of Tumours Thoracic Tumours, 5th ed.; The WHO Classification of Tumours Editorial Board, Ed.; IARC Press: Lyon, France, 2020; pp. 111–114. ISBN 978-92-832-4502-5. [Google Scholar]
- Jelinic, P.; Mueller, J.J.; Olvera, N.; Dao, F.; Scott, S.N.; Shah, R.; Gao, J.; Schultz, N.; Gonen, M.; Soslow, R.A.; et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat. Genet. 2014, 46, 424–426. [Google Scholar] [CrossRef]
- Lin, D.I.; Allen, J.M.; Hecht, J.L.; Killian, J.K.; Ngo, N.T.; Edgerly, C.; Severson, E.A.; Ali, S.M.; Erlich, R.L.; Ramkissoon, S.H.; et al. SMARCA4 inactivation defines a subset of undifferentiated uterine sarcomas with rhabdoid and small cell features and germline mutation association. Mod. Pathol. 2019, 32, 1675–1687. [Google Scholar] [CrossRef]
- Agaimy, A.; Jain, D.; Uddin, N.; Rooper, L.M.; Bishop, J.A. SMARCA4-deficient sinonasal carcinoma: A series of 10 cases expanding the genetic spectrum of SWI/SNF-driven sinonasal malignancies. Am. J. Surg. Pathol. 2020, 44, 703–710. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Priest, J.R.; Duchaine, T.F. DICER1: Mutations, microRNAs and mechanisms. Nat. Rev. Cancer 2014, 14, 662–672. [Google Scholar] [CrossRef] [PubMed]
- McCluggage, W.G.; Foulkes, W.D. DICER1-associated sarcomas: Towards a unified nomenclature. Mod. Pathol. 2021, 34, 1226–1228. [Google Scholar] [CrossRef] [PubMed]
