Ethylene and Jasmonates Signaling Network Mediating Secondary Metabolites under Abiotic Stress
Abstract
1. Introduction
2. Ethylene and Jasmonates
2.1. Ethylene Biosynthesis and Signaling
2.2. Jasmonates Biosynthesis and Signaling
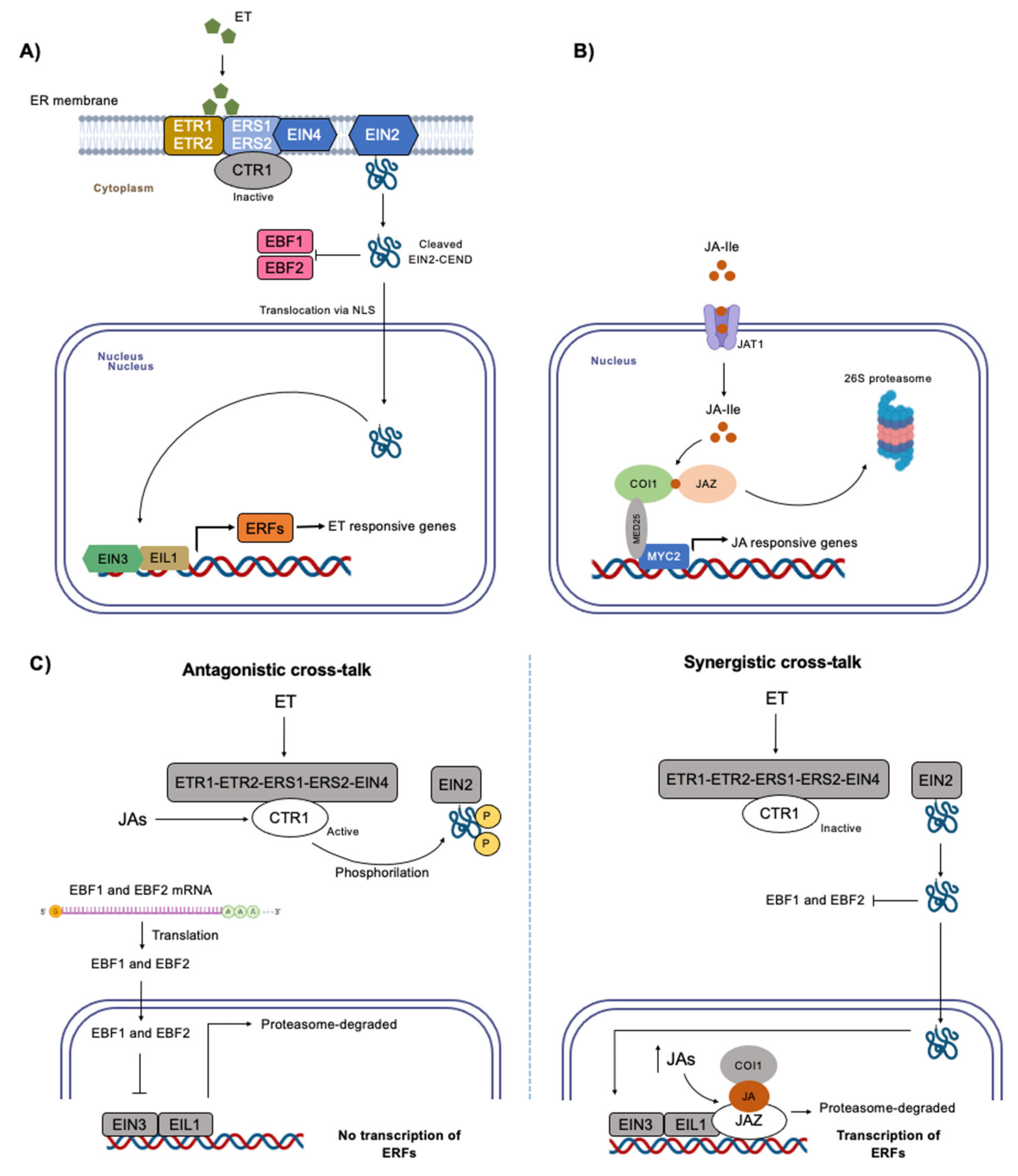
3. Ethylene and Jasmonates Action and Response Mechanism under Abiotic Stress
3.1. Ethylene
3.2. Jasmonates
3.3. Ethylene and Jasmonates Cross-Talk under Abiotic Stress
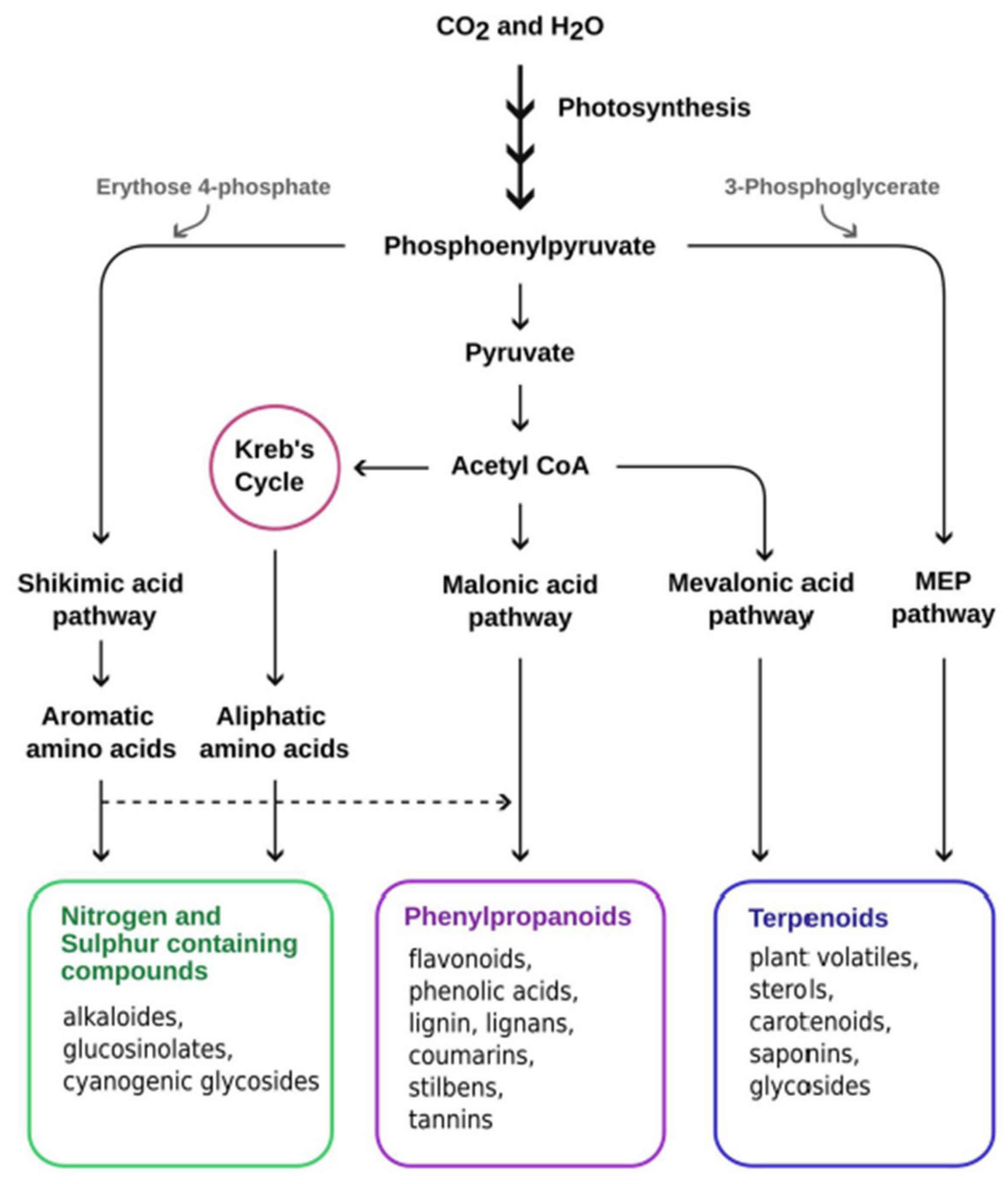
4. Role of Secondary Metabolites under Abiotic Stress
4.1. Secondary Metabolites under Abiotic Stress
4.2. Flavonoids and Polyphenols
4.3. Lignins
4.4. Terpenoids
4.5. Cyanogenic Glucosides
4.6. Amino Acids and Their Derivatives
4.7. Phytoalexins and Glucosinolates
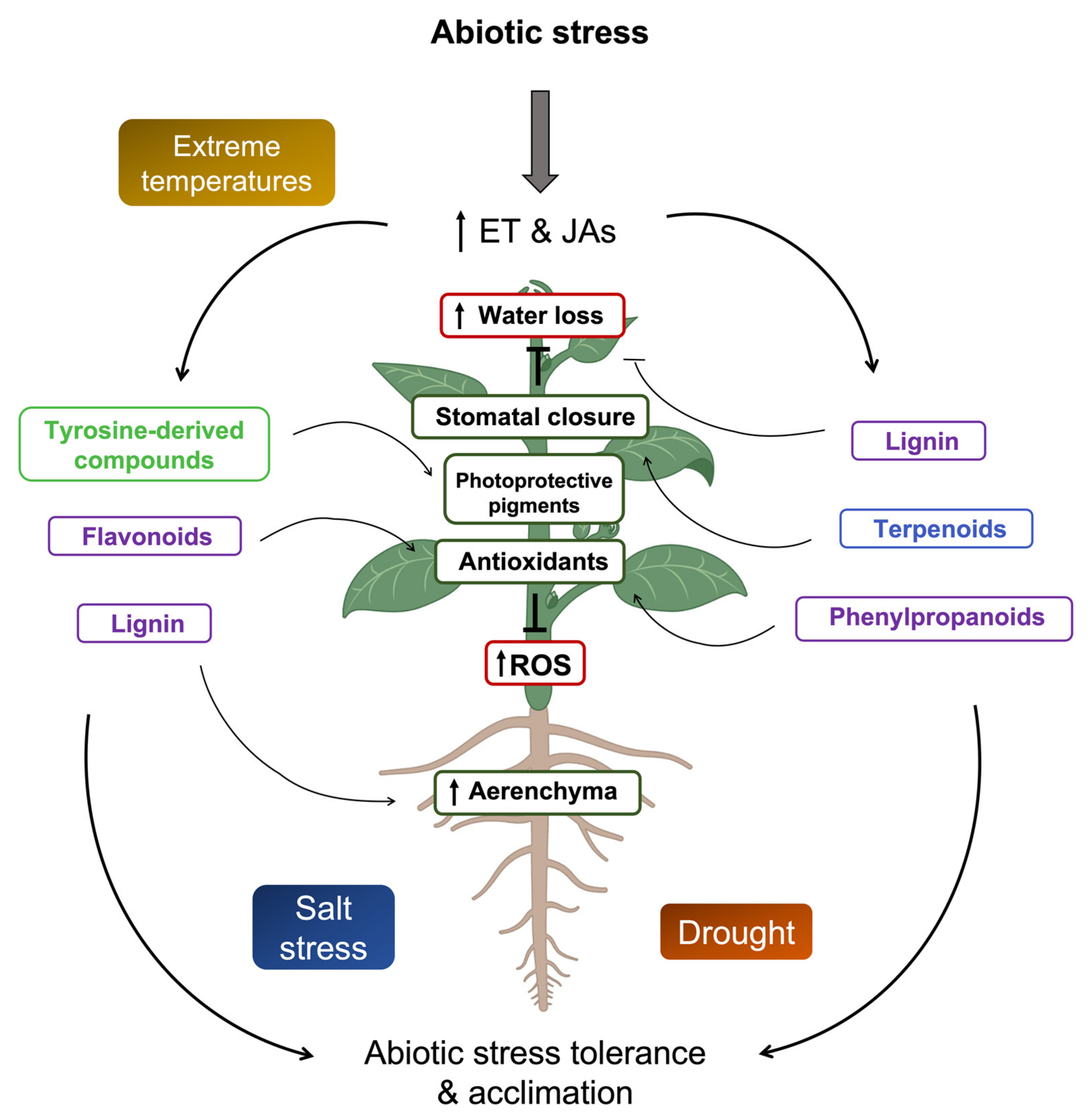
5. Ethylene and Jasmonates Role on Secondary Metabolites under Abiotic Stress
5.1. Ethylene and Secondary Metabolites
5.2. Jasmonates and Secondary Metabolites
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Watts, N.; Amann, M.; Arnell, N.; Ayeb-Karlsson, S.; Beagley, J.; Belesova, K.; Boykoff, M.; Byass, P.; Cai, W.; Campbell-Lendrum, D.; et al. The 2020 Report of The Lancet Countdown on Health and Climate Change: Responding to Converging Crises. Lancet 2021, 397, 129–170. [Google Scholar] [CrossRef]
- Chaudhry, S.; Sidhu, G.P.S. Climate Change Regulated Abiotic Stress Mechanisms in Plants: A Comprehensive Review. Plant Cell Rep. 2022, 41, 1–31. [Google Scholar] [CrossRef]
- IPCC. Climate Change 2022: Impacts, Adaptation and Vulnerability. Working Group II Contribution to the IPCC Sixth Assessment Report; IPCC: Cambridge, UK; New York, NY, USA, 2022. [Google Scholar]
- Van Dijk, M.; Morley, T.; Rau, M.L.; Saghai, Y. A Meta-Analysis of Projected Global Food Demand and Population at Risk of Hunger for the Period 2010–2050. Nat. Food 2021, 2, 494–501. [Google Scholar] [CrossRef]
- St. Clair, S.B.; Lynch, J.P. The Opening of Pandora’s Box: Climate Change Impacts on Soil Fertility and Crop Nutrition in Developing Countries. Plant Soil 2010, 335, 101–115. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic Stress Responses in Plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Sánchez-Bermúdez, M.; del Pozo, J.C.; Pernas, M. Effects of Combined Abiotic Stresses Related to Climate Change on Root Growth in Crops. Front. Plant Sci. 2022, 13, 918537. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic Stress, the Field Environment and Stress Combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive Oxygen Species, Abiotic Stress and Stress Combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Jogawat, A.; Yadav, B.; Chhaya; Lakra, N.; Singh, A.K.; Narayan, O.P. Crosstalk between Phytohormones and Secondary Metabolites in the Drought Stress Tolerance of Crop Plants: A Review. Physiol. Plant. 2021, 172, 1106–1132. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Hormonal Impact on Photosynthesis and Photoprotection in Plants. Plant Physiol. 2021, 185, 1500–1522. [Google Scholar] [CrossRef]
- Tamaoki, M.; Freeman, J.L.; Pilon-Smits, E.A.H. Cooperative Ethylene and Jasmonic Acid Signaling Regulates Selenite Resistance in Arabidopsis. Plant Physiol. 2008, 146, 1219–1230. [Google Scholar] [CrossRef]
- Tuominen, H.; Overmyer, K.; Keinänen, M.; Kollist, H.; Kangasjärvi, J. Mutual Antagonism of Ethylene and Jasmonic Acid Regulates Ozone-Induced Spreading Cell Death in Arabidopsis. Plant J. 2004, 39, 59–69. [Google Scholar] [CrossRef]
- Ku, Y.-S.; Contador, C.A.; Ng, M.-S.; Yu, J.; Chung, G.; Lam, H.-M. The Effects of Domestication on Secondary Metabolite Composition in Legumes. Front. Genet. 2020, 11, 581357. [Google Scholar] [CrossRef] [PubMed]
- EEA Report No 25/2019 Drivers of Change of Relevance for Europe’s Environment and Sustainability. Available online: https://www.eea.europa.eu/publications/drivers-of-change (accessed on 12 February 2023).
- Müller, M. Foes or Friends: ABA and Ethylene Interaction under Abiotic Stress. Plants 2021, 10, 448. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonate Signaling in Plant Stress Responses and Development—Active and Inactive Compounds. New Biotechnol. 2016, 33, 604–613. [Google Scholar] [CrossRef]
- Johnson, P.R.; Ecker, J.R. The ethylene gas signal transduction pathway: A Molecular Perspective. Annu. Rev. Genet. 1998, 32, 227–254. [Google Scholar] [CrossRef] [PubMed]
- Pattyn, J.; Vaughan-Hirsch, J.; Van De Poel, B. The regulation of ethylene biosynthesis: A complex multilevel control circuitry. New Phytol. 2021, 229, 770–782. [Google Scholar] [CrossRef]
- Yang, S.F.; Hoffman, N.E. Ethylene Biosynthesis and its Regulation in Higher Plants. Annu. Rev. Plant Physiol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Kende, H. Ethylene Biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 283–307. [Google Scholar] [CrossRef]
- Park, C.H.; Roh, J.; Youn, J.-H.; Son, S.-H.; Park, J.H.; Kim, S.Y.; Kim, T.-W.; Kim, S.-K. Arabidopsis ACC Oxidase 1 Coordinated by Multiple Signals Mediates Ethylene Biosynthesis and Is Involved in Root Development. Mol. Cells 2018, 41, 923–932. [Google Scholar] [CrossRef]
- Liang, X.; Abel, S.; Keller, J.A.; Shen, N.F.; Theologis, A. The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1992, 89, 11046–11050. [Google Scholar] [CrossRef] [PubMed]
- Houben, M.; Van de Poel, B. 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (ACO): The Enzyme That Makes the Plant Hormone Ethylene. Front. Plant Sci. 2019, 10, 695. [Google Scholar] [CrossRef]
- Vriezen, W.H.; Hulzink, R.; Mariani, C.; Voesenek, L.A. 1-Aminocyclopropane-1-Carboxylate Oxidase Activity Limits Ethylene Biosynthesis in Rumex palustris during Submergence. Plant Physiol. 1999, 121, 189–196. [Google Scholar] [CrossRef]
- English, P.J.; Lycett, G.; Roberts, J.A.; Jackson, M.B. Increased 1-Aminocyclopropane-1-Carboxylic Acid Oxidase Activity in Shoots of Flooded Tomato Plants Raises Ethylene Production to Physiologically Active Levels. Plant Physiol. 1995, 109, 1435–1440. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Fukao, T.; Ronald, P.; Ismail, A.; Heuer, S.; Mackill, D. Submergence Tolerant Rice: SUB1’s Journey from Landrace to Modern Cultivar. Rice 2010, 3, 138–147. [Google Scholar] [CrossRef]
- Tsuchisaka, A.; Theologis, A. Unique and Overlapping Expression Patterns among the Arabidopsis 1-Amino-Cyclopropane-1-Carboxylate Synthase Gene Family Members. Plant Physiol. 2004, 136, 2982–3000. [Google Scholar] [CrossRef] [PubMed]
- Tsuchisaka, A.; Theologis, A. Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc. Natl. Acad. Sci. USA 2004, 101, 2275–2280. [Google Scholar] [CrossRef]
- Skottke, K.R.; Yoon, G.M.; Kieber, J.J.; Delong, A. Protein Phosphatase 2A Controls Ethylene Biosynthesis by Differentially Regulating the Turnover of ACC Synthase Isoforms. PLoS Genet. 2011, 7, e1001370. [Google Scholar] [CrossRef]
- Lyzenga, W.J.; Booth, J.K.; Stone, S.L. The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J. 2012, 71, 23–34. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef]
- Fatma, M.; Iqbal, N.; Gautam, H.; Sehar, Z.; Sofo, A.; D’Ippolito, I.; Khan, N. Ethylene and Sulfur Coordinately Modulate the Antioxidant System and ABA Accumulation in Mustard Plants Under Salt Stress. Plants 2021, 10, 180. [Google Scholar] [CrossRef] [PubMed]
- Merchante, C.; Brumos, J.; Yun, J.; Hu, Q.; Spencer, K.R.; Enríquez, P.; Binder, B.M.; Heber, S.; Stepanova, A.N.; Alonso, J.M. Gene-Specific Translation Regulation Mediated by the Hormone-Signaling Molecule EIN2. Cell 2015, 163, 684–697. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Bullock, D.A.; Alonso, J.M.; Stepanova, A.N. To Fight or to Grow: The Balancing Role of Ethylene in Plant Abiotic Stress Responses. Plants 2021, 11, 33. [Google Scholar] [CrossRef]
- Dolgikh, V.A.; Pukhovaya, E.M.; Zemlyanskaya, E.V. Shaping Ethylene Response: The Role of EIN3/EIL1 Transcription Factors. Front. Plant Sci. 2019, 10, 1030. [Google Scholar] [CrossRef] [PubMed]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef]
