The Crystal Structure of Engineered Nitroreductase NTR 2.0 and Impact of F70A and F108Y Substitutions on Substrate Specificity
Abstract
:1. Introduction
2. Results and Discussion
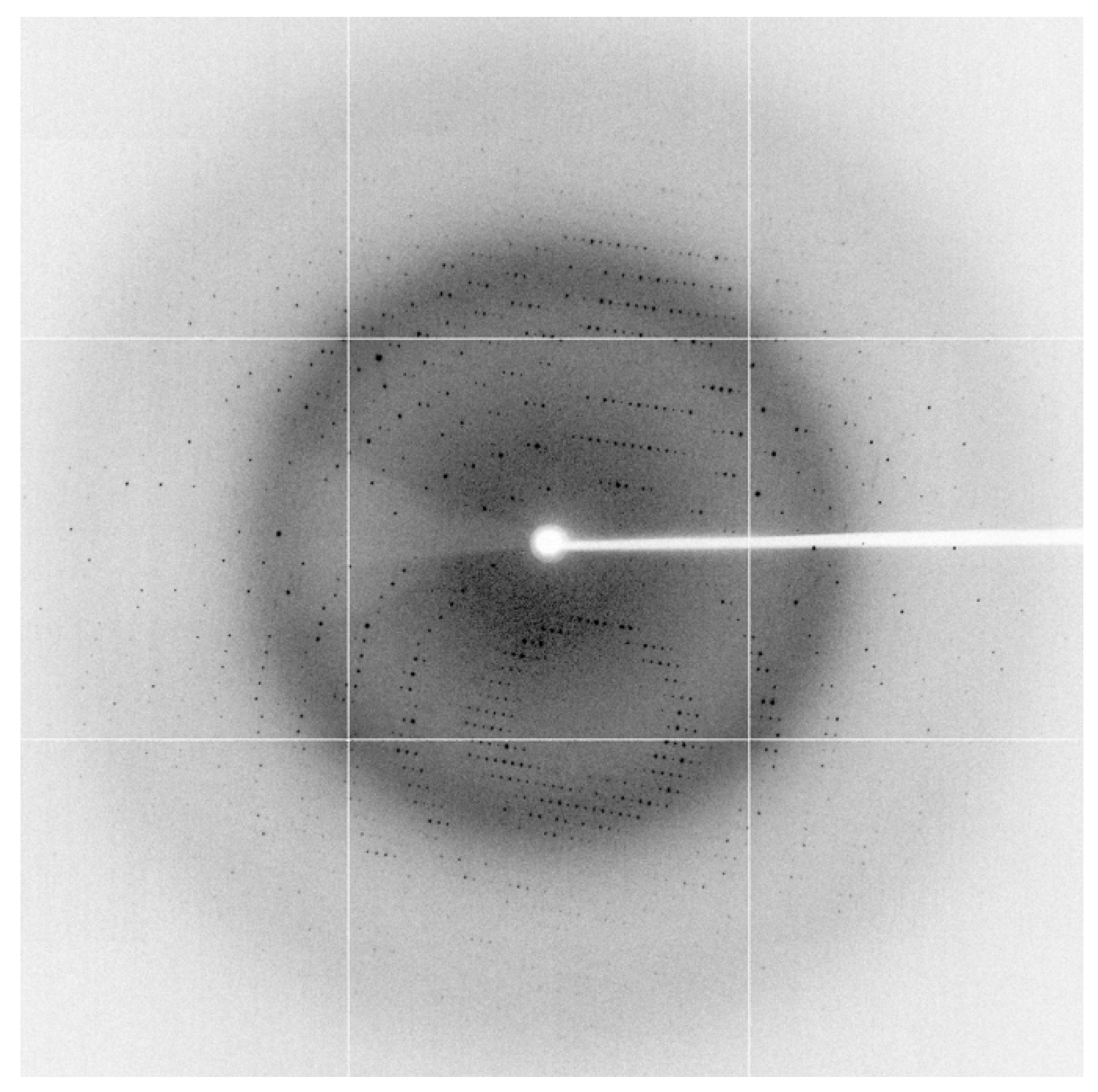
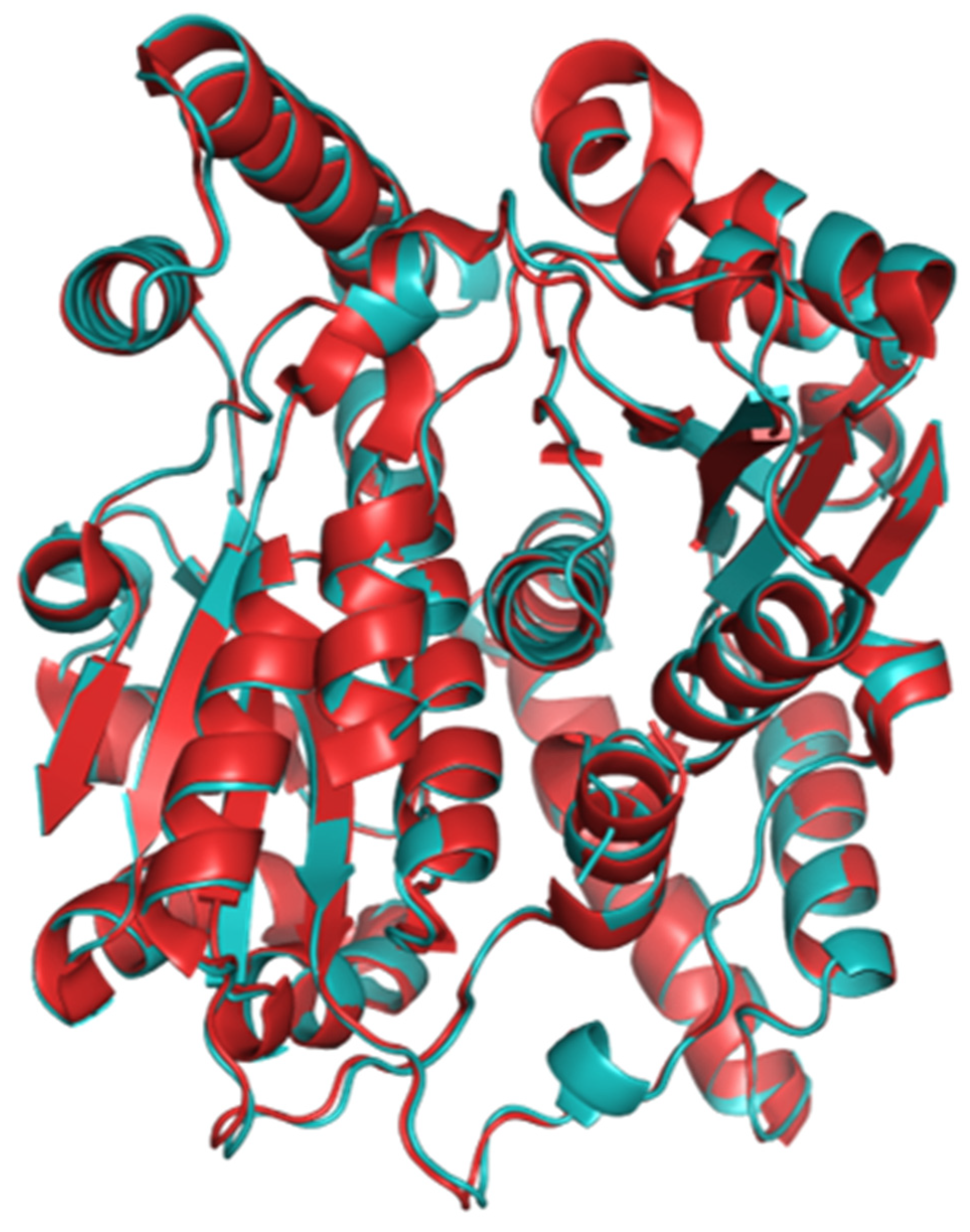
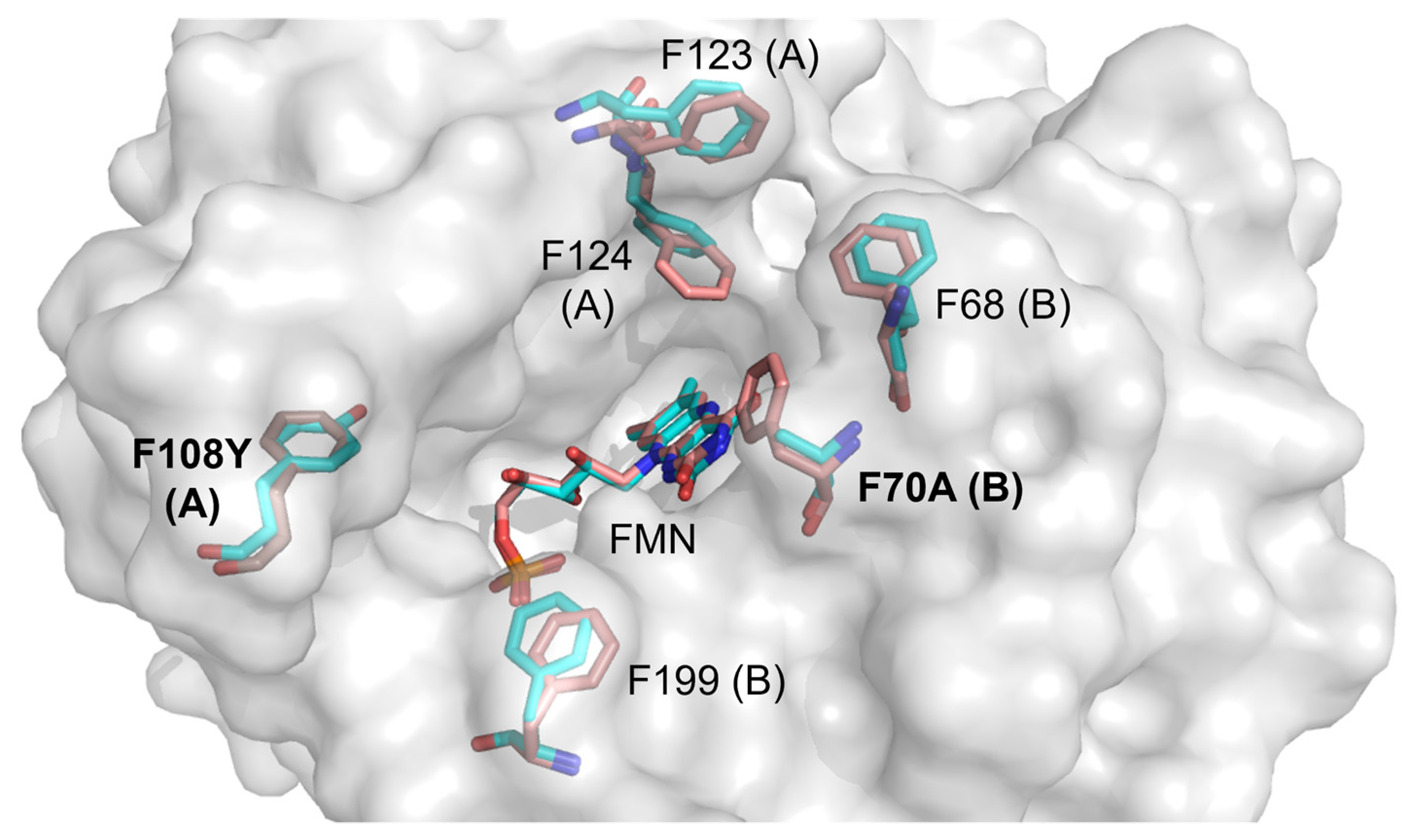
2.1. The Structure of Homodimeric NTR 2.0 and Comparison to Unmodified V. vulnificus NfsB
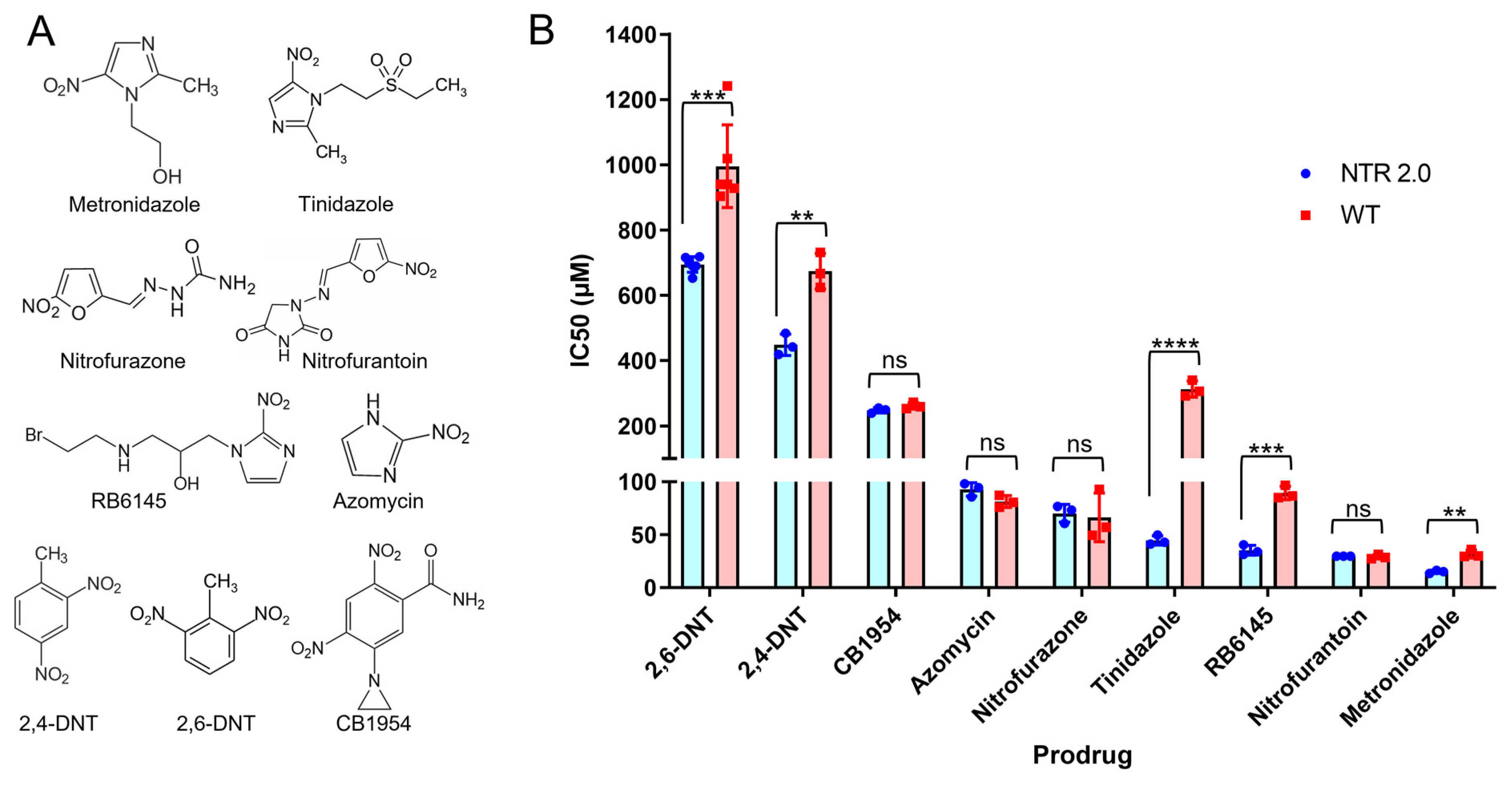
2.2. Surveying NTR 2.0 Activity with Alternative Nitroaromatic Substrates