- Warren, M.; Hiemenz, M.C.; Schmidt, R.; Shows, J.; Cotter, J.; Toll, S.; Parham, D.M.; Biegel, J.A.; Mascarenhas, L.; Shah, R. Expanding the spectrum of dicer1-associated sarcomas. Mod. Pathol. 2020, 33, 164–174. [Google Scholar] [CrossRef]
- McCluggage, W.G.; Foulkes, W.D. DICER1-associated sarcomas at different sites exhibit morphological overlap arguing for a unified nomenclature. Virchows Arch. 2021, 479, 431–433. [Google Scholar] [CrossRef] [PubMed]
- Kommoss, F.K.F.; Stichel, D.; Mora, J.; Esteller, M.; Jones, D.T.W.; Pfister, S.M.; Brack, E.; Wachtel, M.; Bode, P.K.; Sinn, H.P.; et al. Clinicopathologic and molecular analysis of embryonal rhabdomyosarcoma of the genitourinary tract: Evidence for a distinct DICER1-associated subgroup. Mod. Pathol. 2021, 34, 1558–1569. [Google Scholar] [CrossRef] [PubMed]
- McCluggage, W.G.; Foulkes, W.D. DICER1-sarcoma: An emerging entity. Mod. Pathol. 2021, 34, 2096–2097. [Google Scholar] [CrossRef]
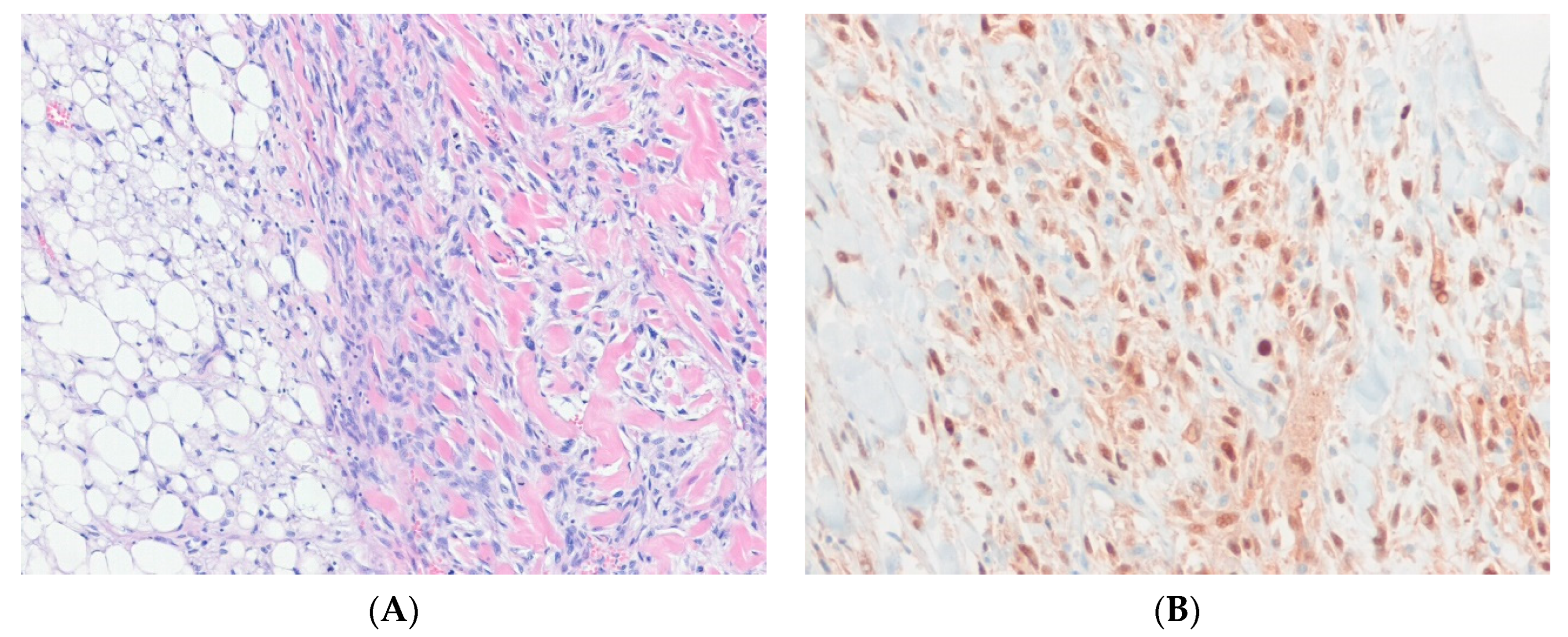
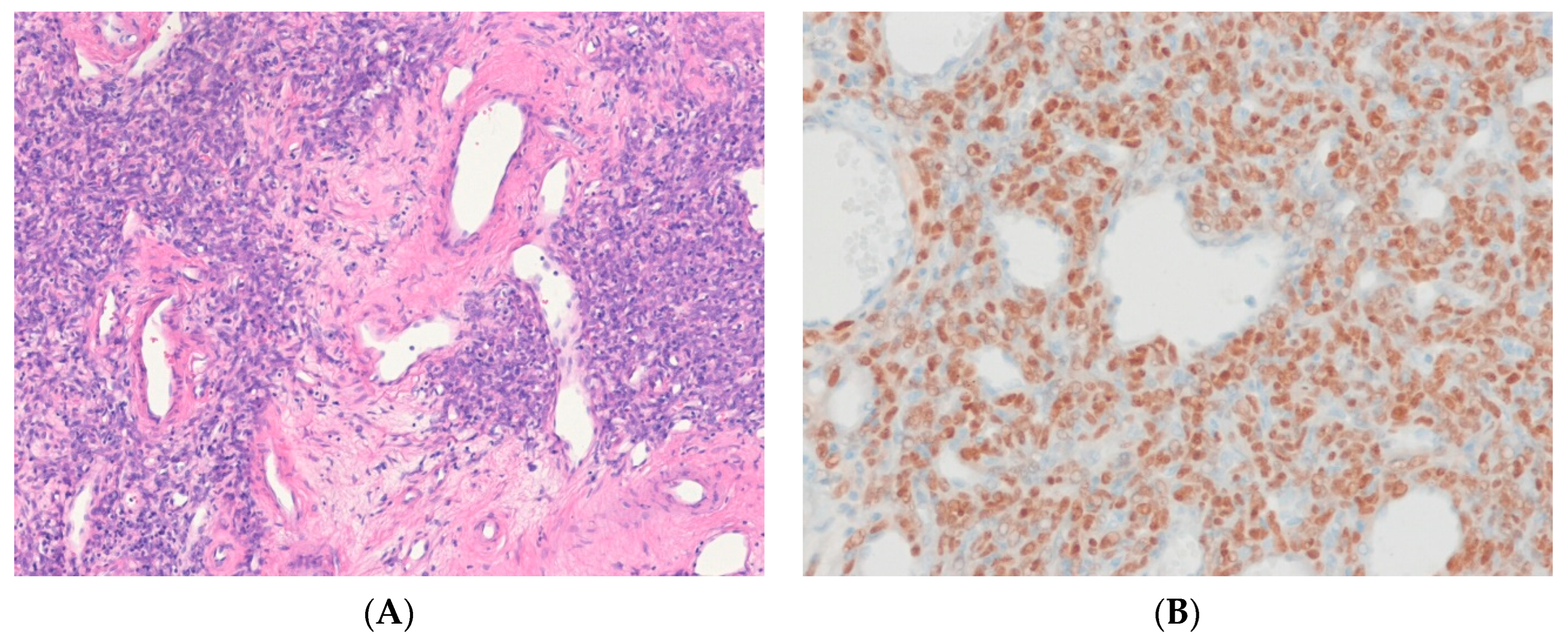
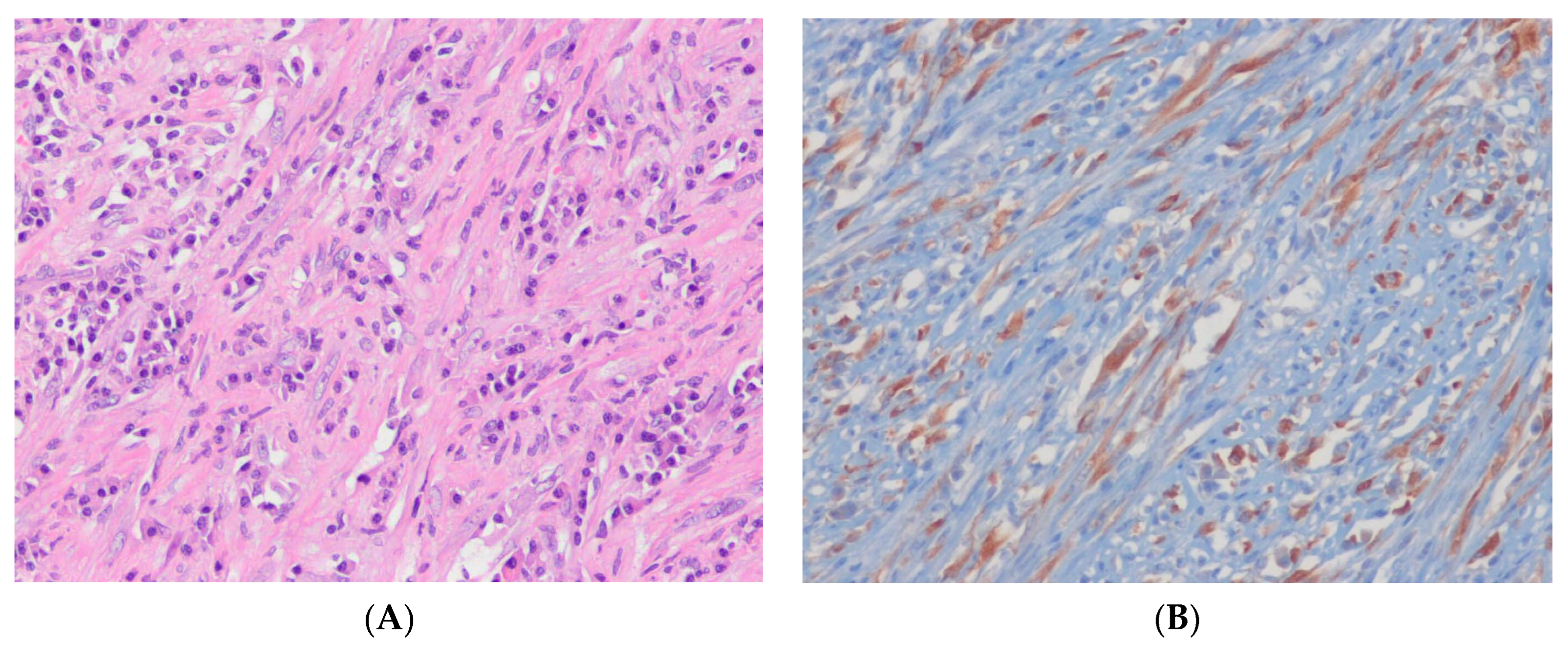
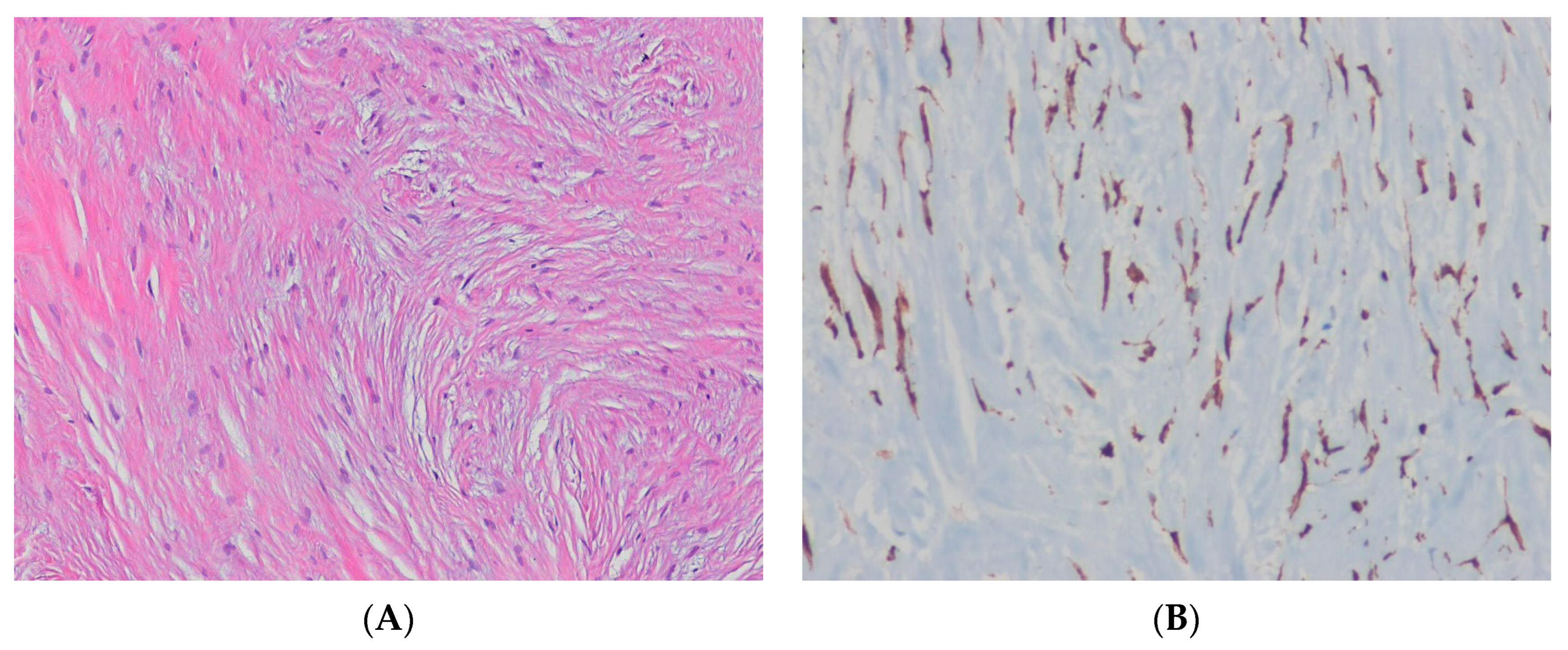
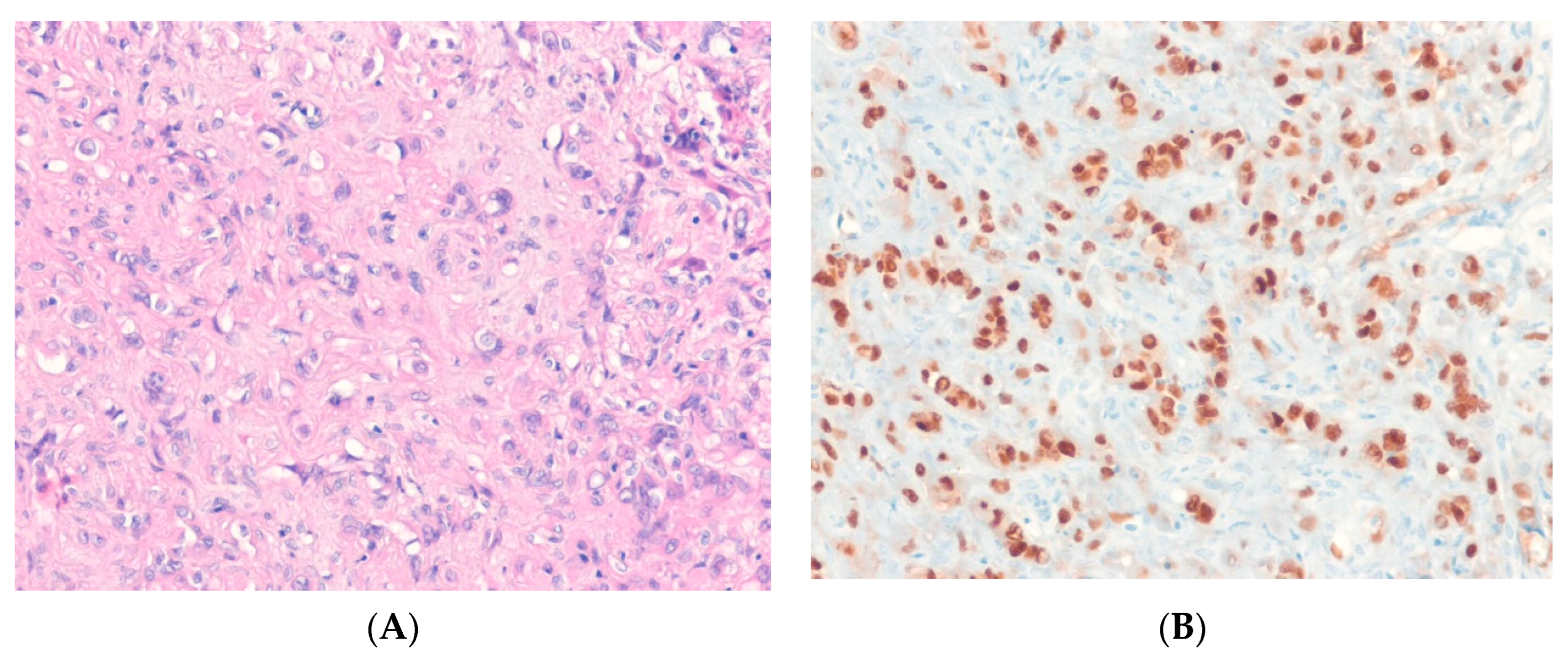

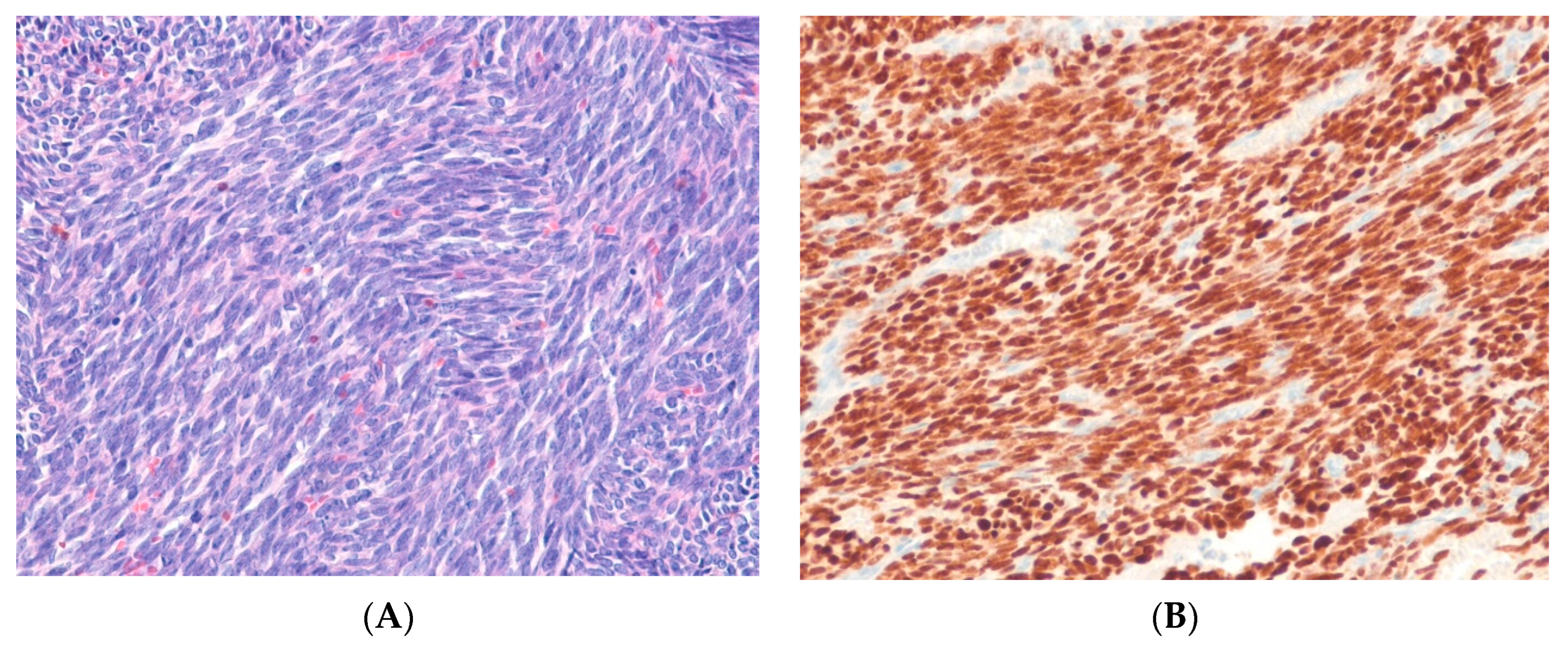