- Demole, E.; Lederer, E.; Mercier, D. Isolement et détermination de la structure du jasmonate de méthyle, constituant odorant caractéristique de l’essence de jasmin. Helv. Chim. Acta 1962, 45, 675–685. [Google Scholar] [CrossRef]
- Ueda, J.; Kato, J. Isolation and Identification of a Senescence-promoting Substance from Wormwood (Artemisia absinthium L.). Plant Physiol. 1980, 66, 246–249. [Google Scholar] [CrossRef]
- Dathe, W.; Rönsch, H.; Preiss, A.; Schade, W.; Sembdner, G.; Schreiber, K. Endogenous plant hormones of the broad bean, Vicia faba L. (-)-jasmonic acid, a plant growth inhibitor in pericarp. Planta 1981, 153, 530–535. [Google Scholar] [CrossRef]
- Poudel, A.N.; Holtsclaw, R.E.; Kimberlin, A.; Sen, S.; Zeng, S.; Joshi, T.; Lei, Z.; Sumner, L.W.; Singh, K.; Matsuura, H.; et al. 12-Hydroxy-Jasmonoyl-l-Isoleucine Is an Active Jasmonate That Signals through CORONATINE INSENSITIVE 1 and Contributes to the Wound Response in Arabidopsis. Plant Cell Physiol. 2019, 60, 2152–2166. [Google Scholar] [CrossRef]
- Schuman, M.C.; Meldau, S.; Gaquerel, E.; Diezel, C.; McGale, E.; Greenfield, S.; Baldwin, I.T. The Active Jasmonate JA-Ile Regulates a Specific Subset of Plant Jasmonate-Mediated Resistance to Herbivores in Nature. Front. Plant Sci. 2018, 9, 787. [Google Scholar] [CrossRef] [PubMed]
- Aleman, G.H.J.; Thirumalaikumar, V.P.; Jander, G.; Fernie, A.R.; Skirycz, A. OPDA, more than just a jasmonate precursor. Phytochemistry 2022, 204, 113432. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Dave, A.; Graham, I.A. Oxylipin Signaling: A Distinct Role for the Jasmonic Acid Precursor cis-(+)-12-Oxo-Phytodienoic Acid (cis-OPDA). Front. Plant Sci. 2012, 3, 42. [Google Scholar] [CrossRef]
- Guan, L.; Denkert, N.; Eisa, A.; Lehmann, M.; Sjuts, I.; Weiberg, A.; Soll, J.; Meinecke, M.; Schwenkert, S. JASSY, a chloroplast outer membrane protein required for jasmonate biosynthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 10568–10575. [Google Scholar] [CrossRef]
- Theodoulou, F.L.; Job, K.; Slocombe, S.P.; Footitt, S.; Holdsworth, M.; Baker, A.; Larson, T.R.; Graham, I.A. Jasmonic Acid Levels Are Reduced in COMATOSE ATP-Binding Cassette Transporter Mutants. Implications for Transport of Jasmonate Precursors into Peroxisomes. Plant Physiol. 2005, 137, 835–840. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, Perception, Signal Transduction and Action in Plant Stress Response, Growth and Development. An Update to the 2007 Review in Annals of Botany. Ann. Bot. 2013, 111, 1021. [Google Scholar] [CrossRef]
- Chini, A.; Monte, I.; Zamarreño, A.M.; Hamberg, M.; Lassueur, S.; Reymond, P.; Weiss, S.; Stintzi, A.; Schaller, A.; Porzel, A.; et al. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 2018, 14, 171–178. [Google Scholar] [CrossRef]
- Stumpe, M.; Göbel, C.; Faltin, B.; Beike, A.K.; Hause, B.; Himmelsbach, K.; Bode, J.; Kramell, R.; Wasternack, C.; Frank, W.; et al. The moss Physcomitrella patens contains cyclopentenones but no jasmonates: Mutations in allene oxide cyclase lead to reduced fertility and altered sporophyte morphology. New Phytol. 2010, 188, 740–749. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Ohshika, J.; Takahashi, T.; Ishizaki, K.; Kohchi, T.; Matusuura, H.; Takahashi, K. Functional analysis of allene oxide cyclase, MpAOC, in the liverwort Marchantia polymorpha. Phytochemistry 2015, 116, 48–56. [Google Scholar] [CrossRef]
- Monte, I.; Ishida, S.; Zamarreño, A.M.; Hamberg, M.; Franco-Zorrilla, J.M.; García-Casado, G.; Gouhier-Darimont, C.; Reymond, P.; Takahashi, K.; García-Mina, J.M.; et al. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 2018, 14, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zheng, J.; Li, S.; Huang, G.; Skilling, S.J.; Wang, L.; Li, L.; Li, M.; Yuan, L.; Liu, P. Transporter-Mediated Nuclear Entry of Jasmonoyl-Isoleucine Is Essential for Jasmonate Signaling. Mol. Plant 2017, 10, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.-X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis Gene Required for Jasmonate-Regulated Defense and Fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.-F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Yan, J.; Yao, R.; Chen, L.; Li, S.; Gu, M.; Nan, F.; Xie, D. Dynamic Perception of Jasmonates by the F-Box Protein COI1. Mol. Plant 2018, 11, 1237–1247. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef]
- Zhai, Q.; Deng, L.; Li, C. Mediator subunit MED25: At the nexus of jasmonate signaling. Curr. Opin. Plant Biol. 2020, 57, 78–86. [Google Scholar] [CrossRef]
- Boter, M.; Ruíz-Rivero, O.; Abdeen, A.; Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004, 18, 1577–1591. [Google Scholar] [CrossRef]
- Lorenzo, O.; Chico, J.M.; Saénchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 Encodes a MYC Transcription Factor Essential to Discriminate between Different Jasmonate-Regulated Defense Responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Monte, I.; Franco-Zorrilla, J.M.; García-Casado, G.; Zamarreño, A.M.; García-Mina, J.M.; Nishihama, R.; Kohchi, T.; Solano, R. A Single JAZ Repressor Controls the Jasmonate Pathway in Marchantia polymorpha. Mol. Plant 2019, 12, 185–198. [Google Scholar] [CrossRef]
- Taki, N.; Sasaki-Sekimoto, Y.; Obayashi, T.; Kikuta, A.; Kobayashi, K.; Ainai, T.; Yagi, K.; Sakurai, N.; Suzuki, H.; Masuda, T.; et al. 12-Oxo-Phytodienoic Acid Triggers Expression of a Distinct Set of Genes and Plays a Role in Wound-Induced Gene Expression in Arabidopsis. Plant Physiol. 2005, 139, 1268–1283. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Qi, F.; Zhang, F. Cell Cycle Regulation in the Plant Response to Stress. Front. Plant Sci. 2020, 10, 1765. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.; Wang, L. Chilling and Drought Stresses in Crop Plants: Implications, Cross Talk, and Potential Management Opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef] [PubMed]
- Sobeih, W.Y.; Dodd, I.C.; Bacon, M.A.; Grierson, D.; Davies, W.J. Long-distance signals regulating stomatal conductance and leaf growth in tomato (Lycopersicon esculentum) plants subjected to partial root-zone drying. J. Exp. Bot. 2004, 55, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sano, T.; Tamaoki, M.; Nakajima, N.; Kondo, N.; Hasezawa, S. Ethylene Inhibits Abscisic Acid-Induced Stomatal Closure in Arabidopsis. Plant Physiol. 2005, 138, 2337–2343. [Google Scholar] [CrossRef]
- Wang, P.; Song, C. Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol. 2008, 178, 703–718. [Google Scholar] [CrossRef]
- Sharp, R.E. Interaction with ethylene: Changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ. 2002, 25, 211–222. [Google Scholar] [CrossRef]
- Shi, C.; Qi, C.; Ren, H.; Huang, A.; Hei, S.; She, X. Ethylene mediates brassinosteroid-induced stomatal closure via Gα protein-activated hydrogen peroxide and nitric oxide production in Arabidopsis. Plant J. 2015, 82, 280–301. [Google Scholar] [CrossRef]
- Hao, D.; Jin, L.; Wen, X.; Yu, F.; Xie, Q.; Guo, H. The RING E3 ligase SDIR1 destabilizes EBF1/EBF2 and modulates the ethylene response to ambient temperature fluctuations in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2024592118. [Google Scholar] [CrossRef]
- Xia, Z.; Liu, Q.; Wu, J.; Ding, J. ZmRFP1, the putative ortholog of SDIR1, encodes a RING-H2 E3 ubiquitin ligase and responds to drought stress in an ABA-dependent manner in maize. Gene 2012, 495, 146–153. [Google Scholar] [CrossRef]
- Liu, J.; Xia, Z.; Wang, M.; Zhang, X.; Yang, T.; Wu, J. Overexpression of a maize E3 ubiquitin ligase gene enhances drought tolerance through regulating stomatal aperture and antioxidant system in transgenic tobacco. Plant Physiol. Biochem. 2013, 73, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Wu, Y.; Zhang, Y.; Liu, L.; Ning, Y.; Wang, D.; Tong, H.; Chen, S.; Chu, C.; Xie, Q. OsSDIR1 overexpression greatly improves drought tolerance in transgenic rice. Plant Mol. Biol. 2011, 76, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, J.; Zhu, P.; Yu, J.; Hou, J.; Wang, C.; Long, D.; Yu, M.; Zhao, A. Mulberry EIL3 confers salt and drought tolerances and modulates ethylene biosynthetic gene expression. PeerJ 2019, 7, e6391. [Google Scholar] [CrossRef]