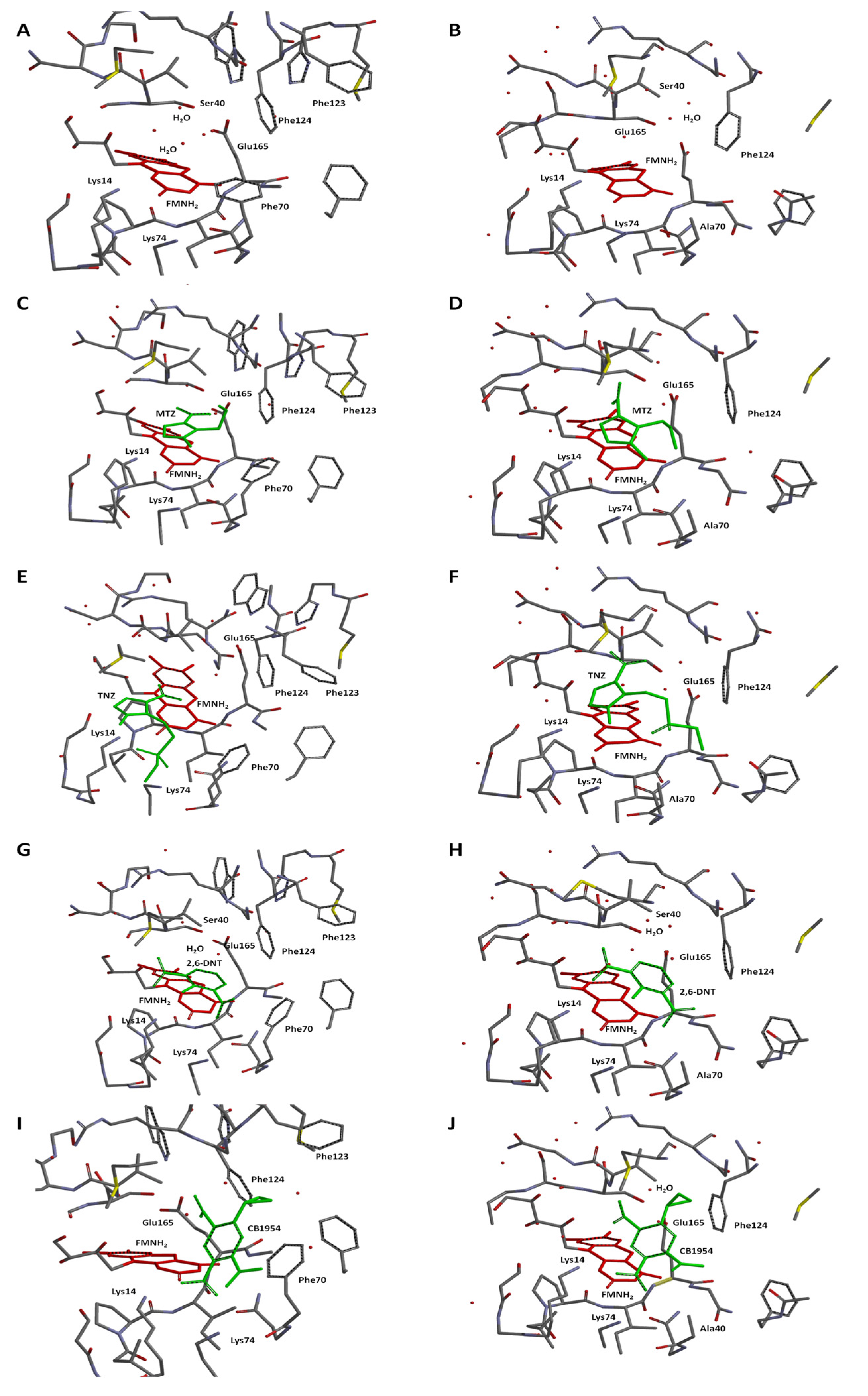
2.3. Molecular Modelling of Prodrug Binding—NTR 2.0 vs. Unmodified V. vulnificus NfsB
- Metronidazole (MTZ). The plane-to-plane distance between the imidazole ring of MTZ and isoalloxazine in both enzyme forms is approximately 3.5–3.8 Å (Figure 8C,D). However, the nature of the H-bonding differs between the enzymes: while both form an H-bond between Lys14 and N3 of imidazole, a second H-bond in V. vulnificus NfsB is formed between the 2-OH of