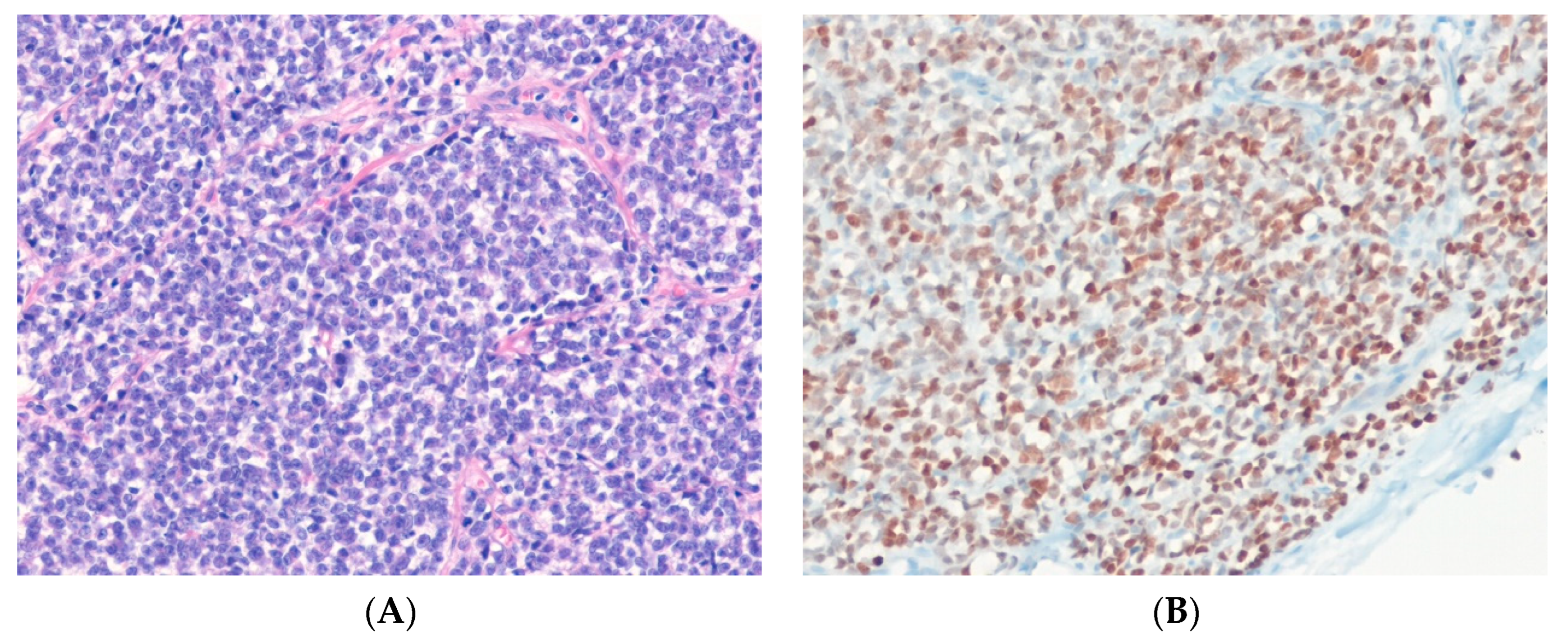
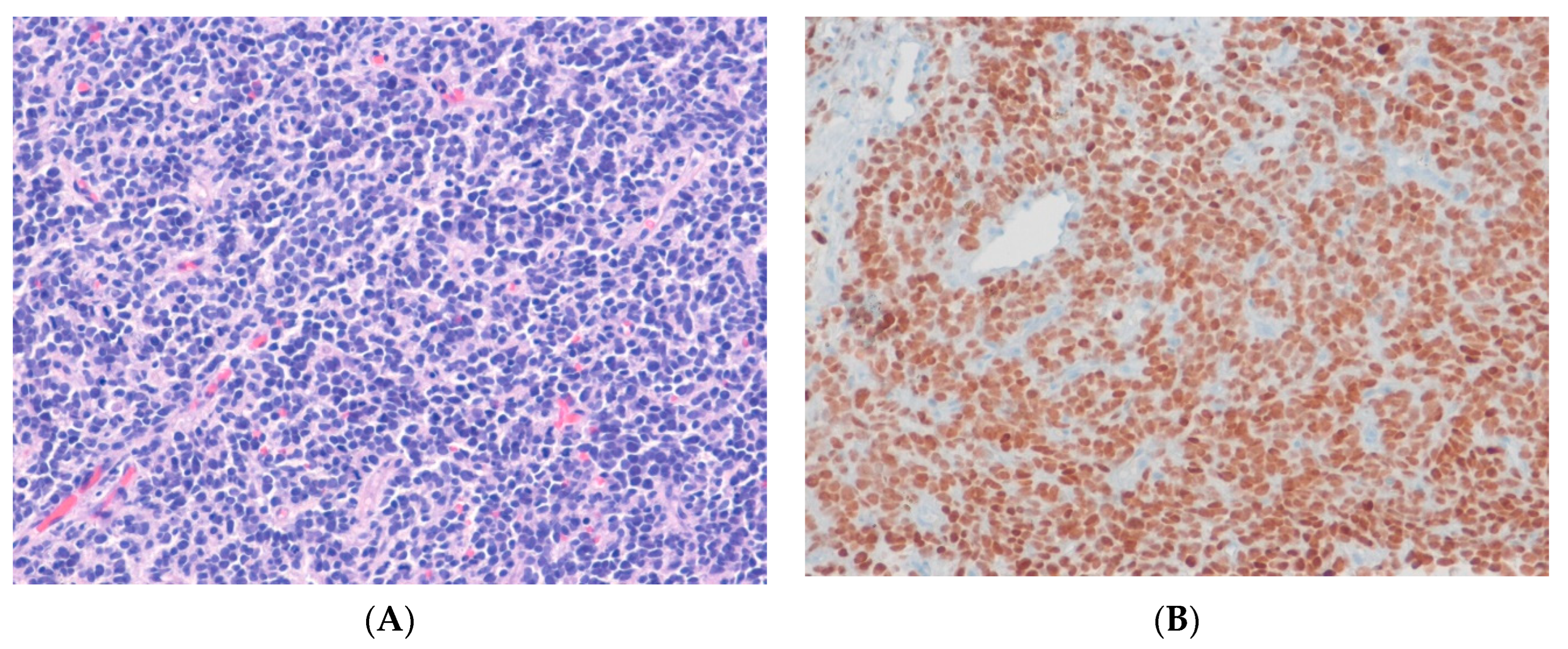
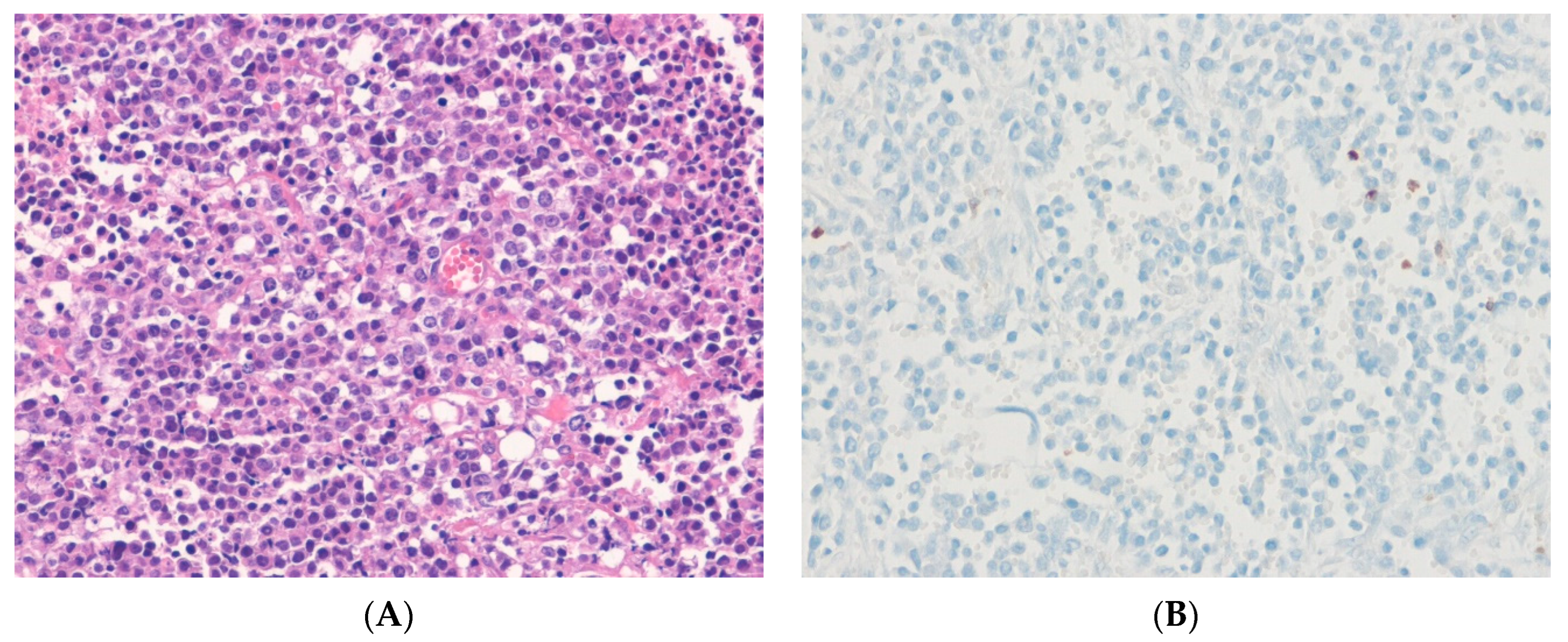
| Tumor Category | Tumor Type | Cytogenetic Alterations | Molecular Alterations | Immunohistochemical Markers | Staining Pattern | References |
|---|---|---|---|---|---|---|
| Adipocytic tumors | Spindle cell/pleomorphic lipoma | Loss of 13q14 Loss of 16q | RB1 deletion | RB1 | Loss | [20,21,22,23] |
| Atypical spindle cell/pleomorphic lipomatous tumor | Loss of 13q14 Monosomy 7 | RB1 deletion (subset) | RB1 | Loss | [28,29,30,31] | |
| Atypical lipomatous tumor/well-differentiated liposarcoma | Gain of 12q13–15 region | MDM2 amplification CDK4 amplification | MDM2, CDK4 | Nuclear staining | [35,36] | |
| Dedifferentiated liposarcoma | Gain of 12q13–15 region | MDM2 amplification CDK4 amplification | MDM2, CDK4 | Nuclear staining | [45,46] | |
| Myxoid liposarcoma | t(12;16)(q13;p11) t(12;22)(q13;q12) | FUS::DDIT3 (>90%) EWSR1::DDIT3 (3%) | DDIT3 | Nuclear staining | [50,56] | |
| Fibroblastic and myofibroblastic tumors | Desmoid fibromatosis | Trisomy 8, trisomy 20 Loss of 5q21 | CTNNB1 mutation APC mutation | β-catenin | Nuclear staining | [60,61,65,66] |
| Solitary fibrous tumor | inv12(q13;q13) | NAB2::STAT6 | STAT6 | Nuclear staining | [72,73,74,80] | |