- Cheng, M.-C.; Liao, P.-M.; Kuo, W.-W.; Lin, T.-P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 Regulates Abiotic Stress-Responsive Gene Expression by Binding to Different cis-Acting Elements in Response to Different Stress Signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed]
- Deokar, A.A.; Kondawar, V.; Kohli, D.; Aslam, M.; Jain, P.K.; Karuppayil, S.M.; Varshney, R.K.; Srinivasan, R. The CarERF genes in chickpea (Cicer arietinum L.) and the identification of CarERF116 as abiotic stress responsive transcription factor. Funct. Integr. Genom. 2015, 15, 27–46. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, L.; Zhang, J.; Chen, J.; Wu, T.; Zhu, S.; Yan, S.; Zhao, X.; Zhong, G. Expressing a Citrus ortholog of Arabidopsis ERF1 enhanced cold-tolerance in tobacco. Sci. Hortic. 2014, 174, 65–76. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef]
- Wu, L.; Chen, X.; Ren, H.; Zhang, Z.; Zhang, H.; Wang, J.; Wang, X.-C.; Huang, R. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta 2007, 226, 815–825. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.; Kim, S.; Gupta, R. Ethylene: A Master Regulator of Salinity Stress Tolerance in Plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Su, C.; Dong, C.-H. Genome-Wide Transcriptomic and Proteomic Exploration of Molecular Regulations in Quinoa Responses to Ethylene and Salt Stress. Plants 2021, 10, 2281. [Google Scholar] [CrossRef] [PubMed]
- Jahan, B.; Iqbal, N.; Fatma, M.; Sehar, Z.; Masood, A.; Sofo, A.; D’Ippolito, I.; Khan, N. Ethylene Supplementation Combined with Split Application of Nitrogen and Sulfur Protects Salt-Inhibited Photosynthesis through Optimization of Proline Metabolism and Antioxidant System in Mustard (Brassica juncea L.). Plants 2021, 10, 1303. [Google Scholar] [CrossRef]
- Khan, S.; Sehar, Z.; Fatma, M.; Mir, I.R.; Iqbal, N.; Tarighat, M.A.; Abdi, G.; Khan, N.A. Involvement of ethylene in melatonin-modified photosynthetic-N use efficiency and antioxidant activity to improve photosynthesis of salt grown wheat. Physiol. Plant. 2022, 174, e13832. [Google Scholar] [CrossRef]
- Singh, D.; Debnath, P.; Sane, A.P.; Sane, V.A. Tomato (Solanum lycopersicum) WRKY23 enhances salt and osmotic stress tolerance by modulating the ethylene and auxin pathways in transgenic Arabidopsis. Plant Physiol. Biochem. 2023, 195, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.A.; Xiang, C.; Farooq, M.; Muhammad, N.; Yan, Z.; Hui, X.; Yuanyuan, K.; Bruno, A.K.; Lele, Z.; Jincai, L. Cold Stress in Wheat: Plant Acclimation Responses and Management Strategies. Front. Plant Sci. 2021, 12, 676884. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene Signaling Negatively Regulates Freezing Tolerance by Repressing Expression of CBF and Type-A ARR Genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, H.; Mao, Z.; Liu, W.; Jiang, S.; Xu, H.; Su, M.; Zhang, J.; Wang, N.; Zhang, Z.; et al. Ethylene increases the cold tolerance of apple via the MdERF1B–MdCIbHLH1 regulatory module. Plant J. 2021, 106, 379–393. [Google Scholar] [CrossRef]
- Zhao, D.; Shen, L.; Fan, B.; Yu, M.; Zheng, Y.; Lv, S.; Sheng, J. Ethylene and cold participate in the regulation of LeCBF1 gene expression in postharvest tomato fruits. FEBS Lett. 2009, 583, 3329–3334. [Google Scholar] [CrossRef]
- Catalá, R.; López-Cobollo, R.; Castellano, M.M.; Angosto, T.; Alonso, J.M.; Ecker, J.R.; Salinas, J. The Arabidopsis 14-3-3 Protein RARE COLD INDUCIBLE 1A Links Low-Temperature Response and Ethylene Biosynthesis to Regulate Freezing Tolerance and Cold Acclimation. Plant Cell 2014, 26, 3326–3342. [Google Scholar] [CrossRef]
- Sehar, Z.; Gautam, H.; Iqbal, N.; Alvi, A.F.; Jahan, B.; Fatma, M.; Albaqami, M.; Khan, N.A. The Functional Interplay between Ethylene, Hydrogen Sulfide, and Sulfur in Plant Heat Stress Tolerance. Biomolecules 2022, 12, 678. [Google Scholar] [CrossRef] [PubMed]
- Jegadeesan, S.; Chaturvedi, P.; Ghatak, A.; Pressman, E.; Meir, S.; Faigenboim, A.; Rutley, N.; Beery, A.; Harel, A.; Weckwerth, W.; et al. Proteomics of Heat-Stress and Ethylene-Mediated Thermotolerance Mechanisms in Tomato Pollen Grains. Front. Plant Sci. 2018, 9, 1558. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, Y.; Wang, R.; Xu, F.; Tong, S.; Song, C.; Shao, Y.; Yi, M.; He, J. Ethylene Response Factor LlERF110 Mediates Heat Stress Response via Regulation of LlHsfA3A Expression and Interaction with LlHsfA2 in Lilies (Lilium longiflorum). Int. J. Mol. Sci. 2022, 23, 16135. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Sarkar, N.K.; Grover, A. Tango between Ethylene and HSFA2 Settles Heat Tolerance. Trends Plant Sci. 2021, 26, 429–432. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Bürger, M.; Wang, Y.; Chory, J. Two interacting ethylene response factors regulate heat stress response. Plant Cell 2021, 33, 338–357. [Google Scholar] [CrossRef]
- Shekhawat, K.; Saad, M.M.; Sheikh, A.; Mariappan, K.; Al-Mahmoudi, H.; Abdulhakim, F.; Eida, A.A.; Jalal, R.; Masmoudi, K.; Hirt, H. Root endophyte induced plant thermotolerance by constitutive chromatin modification at heat stress memory gene loci. EMBO Rep. 2021, 22, e51049. [Google Scholar] [CrossRef]
- Sasidharan, R.; Voesenek, L.A.C.J. Ethylene-Mediated Acclimations to Flooding Stress. Plant Physiol. 2015, 169, 3–12. [Google Scholar] [CrossRef]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The Submergence Tolerance Regulator SUB1A Mediates Crosstalk between Submergence and Drought Tolerance in Rice. Plant Cell 2011, 23, 412–427. [Google Scholar] [CrossRef]
- Peña-Castro, J.M.; van Zanten, M.; Lee, S.C.; Patel, M.R.; Voesenek, L.A.J.C.; Fukao, T.; Bailey-Serres, J. Expression of rice SUB1A and SUB1C transcription factors in Arabidopsis uncovers flowering inhibition as a submergence tolerance mechanism. Plant J. 2011, 67, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Van Veen, H.; Mustroph, A.; Barding, G.A.; Eijk, M.V.-V.; Welschen-Evertman, R.A.M.; Pedersen, O.; Visser, E.J.; Larive, C.; Pierik, R.; Bailey-Serres, J.; et al. Two Rumex Species from Contrasting Hydrological Niches Regulate Flooding Tolerance through Distinct Mechanisms. Plant Cell 2013, 25, 4691–4707. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.-J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Yukiyoshi, K.; Karahara, I. Role of ethylene signalling in the formation of constitutive aerenchyma in primary roots of rice. AoB PLANTS 2014, 6, plu043. [Google Scholar] [CrossRef]
- Liu, Z.; Hartman, S.; van Veen, H.; Zhang, H.; Leeggangers, H.A.C.F.; Martopawiro, S.; Bosman, F.; de Deugd, F.; Su, P.; Hummel, M.; et al. Ethylene augments root hypoxia tolerance via growth cessation and reactive oxygen species amelioration. Plant Physiol. 2022, 190, 1365–1383. [Google Scholar] [CrossRef]
- Riemann, M.; Dhakarey, R.; Hazman, M.; Miro, B.; Kohli, A.; Nick, P. Exploring Jasmonates in the Hormonal Network of Drought and Salinity Responses. Front. Plant Sci. 2015, 6, 1077. [Google Scholar] [CrossRef]
- Avramova, Z. Defence-Related Priming and Responses to Recurring Drought: Two Manifestations of Plant Transcriptional Memory Mediated by the ABA and JA Signalling Pathways. Plant Cell Environ. 2019, 42, 983–997. [Google Scholar] [CrossRef]
- Muñoz, P.; Munné-Bosch, S. Oxylipins in Plastidial Retrograde Signaling. Redox Biol. 2020, 37, 101717. [Google Scholar] [CrossRef]
- Casadesús, A.; Bouchikh, R.; Pérez-Llorca, M.; Munné-Bosch, S. Linking Jasmonates with Vitamin E Accumulation in Plants: A Case Study in the Mediterranean Shrub Cistus Albidus L. Planta 2021, 253, 36. [Google Scholar] [CrossRef]
- Ghaffari, H.; Tadayon, M.R.; Nadeem, M.; Razmjoo, J.; Cheema, M. Foliage Applications of Jasmonic Acid Modulate the Antioxidant Defense under Water Deficit Growth in Sugar Beet. Span. J. Agric. Res. 2019, 17, e0805. [Google Scholar] [CrossRef]
- Munemasa, S.; Oda, K.; Watanabe-Sugimoto, M.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. The Coronatine-Insensitive 1 Mutation Reveals the Hormonal Signaling Interaction between Abscisic Acid and Methyl Jasmonate in Arabidopsis Guard Cells. Specific Impairment of Ion Channel Activation and Second Messenger Production. Plant Physiol. 2007, 143, 1398–1407. [Google Scholar] [CrossRef]