| Inflammatory myofibroblastic tumor | t(1;2)(q21;p23) t(2;19)(p23;p13) t(2;17)(p23;q23) t(6;17)(q22;p13) t(3;6)(q12;q22) | TPM3::ALK TPM4::ALK CLTC::ALK ROS1::YWHAE ROS1::TFG1 | ALK ROS1 | Cytoplasmic staining | [85,86,87,88,89] | |
| Epithelioid inflammatory myofibroblastic sarcoma | t(2;2)(p23;q13) | RANBP2::ALK RRBP1::ALK | ALK | Nuclear membrane or perinuclear accentuation | [90,91] | |
| Low-grade fibromyxoid sarcoma | t(7:16)(q33;p11) t(11;16)(p11;p11) | FUS::CREB3L2 (>90%) FUS::CREB3L1 EWSR1::CREB3L1 | MUC4 | Cytoplasmic staining | [94,95,96,97,98,99,100] | |
| Sclerosing epithelioid fibrosarcoma | t(11;22)(p11;q12) t(11;16)(p11;p11) t(7;16)(q34;p11) | EWSR1::CREB3L1 (80–90%) EWSR1::CREB3L2 FUS::CREB3L2 | MUC4 | Cytoplasmic staining | [102,103,104,105,106,107] | |
| Infantile fibrosarcoma | t(12;15)(p13;q25) | ETV6::NTRK3 EML4::NTRK3 | Pan-TRK | Cytoplasmic or membranous staining | [112,113,114,115] | |
| Vascular tumors | Epithelioid hemangioma | t(19;19)(q13;q13) t(7;19)(q22;q13) | FOS::VIM, FOS::LMNA, ZFP36::FOSB, WWTR1::FOSB | FOS (subset) FOSB (subset) | Nuclear staining | [120,121,122,123] |
| Pseudomyogenic hemangioendothelioma | t(7;19)(q22;q13) | SERPINE1::FOSB ACTB::FOSB | FOSB | Nuclear staining | [126,127,128] | |