- Hossain, M.A.; Munemasa, S.; Uraji, M.; Nakamura, Y.; Mori, I.C.; Murata, Y. Involvement of Endogenous Abscisic Acid in Methyl Jasmonate-Induced Stomatal Closure in Arabidopsis. Plant Physiol. 2011, 156, 430. [Google Scholar] [CrossRef]
- Sarwat, M.; Tuteja, N. Hormonal Signaling to Control Stomatal Movement during Drought Stress. Plant Gene 2017, 11, 143–153. [Google Scholar] [CrossRef]
- Savchenko, T.; Kolla, V.A.; Wang, C.Q.; Nasafi, Z.; Hicks, D.R.; Phadungchob, B.; Chehab, W.E.; Brandizzi, F.; Froehlich, J.; Dehesh, K. Functional Convergence of Oxylipin and Abscisic Acid Pathways Controls Stomatal Closure in Response to Drought. Plant Physiol. 2014, 164, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Simeoni, F.; Skirycz, A.; Simoni, L.; Castorina, G.; de Souza, L.P.; Fernie, A.R.; Alseekh, S.; Giavalisco, P.; Conti, L.; Tonelli, C.; et al. The AtMYB60 Transcription Factor Regulates Stomatal Opening by Modulating Oxylipin Synthesis in Guard Cells. Sci. Rep. 2022, 12, 533. [Google Scholar] [CrossRef]
- Merlaen, B.; de Keyser, E.; van Labeke, M.C. The Jasmonic Acid Pathway, Rather than Abscisic Acid, May Partly Explain Contrasting Stomatal Responses in Two Strawberry Cultivars under Osmotic Stress. Plant Physiol. Biochem. 2020, 151, 21–33. [Google Scholar] [CrossRef]
- Liu, W.; Park, S.W. 12-Oxo-Phytodienoic Acid: A Fuse and/or Switch of Plant Growth and Defense Responses? Front. Plant Sci. 2021, 12, 1687. [Google Scholar] [CrossRef]
- Pérez-Llorca, M.; Caselles, V.; Müller, M.; Munné-Bosch, S. The Threshold between Life and Death in Cistus Albidus L. Seedlings: Mechanisms Underlying Drought Tolerance and Resilience. Tree Physiol. 2021, 41, 1861–1876. [Google Scholar] [CrossRef] [PubMed]
- Cotado, A.; Müller, M.; Morales, M.; Munné-Bosch, S. Linking Jasmonates with Pigment Accumulation and Photoprotection in a High-Mountain Endemic Plant, Saxifraga Longifolia. Environ. Exp. Bot. 2018, 154, 56–65. [Google Scholar] [CrossRef]
- de Ollas, C.; Arbona, V.; Gómez-Cadenas, A.; Dodd, I.C. Attenuated Accumulation of Jasmonates Modifies Stomatal Responses to Water Deficit. J. Exp. Bot. 2018, 69, 2103–2116. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. OPDA-Ile—A New JA-Ile-Independent Signal? Plant Signal. Behav. 2016, 11, e1253646. [Google Scholar] [CrossRef]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-Iso-Jasmonoyl-L-Isoleucine Is the Endogenous Bioactive Jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Floková, K.; Feussner, K.; Herrfurth, C.; Miersch, O.; Mik, V.; Tarkowská, D.; Strnad, M.; Feussner, I.; Wasternack, C.; Novák, O. A Previously Undescribed Jasmonate Compound in Flowering Arabidopsis Thaliana—The Identification of Cis-(+)-OPDA-Ile. Phytochemistry 2016, 122, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.D.; Gruber, C.; Flokova, K.; Miersch, O.; Strnad, M.; Novak, O.; Wasternack, C.; Hause, B. The Recently Identified Isoleucine Conjugate of Cis-12-Oxo-Phytodienoic Acid Is Partially Active in Cis-12-Oxo-Phytodienoic Acid-Specific Gene Expression of Arabidopsis Thaliana. PLoS ONE 2016, 11, e0162829. [Google Scholar] [CrossRef] [PubMed]
- Delgado, C.; Mora-Poblete, F.; Ahmar, S.; Chen, J.-T.; Figueroa, C. Jasmonates and Plant Salt Stress: Molecular Players, Physiological Effects, and Improving Tolerance by Using Genome-Associated Tools. Int. J. Mol. Sci. 2021, 22, 3082. [Google Scholar] [CrossRef]
- Valenzuela, C.E.; Acevedo-Acevedo, O.; Miranda, G.S.; Vergara-Barros, P.; Holuigue, L.; Figueroa, C.R.; Figueroa, P.M. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 2016, 67, 4209–4220. [Google Scholar] [CrossRef]
- Chini, A.; Ben-Romdhane, W.; Hassairi, A.; Aboul-Soud, M.A.M. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef]
- Yao, D.; Zhang, X.; Zhao, X.; Liu, C.; Wang, C.; Zhang, Z.; Zhang, C.; Wei, Q.; Wang, Q.; Yan, H.; et al. Transcriptome analysis reveals salt-stress-regulated biological processes and key pathways in roots of cotton (Gossypium hirsutum L.). Genomics 2011, 98, 47–55. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Q.; Zhai, H.; Li, Y.; Wang, X.; Liu, Q.; He, S. Transcript profile analysis reveals important roles of jasmonic acid signalling pathway in the response of sweet potato to salt stress. Sci. Rep. 2017, 7, 40819. [Google Scholar] [CrossRef]
- Ding, H.; Lai, J.; Wu, Q.; Zhang, S.; Chen, L.; Dai, Y.-S.; Wang, C.; Du, J.; Xiao, S.; Yang, C. Jasmonate complements the function of Arabidopsis lipoxygenase3 in salinity stress response. Plant Sci. 2016, 244, 1–7. [Google Scholar] [CrossRef]
- Verma, D.; Jalmi, S.; Bhagat, P.K.; Verma, N.; Sinha, A.K. A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 2020, 287, 2560–2576. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Zhao, P.; Jung, C.; Chua, N.-H. PLANT U-BOX PROTEIN 10 negatively regulates abscisic acid response in Arabidopsis. Appl. Biol. Chem. 2019, 62, 39. [Google Scholar] [CrossRef]
- Qiu, Z.; Guo, J.; Zhu, A.; Zhang, L.; Zhang, M. Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicol. Environ. Saf. 2014, 104, 202–208. [Google Scholar] [CrossRef]
- Hussain, S.; Zhang, R.; Liu, S.; Li, R.; Wang, Y.; Chen, Y.; Hou, H.; Dai, Q. Methyl Jasmonate Alleviates the Deleterious Effects of Salinity Stress by Augmenting Antioxidant Enzyme Activity and Ion Homeostasis in Rice (Oryza sativa L.). Agronomy 2022, 12, 2343. [Google Scholar] [CrossRef]
- Faghih, S.; Ghobadi, C.; Zarei, A. Response of Strawberry Plant Cv. ‘Camarosa’ to Salicylic Acid and Methyl Jasmonate Application Under Salt Stress Condition. J. Plant Growth Regul. 2017, 36, 651–659. [Google Scholar] [CrossRef]
- Jiang, M.; Xu, F.; Peng, M.; Huang, F.; Meng, F. Methyl jasmonate regulated diploid and tetraploid black locust (Robinia pseudoacacia L.) tolerance to salt stress. Acta Physiol. Plant. 2016, 38, 106. [Google Scholar] [CrossRef]
- Song, R.-F.; Li, T.-T.; Liu, W.-C. Jasmonic Acid Impairs Arabidopsis Seedling Salt Stress Tolerance Through MYC2-Mediated Repression of CAT2 Expression. Front. Plant Sci. 2021, 12, 2331. [Google Scholar] [CrossRef]
- Sheteiwy, M.S.; Ulhassan, Z.; Qi, W.; Lu, H.; AbdElgawad, H.; Minkina, T.; Sushkova, S.; Rajput, V.D.; El-Keblawy, A.; Jośko, I.; et al. Association of jasmonic acid priming with multiple defense mechanisms in wheat plants under high salt stress. Front. Plant Sci. 2022, 13, 2614. [Google Scholar] [CrossRef]
- Karimi, R.; Gavili-Kilaneh, K.; Khadivi, A. Methyl jasmonate promotes salinity adaptation responses in two grapevine (Vitis vinifera L.) cultivars differing in salt tolerance. Food Chem. 2022, 375, 131667. [Google Scholar] [CrossRef]
- Noor, J.; Ullah, A.; Saleem, M.H.; Tariq, A.; Ullah, S.; Waheed, A.; Okla, M.K.; Al-Hashimi, A.; Chen, Y.; Ahmed, Z.; et al. Effect of Jasmonic Acid Foliar Spray on the Morpho-Physiological Mechanism of Salt Stress Tolerance in Two Soybean Varieties (Glycine max L.). Plants 2022, 11, 651. [Google Scholar] [CrossRef]
- Taheri, Z.; Vatankhah, E.; Jafarian, V. Methyl jasmonate improves physiological and biochemical responses of Anchusa italica under salinity stress. South Afr. J. Bot. 2020, 130, 375–382. [Google Scholar] [CrossRef]
- Yuan, F.; Liang, X.; Li, Y.; Yin, S.; Wang, B. Methyl jasmonate improves tolerance to high salt stress in the recretohalophyte Limonium bicolor. Funct. Plant Biol. 2018, 46, 82–92. [Google Scholar] [CrossRef]
- Shahzad, A.N.; Pitann, B.; Ali, H.; Qayyum, M.F.; Fatima, A.; Bakhat, H.F. Maize Genotypes Differing in Salt Resistance Vary in Jasmonic Acid Accumulation During the First Phase of Salt Stress. J. Agron. Crop. Sci. 2015, 201, 443–451. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate Regulates the inducer of cbf expression–c-repeat bind-ing factor/dre binding factor1 Cascade and Freezing Tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold Stress Regulation of Gene Expression in Plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef]