| Epithelioid hemangioendothelioma | t(1;3)(p36;q25) t(X;11)(p11;q13) | WWTR1::CAMTA1 (85–90%) YAP1::TFE3 (5%) | CAMTA1 TFE3 | Nuclear staining | [134,135,136,137,138,139,140] | |
| Skeletal muscle tumors | Alveolar rhabdomyosarcoma | t(2;13)(q35;q14) t(1;13)(p36;q14) | PAX3::FOXO1 (70–90%) PAX7::FOXO1 (10–30%) | Myogenin MyoD1 | Nuclear staining | [142,143,148,149] |
| Congenital/infantile spindle cell rhabdomyosarcoma | SRF::NCOA2, TEAD1::NCOA2, VGLL2::NCOA2, VGLL2::CITED2 | Myogenin MyoD1 | Nuclear staining | [153,154] | ||
| MYOD1-mutant spindle cell/sclerosing rhabdomyosarcoma | MYOD1 mutations | Myogenin MyoD1 | Nuclear staining | [154,155] | ||
| Intraosseous spindle cell rhabdomyosarcoma | EWSR1/FUS::TFCP2 MEIS1::NCOA2 | Myogenin MyoD1 | Nuclear staining | [155,156] | ||
| Gastrointestinal stromal tumors | Gastrointestinal stromal tumor | KIT mutations (75%) PDGFRA mutations (10%) | KIT (CD117) DOG1 | Cytoplasmic or membranous staining | [160,161,162,163,169] | |
| SDH-deficient gastrointestinal stromal tumor | SDH mutations | SDHB SDHA | Loss | [165,166,168] | ||
| Peripheral nerve sheath tumor | MPNST | Complex karyotype with numerical and structural abnormalities | NF1 inactivation; PRC2 components (EED or SUZ12) inactivation | H3K27me3 | Loss | [177,178,179] |