- Li, Q.; Lei, S.; Du, K.; Li, L.; Pang, X.; Wang, Z.; Wei, M.; Fu, S.; Hu, L.; Xu, L. RNA-seq based transcriptomic analysis uncovers α-linolenic acid and jasmonic acid biosynthesis pathways respond to cold acclimation in Camellia japonica. Sci. Rep. 2016, 6, 36463. [Google Scholar] [CrossRef]
- An, J.; Wang, X.; Zhang, X.; You, C.; Hao, Y. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2021, 229, 2707–2729. [Google Scholar] [CrossRef]
- Ba, L.-J.; Kuang, J.-F.; Chen, J.Y.; Lu, W.-J. MaJAZ1 Attenuates the MaLBD5-Mediated Transcriptional Activation of Jasmonate Biosynthesis Gene MaAOC2 in Regulating Cold Tolerance of Banana Fruit. J. Agric. Food Chem. 2016, 64, 738–745. [Google Scholar] [CrossRef]
- González-Aguilar, G.A.; Fortiz, J.; Cruz, R.; Baez, R.; Wang, C.Y. Methyl Jasmonate Reduces Chilling Injury and Maintains Postharvest Quality of Mango Fruit. J. Agric. Food Chem. 2000, 48, 515–519. [Google Scholar] [CrossRef]
- González-Aguilar, G.A.; Tiznado-Hernández, M.E.; Zavaleta-Gatica, R.; Martínez-Téllez, M.Á. Methyl jasmonate treatments reduce chilling injury and activate the defense response of guava fruits. Biochem. Biophys. Res. Commun. 2004, 313, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Zheng, Y.; Wang, K.; Jin, P.; Rui, H. Methyl jasmonate reduces chilling injury and enhances antioxidant enzyme activity in postharvest loquat fruit. Food Chem. 2009, 115, 1458–1463. [Google Scholar] [CrossRef]
- Jin, P.; Zhu, H.; Wang, J.; Chen, J.; Wang, X.; Zheng, Y. Effect of methyl jasmonate on energy metabolism in peach fruit during chilling stress. J. Sci. Food Agric. 2013, 93, 1827–1832. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sheng, J.; Li, F.; Meng, D.; Shen, L. Methyl jasmonate alters arginine catabolism and improves postharvest chilling tolerance in cherry tomato fruit. Postharvest Biol. Technol. 2012, 64, 160–167. [Google Scholar] [CrossRef]
- Duan, W.; Yang, C.; Cao, X.; Zhang, C.; Liu, H.; Chen, K.; Li, X.; Zhang, B. Transcriptome and DNA methylome analysis reveal new insights into methyl jasmonate-alleviated chilling injury of peach fruit after cold storage. Postharvest Biol. Technol. 2022, 189, 111915. [Google Scholar] [CrossRef]
- Clarke, S.M.; Cristescu, S.M.; Miersch, O.; Harren, F.J.M.; Wasternack, C.; Mur, L.A.J. Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 2009, 182, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Balfagón, D.; Sengupta, S.; Gómez-Cadenas, A.; Fritschi, F.B.; Azad, R.K.; Mittler, R.; Zandalinas, S.I. Jasmonic Acid Is Required for Plant Acclimation to a Combination of High Light and Heat Stress. Plant Physiol. 2019, 181, 1668–1682. [Google Scholar] [CrossRef]
- Hu, T.; Zeng, H.; Hu, Z.; Qv, X.; Chen, G. Overexpression of the Tomato 13-Lipoxygenase Gene TomloxD Increases Generation of Endogenous Jasmonic Acid and Resistance to Cladosporium fulvum and High Temperature. Plant Mol. Biol. Rep. 2013, 31, 1141–1149. [Google Scholar] [CrossRef]
- Fatma, M.; Iqbal, N.; Sehar, Z.; Alyemeni, M.; Kaushik, P.; Khan, N.; Ahmad, P. Methyl Jasmonate Protects the PS II System by Maintaining the Stability of Chloroplast D1 Protein and Accelerating Enzymatic Antioxidants in Heat-Stressed Wheat Plants. Antioxidants 2021, 10, 1216. [Google Scholar] [CrossRef]
- Yang, J.; Fei, K.; Chen, J.; Wang, Z.; Zhang, W.; Zhang, J. Jasmonates alleviate spikelet-opening impairment caused by high temperature stress during anthesis of photo-thermo-sensitive genic male sterile rice lines. Food Energy Secur. 2020, 9, e233. [Google Scholar] [CrossRef]
- Chen, J.; Miao, W.; Fei, K.; Shen, H.; Zhou, Y.; Shen, Y.; Li, C.; He, J.; Zhu, K.; Wang, Z.; et al. Jasmonates Alleviate the Harm of High-Temperature Stress During Anthesis to Stigma Vitality of Photothermosensitive Genetic Male Sterile Rice Lines. Front. Plant Sci. 2021, 12, 412. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Yang, D.; Zhao, X.; Jiao, C.; Yan, Y.; Lamin-Samu, A.T.; Wang, Q.; Xu, X.; Fei, Z.; Lu, G. Tomato stigma exsertion induced by high temperature is associated with the jasmonate signalling pathway. Plant Cell Environ. 2019, 42, 1205–1221. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-C.; Millet, Y.A.; Cheng, Z.; Bush, J.; Ausubel, F.M. Jasmonate signalling in Arabidopsis involves SGT1b–HSP70–HSP90 chaperone complexes. Nat. Plants 2015, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.-F.; Wang, Y.-N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.-Q.; Wang, X.; Qiu, A.-L.; Zhang, T.-X.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, G.; Yuan, M.; Guo, S.; Wang, Y.; Sun, J. CsbZIP2-miR9748-CsNPF4.4 Module Mediates High Temperature Tolerance of Cucumber Through Jasmonic Acid Pathway. Front. Plant Sci. 2022, 13, 1365. [Google Scholar] [CrossRef]
- Hinz, M.; Wilson, I.W.; Yang, J.; Buerstenbinder, K.; Llewellyn, D.; Dennis, E.S.; Sauter, M.; Dolferus, R. Arabidopsis RAP2.2: An Ethylene Response Transcription Factor That Is Important for Hypoxia Survival. Plant Physiol. 2010, 153, 757–772. [Google Scholar] [CrossRef]
- Voesenek, L.A.C.J.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef]
- Shukla, V.; Lombardi, L.; Pencik, A.; Novak, O.; Weits, D.; Loreti, E.; Perata, P.; Giuntoli, B.; Licausi, F. Jasmonate Signalling Contributes to Primary Root Inhibition Upon Oxygen Deficiency in Arabidopsis thaliana. Plants 2020, 9, 1046. [Google Scholar] [CrossRef]
- Yuan, L.-B.; Dai, Y.-S.; Xie, L.-J.; Yu, L.-J.; Zhou, Y.; Lai, Y.-X.; Yang, Y.-C.; Xu, L.; Chen, Q.-F.; Xiao, S. Jasmonate Regulates Plant Responses to Postsubmergence Reoxygenation through Transcriptional Activation of Antioxidant Synthesis. Plant Physiol. 2017, 173, 1864–1880. [Google Scholar] [CrossRef]
- Yeung, E.; Bailey-Serres, J.; Sasidharan, R. After The Deluge: Plant Revival Post-Flooding. Trends Plant Sci. 2019, 24, 443–454. [Google Scholar] [CrossRef]
- Savchenko, T.; Rolletschek, H.; Heinzel, N.; Tikhonov, K.; Dehesh, K. Waterlogging tolerance rendered by oxylipin-mediated metabolic reprogramming in Arabidopsis. J. Exp. Bot. 2019, 70, 2919–2932. [Google Scholar] [CrossRef]
- Sreeratree, J.; Butsayawarapat, P.; Chaisan, T.; Somta, P.; Juntawong, P. RNA-Seq Reveals Waterlogging-Triggered Root Plasticity in Mungbean Associated with Ethylene and Jasmonic Acid Signal Integrators for Root Regeneration. Plants 2022, 11, 930. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Diao, P.; Chen, C.; Zhang, Y.; Meng, Q.; Lv, W.; Ma, N. The role of NAC transcription factor in plant cold response. Plant Signal. Behav. 2020, 15, 1785668. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Lee, B. Friends or Foes: New Insights in Jasmonate and Ethylene Co-Actions. Plant Cell Physiol. 2015, 56, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Larkindale, J.; Hall, J.D.; Knight, M.; Vierling, E. Heat Stress Phenotypes of Arabidopsis Mutants Implicate Multiple Signaling Pathways in the Acquisition of Thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef]
- Cebrián, G.; Segura, M.; Martínez, J.; Iglesias-Moya, J.; Martínez, C.; Garrido, D.; Jamilena, M. Jasmonate-deficient mutant lox3a reveals crosstalk between jasmonate and ethylene in the differential regulation of male and female flower opening and early fruit development in Cucurbita pepo. J. Exp. Bot. 2023, 74, 1258–1274. [Google Scholar] [CrossRef] [PubMed]
- Fattorini, L.; Hause, B.; Gutierrez, L.; Veloccia, A.; Della Rovere, F.; Piacentini, D.; Falasca, G.; Altamura, M.M. Jasmonate promotes auxin-induced adventitious rooting in dark-grown Arabidopsis thaliana seedlings and stem thin cell layers by a cross-talk with ethylene signalling and a modulation of xylogenesis. BMC Plant Biol. 2018, 18, 182. [Google Scholar] [CrossRef]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; Zhang, X.; et al. Derepression of Ethylene-Stabilized Transcription Factors (EIN3/EIL1) Mediates Jasmonate and Ethylene Signaling Synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, Z.; An, F.; Hao, D.; Li, P.; Song, J.; Yi, C.; Guo, H. Jasmonate-Activated MYC2 Represses ETHYLENE INSENSITIVE3 Activity to Antagonize Ethylene-Promoted Apical Hook Formation in Arabidopsis. Plant Cell 2014, 26, 1105–1117. [Google Scholar] [CrossRef]
- Song, S.; Huang, H.; Gao, H.; Wang, J.; Wu, D.; Liu, X.; Yang, S.; Zhai, Q.; Li, C.; Qi, T.; et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 Modulates Antagonism between Jasmonate and Ethylene Signaling in Arabidopsis. Plant Cell 2014, 26, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef]
- Wink, M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 2003, 64, 3–19. [Google Scholar] [CrossRef]
- Noel, J.P.; Austin, M.B.; Bomati, E.K. Structure–function relationships in plant phenylpropanoid biosynthesis. Curr. Opin. Plant Biol. 2005, 8, 249–253. [Google Scholar] [CrossRef]
- Świeca, M. Elicitation with abiotic stresses improves pro-health constituents, antioxidant potential and nutritional quality of lentil sprouts. Saudi J. Biol. Sci. 2015, 22, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, H.J.; Aaby, K.; Lea, P. Influence of temperature and plant density on sensory quality and volatile terpenoids of carrot (Daucus carota L.) root. J. Sci. Food Agric. 2002, 82, 1384–1390. [Google Scholar] [CrossRef]
- Yeshi, K.; Crayn, D.; Ritmejerytė, E.; Wangchuk, P. Plant Secondary Metabolites Produced in Response to Abiotic Stresses Has Potential Application in Pharmaceutical Product Development. Molecules 2022, 27, 313. [Google Scholar] [CrossRef]
- Verma, N.; Shukla, S. Impact of various factors responsible for fluctuation in plant secondary metabolites. J. Appl. Res. Med. Aromat. Plants 2015, 2, 105–113. [Google Scholar] [CrossRef]
- Hausman, J.F.; Evers, D.; Thiellement, H.; Jouve, L. Compared responses of poplar cuttings and in vitro raised shoots to short-term chilling treatments. Plant Cell Rep. 2000, 19, 954–960. [Google Scholar] [CrossRef]
- Yulvianti, M.; Zidorn, C. Chemical Diversity of Plant Cyanogenic Glycosides: An Overview of Reported Natural Products. Molecules 2021, 26, 719. [Google Scholar] [CrossRef]
- Vaughan, M.M.; Christensen, S.; Schmelz, E.; Huffaker, A.; McAuslane, H.J.; Alborn, H.T.; Romero, M.; Allen, L.H.; Teal, P.E.A. Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ. 2015, 38, 2195–2207. [Google Scholar] [CrossRef]
- Tungmunnithum, D.; Thongboonyou, A.; Pholboon, A.; Yangsabai, A. Flavonoids and Other Phenolic Compounds from Medicinal Plants for Pharmaceutical and Medical Aspects: An Overview. Medicines 2018, 5, 93. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid Pathway and the Role of Polyphenols in Plants under Abiotic Stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef]
- André, C.M.; Schafleitner, R.; Legay, S.; Lefèvre, I.; Aliaga, C.A.A.; Nomberto, G.; Hoffmann, L.; Hausman, J.-F.; Larondelle, Y.; Evers, D. Gene expression changes related to the production of phenolic compounds in potato tubers grown under drought stress. Phytochemistry 2009, 70, 1107–1116. [Google Scholar] [CrossRef]
- Ma, D.Y.; Sun, D.X.; Wang, C.Y.; Li, Y.G.; Guo, T.C. Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol. Biochem. 2014, 80, 60–66. [Google Scholar] [CrossRef]
- Gharibi, S.; Tabatabaei, B.E.S.; Saeidi, G.; Talebi, M.; Matkowski, A. The effect of drought stress on polyphenolic compounds and expression of flavonoid biosynthesis related genes in Achillea pachycephala Rech.f. Phytochemistry 2019, 162, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Rodríguez, E.; Moreno, D.A.; Ferreres, F.; Rubio-Wilhelmi, M.D.M.; Ruiz, J.M. Differential responses of five cherry tomato varieties to water stress: Changes on phenolic metabolites and related enzymes. Phytochemistry 2011, 72, 723–729. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Drought stress enhances nutritional and bioactive compounds, phenolic acids and antioxidant capacity of Amaranthus leafy vegetable. BMC Plant Biol. 2018, 18, 258. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. (PCTOC) 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Chen, S.; Wu, F.; Li, Y.; Qian, Y.; Pan, X.; Li, F.; Wang, Y.; Wu, Z.; Fu, C.; Lin, H.; et al. NtMYB4 and NtCHS1 Are Critical Factors in the Regulation of Flavonoid Biosynthesis and Are Involved in Salinity Responsiveness. Front. Plant Sci. 2019, 10, 178. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Wang, B.; Jiang, Y.; Cheng, L.; Wu, T. GmFNSII-Controlled Soybean Flavone Metabolism Responds to Abiotic Stresses and Regulates Plant Salt Tolerance. Plant Cell Physiol. 2014, 55, 74–86. [Google Scholar] [CrossRef]
- Birhanie, Z.M.; Yang, D.; Luan, M.; Xiao, A.; Liu, L.; Zhang, C.; Biswas, A.; Dey, S.; Deng, Y.; Li, D. Salt Stress Induces Changes in Physiological Characteristics, Bioactive Constituents, and Antioxidants in Kenaf (Hibiscus cannabinus L.). Antioxidants 2022, 11, 2005. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shan, T.; Xie, B.; Ling, C.; Shao, S.; Jin, P.; Zheng, Y. Glycine betaine reduces chilling injury in peach fruit by enhancing phenolic and sugar metabolisms. Food Chem. 2019, 272, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Martinez, V.; Mestre, T.C.; Rubio, F.; Girones-Vilaplana, A.; Moreno, D.A.; Mittler, R.; Rivero, R.M. Accumulation of Flavonols over Hydroxycinnamic Acids Favors Oxidative Damage Protection under Abiotic Stress. Front. Plant Sci. 2016, 7, 838. [Google Scholar] [CrossRef]
- Huang, D.; Sun, M.; Zhang, A.; Chen, J.; Zhang, J.; Lin, C.; Zhang, H.; Lu, X.; Wang, X.; Yan, H.; et al. Transcriptional Changes in Pearl Millet Leaves under Heat Stress. Genes 2021, 12, 1716. [Google Scholar] [CrossRef]
- Wang, J.; Yuan, B.; Huang, B. Differential Heat-Induced Changes in Phenolic Acids Associated with Genotypic Variations in Heat Tolerance for Hard Fescue. Crop. Sci. 2019, 59, 667–674. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J.K.; Wu, Q.; Park, S.U. Effects of cold stress on transcripts and metabolites in tartary buckwheat (Fagopyrum tataricum). Environ. Exp. Bot. 2018, 155, 488–496. [Google Scholar] [CrossRef]
- Wang, T.; Zou, Q.; Guo, Q.; Yang, F.; Wu, L.; Zhang, W. Widely Targeted Metabolomics Analysis Reveals the Effect of Flooding Stress on the Synthesis of Flavonoids in Chrysanthemum morifolium. Molecules 2019, 24, 3695. [Google Scholar] [CrossRef]
- Jaksomsak, P.; Konseang, S.; Dell, B.; Rouached, H.; Prom-U-Thai, C. Grain and Leaf Anthocyanin Concentration Varies among Purple Rice Varieties and Growing Condition in Aerated and Flooded Soil. Molecules 2022, 27, 8355. [Google Scholar] [CrossRef] [PubMed]
- Ralph, J.; Lundquist, K.; Brunow, G.; Lu, F.; Kim, H.; Schatz, P.F.; Marita, J.M.; Hatfield, R.D.; Ralph, S.A.; Christensen, J.H.; et al. Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem. Rev. 2004, 3, 29–60. [Google Scholar] [CrossRef]
- Lei, L. Lignin evolution: Invasion of land. Nat. Plants 2017, 3, 17042. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Kim, J.I.; Tobimatsu, Y.; Ciesielski, P.N.; Anderson, N.A.; Ximenes, E.; Maeda, J.; Ralph, J.; Donohoe, B.S.; Ladisch, M.; et al. Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 2014, 509, 376–380. [Google Scholar] [CrossRef]
- Schilmiller, A.L.; Stout, J.; Weng, J.-K.; Humphreys, J.; Ruegger, M.O.; Chapple, C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009, 60, 771–782. [Google Scholar] [CrossRef]
- Cesarino, I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L.Q. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Hu, Y.; Li, W.C.; Xu, Y.Q.; Li, G.J.; Liao, Y.; Fu, F.L. Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J. Appl. Genet. 2009, 50, 213–223. [Google Scholar] [CrossRef]
- Fan, L.; Linker, R.; Gepstein, S.; Tanimoto, E.; Yamamoto, R.; Neumann, P.M. Progressive Inhibition by Water Deficit of Cell Wall Extensibility and Growth along the Elongation Zone of Maize Roots Is Related to Increased Lignin Metabolism and Progressive Stelar Accumulation of Wall Phenolics. Plant Physiol. 2006, 140, 603–612. [Google Scholar] [CrossRef]
- Srivastava, S.; Vishwakarma, R.K.; Arafat, Y.A.; Gupta, S.K.; Khan, B.M. Abiotic stress induces change in Cinnamoyl CoA Reductase (CCR) protein abundance and lignin deposition in developing seedlings of Leucaena leucocephala. Physiol. Mol. Biol. Plants 2015, 21, 197–205. [Google Scholar] [CrossRef]