| Epithelioid MPNST | 22q deletion | SMARCB1 inactivation | SMARCB1 | Loss | [180] | |
| MMNST | Mutation and/or loss of heterozygosity of 17q | PRKAR1A inactivation | PRKAR1A | Loss | [183,184] | |
| Tumors of uncertain differentiation | Synovial sarcoma | t(X;18)(p11.2;q11.2) | SS18::SS1, SS2, or SS4 fusion; SS18L1::SSX1 fusion (rare) | SS18-SSX | Nuclear staining | [189,190,193,194] |
| Epithelioid sarcoma | 22q11.2 deletion | SMARCB1 inactivation | SMARCB1 (INI1) | Loss | [197,199,200,201] | |
| Extrarenal rhabdoid tumor | 22q11.2 deletion | SMARCB1 inactivation | SMARCB1 (INI1) | Loss | [206,207] | |
| Alveolar soft part sarcoma | der(17)t(X;17)(p11;q25) | ASPSCR1::TFE3 | TFE3 | Nuclear staining | [217,218] | |
| DSRCT | t(11;22)(p13;q12) | EWSR1::WT1 | WT1 (C-erminus) | Nuclear staining | [225,226,227,228,231] | |
| Intimal sarcoma | 12q12-15 amplification | MDM2 amplification PDGFRA amplification | MDM2 | Nuclear staining | [237,238,239,240] |
| Tumor Type | Cytogenetic Alterations | Molecular Alterations | Immunohistochemical Markers | Staining Pattern | References |
|---|---|---|---|---|---|
| Ewing sarcoma | t(11;22)(q24;q12) t(21;22)(q22;q12) t(2;22)(q33;q12) t(7;22)(p22;q12) t(17;22)(q21;q12) | EWSR1::FLI1 (85–90%) EWSR1::ERG (5–10%) EWSR1::FEV EWSR1::ETV1 EWSR1::ETV4 | NKX2-2 FLI1 ERG CD99 | NKX2-2, FLI1, ERG, nuclear staining; CD99, membranous staining | [244,245,246,247,248] |