- Chun, H.J.; Baek, D.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Yun, D.-J.; Hong, Y.-S.; Kim, M.C. Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress. Plant Signal. Behav. 2019, 14, 1625697. [Google Scholar] [CrossRef] [PubMed]
- Shafi, A.; Chauhan, R.; Gill, T.; Swarnkar, M.K.; Sreenivasulu, Y.; Kumar, S.; Kumar, N.; Shankar, R.; Ahuja, P.S.; Singh, A.K. Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol. Biol. 2015, 87, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Dhanaraj, A.L.; Arora, R.; Rowland, L.J.; Fu, Y.; Sun, L. Identification of cold acclimation-responsive Rhododendron genes for lipid metabolism, membrane transport and lignin biosynthesis: Importance of moderately abundant ESTs in genomic studies. Plant Cell Environ. 2006, 29, 558–570. [Google Scholar] [CrossRef]
- Chen, B.; Guo, Y.; Zhang, X.; Wang, L.; Cao, L.; Zhang, T.; Zhang, Z.; Zhou, W.; Xie, L.; Wang, J.; et al. Climate-responsive DNA methylation is involved in the biosynthesis of lignin in birch. Front. Plant Sci. 2022, 13, 1090967. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.-N.; Son, S.; Jordan, M.C.; Levin, D.B.; Ayele, B.T. Lignin biosynthesis in wheat (Triticum aestivum L.): Its response to waterlogging and association with hormonal levels. BMC Plant Biol. 2016, 16, 28. [Google Scholar] [CrossRef]
- Kubeczka, K.-H. History and Sources of Essential Oil Research. In Handbook of Essential Oils; CRC Press: Boca Raton, FL, USA, 2020; pp. 3–39. [Google Scholar] [CrossRef]
- Šamec, D.; Karalija, E.; Šola, I.; Vujčić Bok, V.; Salopek-Sondi, B. The role of polyphenols in abiotic stress response: The influence of molecular structure. Plants 2021, 10, 118. [Google Scholar] [CrossRef]
- Pérez-Llorca, M.; Casadesús, A.; Munné-Bosch, S.; Müller, M. Contrasting Patterns of Hormonal and Photoprotective Isoprenoids in Response to Stress in Cistus Albidus during a Mediterranean Winter. Planta 2019, 250, 1409–1422. [Google Scholar] [CrossRef] [PubMed]
- Griesser, M.; Weingart, G.; Schoedl-Hummel, K.; Neumann, N.; Becker, M.; Varmuza, K.; Liebner, F.; Schuhmacher, R.; Forneck, A. Severe drought stress is affecting selected primary metabolites, polyphenols, and volatile metabolites in grapevine leaves (Vitis vinifera cv. Pinot noir). Plant Physiol. Biochem. 2015, 88, 17–26. [Google Scholar] [CrossRef]
- Bertamini, M.; Grando, M.S.; Zocca, P.; Pedrotti, M.; Lorenzi, S.; Cappellin, L. Linking monoterpenes and abiotic stress resistance in grapevines. BIO Web Conf. 2019, 13, 01003. [Google Scholar] [CrossRef]
- Rogowska, A.; Szakiel, A. The role of sterols in plant response to abiotic stress. Phytochem. Rev. 2020, 19, 1525–1538. [Google Scholar] [CrossRef]
- Rudolf, J.D.; Chang, C.-Y. Terpene synthases in disguise: Enzymology, structure, and opportunities of non-canonical terpene synthases. Nat. Prod. Rep. 2020, 37, 425–463. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-C.; Shamala, L.F.; Yi, X.-K.; Yan, Z.; Wei, S. Analysis of Terpene Synthase Family Genes in Camellia sinensis with an Emphasis on Abiotic Stress Conditions. Sci. Rep. 2020, 10, 933. [Google Scholar] [CrossRef]
- Yan, Y.; Li, M.; Zhang, X.; Kong, W.; Bendahmane, M.; Bao, M.; Fu, X. Tissue-Specific Expression of the Terpene Synthase Family Genes in Rosa chinensis and Effect of Abiotic Stress Conditions. Genes 2022, 13, 547. [Google Scholar] [CrossRef]
- Zhan, X.; Qian, Y.; Mao, B. Metabolic Profiling of Terpene Diversity and the Response of Prenylsynthase-Terpene Synthase Genes during Biotic and Abiotic Stresses in Dendrobium catenatum. Int. J. Mol. Sci. 2022, 23, 6398. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, J.S.; Neto, V.G.; Loureiro, M.B.; Ribeiro, P.R. Genome-wide characterization of the terpene synthase gene family in Ricinus communis and its transcriptional regulation under heat stress. Agron. J. 2022, 114, 3272–3282. [Google Scholar] [CrossRef]
- Zagrobelny, M.; Bak, S.; Møller, B.L. Cyanogenesis in plants and arthropods. Phytochemistry 2008, 69, 1457–1468. [Google Scholar] [CrossRef]
- Halkiers, B.A.; Olsens, C.E.; Lindberg Møller, B. The Biosynthesis of Cyanogenic Glucosides in Higher Plants the (e)-and (2)-isomers of p-hydroxyphenylacetaldehyde oxime as intermediates in the biosynthesis of dhurrin in sorghum bicolor (L.) moench*. J. Biol. Chem. 1989, 264, 19487–19494. [Google Scholar]
- Gleadow, R.M.; Møller, B.L. Cyanogenic Glycosides: Synthesis, Physiology, and Phenotypic Plasticity. Annu. Rev. Plant Biol. 2014, 65, 155–185. [Google Scholar] [CrossRef]
- Adewusi, S.R.A. Turnover of Dhurrin in Green Sorghum Seedlings. Plant Physiol. 1990, 94, 1219–1224. [Google Scholar] [CrossRef]
- O’Donnell, N.H.; Møller, B.L.; Neale, A.D.; Hamill, J.D.; Blomstedt, C.K.; Gleadow, R.M. Effects of PEG-induced osmotic stress on growth and dhurrin levels of forage sorghum. Plant Physiol. Biochem. 2013, 73, 83–92. [Google Scholar] [CrossRef]
- Emendack, Y.; Burke, J.; Laza, H.; Sanchez, J.; Hayes, C. Abiotic Stress Effects on Sorghum Leaf Dhurrin and Soluble Sugar Contents throughout Plant Development. Crop. Sci. 2018, 58, 1706–1716. [Google Scholar] [CrossRef]
- Sohail, M.N.; Quinn, A.A.; Blomstedt, C.K.; Gleadow, R.M. Dhurrin increases but does not mitigate oxidative stress in droughted Sorghum bicolor. Planta 2022, 255, 74. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.; Valadier, M.-H.; Migge, A.; Becker, T.W. Drought-Induced Effects on Nitrate Reductase Activity and mRNA and on the Coordination of Nitrogen and Carbon Metabolism in Maize Leaves. Plant Physiol. 1998, 117, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.L.; Cavagnaro, T.R.; Gleadow, R.; Miller, R.E. Interactive effects of temperature and drought on cassava growth and toxicity: Implications for food security? Glob. Chang. Biol. 2016, 22, 3461–3473. [Google Scholar] [CrossRef]
- Gleadow, R.; Pegg, A.; Blomstedt, C.K. Resilience of cassava ( Manihot esculenta Crantz) to salinity: Implications for food security in low-lying regions. J. Exp. Bot. 2016, 67, 5403–5413. [Google Scholar] [CrossRef]
- Vranova, V.; Rejsek, K.; Skene, K.R.; Formanek, P. Non-protein amino acids: Plant, soil and ecosystem interactions. Plant Soil 2011, 342, 31–48. [Google Scholar] [CrossRef]
- Akashi, K.; Miyake, C.; Yokota, A. Citrulline, a novel compatible solute in drought-tolerant wild watermelon leaves, is an efficient hydroxyl radical scavenger. FEBS Lett. 2001, 508, 438–442. [Google Scholar] [CrossRef]
- Abid, G.; Ouertani, R.N.; Jebara, S.H.; Boubakri, H.; Muhovski, Y.; Ghouili, E.; Abdelkarim, S.; Chaieb, O.; Hidri, Y.; Kadri, S.; et al. Alleviation of drought stress in faba bean (Vicia faba L.) by exogenous application of β-aminobutyric acid (BABA). Physiol. Mol. Biol. Plants 2020, 26, 1173–1186. [Google Scholar] [CrossRef]
- Mekonnen, D.W.; Flügge, U.-I.; Ludewig, F. Gamma-aminobutyric acid depletion affects stomata closure and drought tolerance of Arabidopsis thaliana. Plant Sci. 2016, 245, 25–34. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Z.; Fan, Y.; Liu, C.; Wang, H.; Li, Y.; Xin, Y.; Gai, Y.; Ji, X. Characterization of GABA-Transaminase Gene from Mulberry (Morus multicaulis) and Its Role in Salt Stress Tolerance. Genes 2022, 13, 501. [Google Scholar] [CrossRef]
- Shabala, S.; Shabala, L.; Barceló, J.; Poschenrieder, C. Membrane transporters mediating root signalling and adaptive responses to oxygen deprivation and soil flooding. Plant Cell Environ. 2014, 37, 2216–2233. [Google Scholar] [CrossRef] [PubMed]
- Kuć, J.; Rush, J.S. Phytoalexins. Arch. Biochem. Biophys. 1985, 236, 455–472. [Google Scholar] [CrossRef]
- Jeandet, P.; Hébrard, C.; Deville, M.-A.; Cordelier, S.; Dorey, S.; Aziz, A.; Crouzet, J. Deciphering the Role of Phytoalexins in Plant-Microorganism Interactions and Human Health. Molecules 2014, 19, 18033–18056. [Google Scholar] [CrossRef]
- Klein, A.P.; Sattely, E.S. Biosynthesis of cabbage phytoalexins from indole glucosinolate. Proc. Natl. Acad. Sci. USA 2017, 114, 1910–1915. [Google Scholar] [CrossRef]