| Round cell sarcoma with EWSR1–non-ETS fusions | EWSR1/FUS::FATC2 sarcoma; t(20;22)(q13;q12) t(16;20)(p11;q13) EWSR1::PATZ1 sarcoma; t(22;22)(q12;q12) | EWSR1/FUS::FATC2 sarcoma; EWSR1::NFATC2 FUS::NFATC2 EWSR1::PATZ1 sarcoma; EWSR1::PATZ1 | EWSR1/FUS::FATC2 sarcoma; AGGRECAN NKX3-1 EWSR1::PATZ1 sarcoma; Myogenic (myogenin, MyoD1) and neurogenic markers (S100 protein) | AGGRECAN, cytoplasmic staining; NKX3-1, myogenin, myoD1, nuclear staining; S100 protein, nuclear and cytoplasmic staining | [230,255,259] |
| CIC-rearranged sarcoma | t(4;19)(q35;q13) t(10;19)(q26;q13) t(X;19)(q13;q13) t(15;19)(q14;q13) | CIC::DUX4 (95%) CIC::FOXO4 CIC::LEUTX CIC::NUTM1 CIC::NUTM2A | DUX4 ETV4 WT1 (N-terminus) NUT | Nuclear staining | [265,266,267,268,269,272,273,274] |
| Sarcoma with BCOR genetic alterations | inv(X)(p11.4p11.22) t(X;22)(p11q13) t(10;17)(q22;p13) | BCOR::CCNB3, BCOR::MAML3, BCOR::ZC3H7B; BCOR internal tandem duplications | BCOR CCNB3 SATB2 | Nuclear staining | [282,283,284,285] |
| Tumor Type | Cytogenetic Alterations | Molecular Alterations | Immunohistochemical Markers | Staining Pattern | References |
|---|---|---|---|---|---|
| EWSR1::SMAD3–positive fibroblastic tumor | t(15;22)(q22.33;q12.2) | EWSR1::SMAD3 fusion | ERG | Nuclear staining | [289] |
| NTRK-rearranged spindle cell neoplasm | NTRK1 fusions with LMNA, TPR, or TPM3; NTRK2, NTRK3 fusions | Pan-TRK TRK-A | Cytoplasmic or nuclear staining | [115,117,293,294,295,296,298], | |
| Thoracic SMARCA4-deficient undifferentiated tumor | Biallelic inactivation of SMARCA4 | SMARCA4 (BRG1) | Loss | [307] | |
| DICER1-associated sarcoma | DICER1 mutations | Myogenin MyoD1 | Nuclear staining | [315] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, J.H.; Ro, J.Y. The Recent Advances in Molecular Diagnosis of Soft Tissue Tumors. Int. J. Mol. Sci. 2023, 24, 5934. https://doi.org/10.3390/ijms24065934
Choi JH, Ro JY. The Recent Advances in Molecular Diagnosis of Soft Tissue Tumors. International Journal of Molecular Sciences. 2023; 24(6):5934. https://doi.org/10.3390/ijms24065934
Chicago/Turabian StyleChoi, Joon Hyuk, and Jae Y. Ro. 2023. "The Recent Advances in Molecular Diagnosis of Soft Tissue Tumors" International Journal of Molecular Sciences 24, no. 6: 5934. https://doi.org/10.3390/ijms24065934
APA StyleChoi, J. H., & Ro, J. Y. (2023). The Recent Advances in Molecular Diagnosis of Soft Tissue Tumors. International Journal of Molecular Sciences, 24(6), 5934. https://doi.org/10.3390/ijms24065934