- Hatmi, S.; Villaume, S.; Trotel-Aziz, P.; Barka, E.A.; Clément, C.; Aziz, A. Osmotic Stress and ABA Affect Immune Response and Susceptibility of Grapevine Berries to Gray Mold by Priming Polyamine Accumulation. Front. Plant Sci. 2018, 9, 1010. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Williams, C.C.; Last, R. Induction of Arabidopsis Tryptophan Pathway Enzymes and Camalexin by Amino Acid Starvation, Oxidative Stress, and an Abiotic Elicitor. Plant Cell 1998, 10, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Pedras, M.S.C.; Zheng, Q.-A.; Gadagi, R.S.; Rimmer, S.R. Phytoalexins and polar metabolites from the oilseeds canola and rapeseed: Differential metabolic responses to the biotroph Albugo candida and to abiotic stress. Phytochemistry 2008, 69, 894–910. [Google Scholar] [CrossRef] [PubMed]
- Clausen, M.; Kannangara, R.M.; Olsen, C.E.; Blomstedt, C.K.; Gleadow, R.M.; Jørgensen, K.; Bak, S.; Motawie, M.S.; Møller, B.L. The bifurcation of the cyanogenic glucoside and glucosinolate biosynthetic pathways. Plant J. 2015, 84, 558–573. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Bodenhausen, N.; Buchala, A.; Mauch, F.; Reymond, P. The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J. 2008, 55, 774–786. [Google Scholar] [CrossRef]
- Bejai, S.; Fridborg, I.; Ekbom, B. Varied response of Spodoptera littoralis against Arabidopsis thaliana with metabolically engineered glucosinolate profiles. Plant Physiol. Biochem. 2012, 50, 72–78. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, B.W.; Schroeder, F.C.; Jander, G. Identification of indole glucosinolate breakdown products with antifeedant effects on Myzus persicae (green peach aphid). Plant J. 2008, 54, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Del Carmen Martinez-Ballesta, M.; Moreno, D.A.; Carvajal, M. The Physiological Importance of Glucosinolates on Plant Response to Abiotic Stress in Brassica. Int. J. Mol. Sci. 2013, 14, 11607–11625. [Google Scholar] [CrossRef] [PubMed]
- Abuyusuf, M.; Rubel, M.H.; Kim, H.-T.; Jung, H.-J.; Nou, I.-S.; Park, J.-I. Glucosinolates and Biotic Stress Tolerance in Brassicaceae with Emphasis on Cabbage: A Review. Biochem. Genet. 2022, 1–20. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, H.; Xie, J.; Lv, J.; Zhang, G.; Hu, L.; Luo, S.; Li, L.; Yu, J. The Roles of Cruciferae Glucosinolates in Disease and Pest Resistance. Plants 2021, 10, 1097. [Google Scholar] [CrossRef] [PubMed]
- Balestrini, R.; Brunetti, C.; Cammareri, M.; Caretto, S.; Cavallaro, V.; Cominelli, E.; De Palma, M.; Docimo, T.; Giovinazzo, G.; Grandillo, S.; et al. Strategies to Modulate Specialized Metabolism in Mediterranean Crops: From Molecular Aspects to Field. Int. J. Mol. Sci. 2021, 22, 2887. [Google Scholar] [CrossRef] [PubMed]
- Variyar, P.; Banerjee, A.; Akkarakaran, J.J.; Suprasanna, P. Role of Glucosinolates in Plant Stress Tolerance. Emerg. Technol. Manag. Crop Stress Toler. Biol. Tech. 2014, 1, 271–291. [Google Scholar] [CrossRef]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 2019, 10, 4021. [Google Scholar] [CrossRef]
- Coleto, I.; Bejarano, I.; Marín-Peña, A.J.; Medina, J.; Rioja, C.; Burow, M.; Marino, D. Arabidopsis thaliana transcription factors MYB28 and MYB29 shape ammonium stress responses by regulating Fe homeostasis. New Phytol. 2021, 229, 1021–1035. [Google Scholar] [CrossRef]
- Coleto, I.; De La Peña, M.; Rodríguez-Escalante, J.; Bejarano, I.; Glauser, G.; Aparicio-Tejo, P.M.; González-Moro, M.B.; Marino, D. Leaves play a central role in the adaptation of nitrogen and sulfur metabolism to ammonium nutrition in oilseed rape (Brassica napus). BMC Plant Biol. 2017, 17, 157. [Google Scholar] [CrossRef]
- Shkryl, Y.; Yugay, Y.; Avramenko, T.; Grigorchuk, V.; Gorpenchenko, T.; Grischenko, O.; Bulgakov, V. CRISPR/Cas9-Mediated Knockout of HOS1 Reveals Its Role in the Regulation of Secondary Metabolism in Arabidopsis thaliana. Plants 2021, 10, 104. [Google Scholar] [CrossRef]
- Lee, D.-K.; Yoon, S.; Kim, Y.S.; Kim, J.-K. Rice OsERF71-mediated root modification affects shoot drought tolerance. Plant Signal. Behav. 2017, 12, e1268311. [Google Scholar] [CrossRef]
- Watkins, J.M.; Hechler, P.J.; Muday, G.K. Ethylene-Induced Flavonol Accumulation in Guard Cells Suppresses Reactive Oxygen Species and Moderates Stomatal Aperture. Plant Physiol. 2014, 164, 1707–1717. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Zhang, H.; Song, Z.; Gao, Y.; Li, W.; Li, M.; Zhao, L.; Li, Y.; Wang, B. Ethylene response factor NtERF91 positively regulates alkaloid accumulations in tobacco (Nicotiana tabacum L.). Biochem. Biophys. Res. Commun. 2019, 517, 164–171. [Google Scholar] [CrossRef]
- Papon, N.; Bremer, J.; Vansiri, A.; Andreu, F.; Rideau, M.; Crèche, J. Cytokinin and Ethylene Control Indole Alkaloid Production at the Level of the MEP/Terpenoid Pathway in Catharanthus roseus Suspension Cells. Planta Medica 2005, 71, 572–574. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Xu, L.; Gao, S.; Lyu, X.; Cao, X.; Yao, Y. Melatonin alters the secondary metabolite profile of grape berry skin by promoting VvMYB14-mediated ethylene biosynthesis. Hortic. Res. 2021, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Akula, R.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [CrossRef]
- Ndiaye, A.; Diallo, A.O.; Fall, N.C.; Diouf, R.D.; Diouf, D.; Kane, N.A. Transcriptomic analysis of methyl jasmonate treatment reveals gene networks involved in drought tolerance in pearl millet. Sci. Rep. 2022, 12, 5158. [Google Scholar] [CrossRef]
- Gu, X.-C.; Chen, J.-F.; Xiao, Y.; Di, P.; Xuan, H.-J.; Zhou, X.; Zhang, L.; Chen, W.-S. Overexpression of allene oxide cyclase promoted tanshinone/phenolic acid production in Salvia miltiorrhiza. Plant Cell Rep. 2012, 31, 2247–2259. [Google Scholar] [CrossRef]
- Kim, O.T.; Bang, K.H.; Kim, Y.C.; Hyun, D.Y.; Kim, M.Y.; Cha, S.W. Upregulation of ginsenoside and gene expression related to triterpene biosynthesis in ginseng hairy root cultures elicited by methyl jasmonate. Plant Cell Tissue Organ Cult. 2009, 98, 25–33. [Google Scholar] [CrossRef]
- Farooq, M.A.; Gill, R.A.; Islam, F.; Ali, B.; Liu, H.; Xu, J.; He, S.; Zhou, W. Methyl Jasmonate Regulates Antioxidant Defense and Suppresses Arsenic Uptake in Brassica napus L. Front. Plant Sci. 2016, 7, 468. [Google Scholar] [CrossRef]
- Choudhri, P.; Rani, M.; Sangwan, R.S.; Kumar, R.; Kumar, A.; Chhokar, V. De novo sequencing, assembly and characterisation of Aloe vera transcriptome and analysis of expression profiles of genes related to saponin and anthraquinone metabolism. BMC Genom. 2018, 19, 427. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.J.; An, C.H.; Park, S.-C.; Pyun, J.W.; Lee, J.-Y.; Kim, S.W.; Kim, H.-S.; Kim, H.; Jeong, J.C.; Kim, C.Y. Methyl Jasmonate Increases Isoflavone Production in Soybean Cell Cultures by Activating Structural Genes Involved in Isoflavonoid Biosynthesis. J. Agric. Food Chem. 2018, 66, 4099–4105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Logacheva, M.D.; Meng, Y.; Hu, J.; Wan, D.; Li, L.; Janovská, D.; Wang, Z.; Georgiev, M.I.; Yu, Z.; et al. Jasmonate-responsive MYB factors spatially repress rutin biosynthesis in Fagopyrum tataricum. J. Exp. Bot. 2018, 69, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Llorca, M.; Pollmann, S.; Müller, M. Ethylene and Jasmonates Signaling Network Mediating Secondary Metabolites under Abiotic Stress. Int. J. Mol. Sci. 2023, 24, 5990. https://doi.org/10.3390/ijms24065990
Pérez-Llorca M, Pollmann S, Müller M. Ethylene and Jasmonates Signaling Network Mediating Secondary Metabolites under Abiotic Stress. International Journal of Molecular Sciences. 2023; 24(6):5990. https://doi.org/10.3390/ijms24065990
Chicago/Turabian StylePérez-Llorca, Marina, Stephan Pollmann, and Maren Müller. 2023. "Ethylene and Jasmonates Signaling Network Mediating Secondary Metabolites under Abiotic Stress" International Journal of Molecular Sciences 24, no. 6: 5990. https://doi.org/10.3390/ijms24065990
APA StylePérez-Llorca, M., Pollmann, S., & Müller, M. (2023). Ethylene and Jasmonates Signaling Network Mediating Secondary Metabolites under Abiotic Stress. International Journal of Molecular Sciences, 24(6), 5990. https://doi.org/10.3390/ijms24065990