the ribityl group of FMN and the nitro group, while in NTR 2.0, the second bond is formed between N3-H of isoalloxazine and the ethanol OH group of metronidazole (Figure 8C,D). The calculated distances between the isoalloxazine N5-H and the nitro group oxygen are equal to 4.2 Å (V. vulnificus NfsB) and 4.3 Å (NTR 2.0). The calculated Ein are −80.9 kJ·mol−1 and −84.1 kJ·mol−1, respectively.
- Tinidazole (TNZ). As with MTZ, differences in H-bond formation are observed: in unmodified V. vulnificus NfsB, the H-bonds are formed between the tinidazole sulfonyl group and Lys14 and Lys74, whereas in NTR 2.0, Lys14 interacts with N3 of the imidazole ring, and the sulfonyl interacts with N3-H of isoalloxazine (Figure 8E,F). The distances between the oxygen of the nitro group and the isoalloxazine N5-H are equal to 4.4 Å (V. vulnificus NfsB) and 4.7 Å (NTR 2.0), respectively. The calculated Ein are −32.7 kJ·mol−1 and −80.9 kJ·mol−1, respectively.
- 2,6-Dinitrotoluene (2,6-DNT). When bound to both enzymes forms, the compound is nearly parallel with respect to the isoalloxazine ring of FMNH2, but is not involved in the π–π interaction with the isoalloxazine or Phe70 of V. vulnificus NfsB (Figure 8G,H). The nitro groups of 2,6-DNT form H-bonds with Lys14 and Lys74, but do not interact with other amino acids. The calculated distances between the isoalloxazine N5–8 and the nitro group oxygen are equal to 6.7 Å (parent enzyme) and 6.2 Å (NTR 2.0), whereas Ein are equal to −50.6 kJ·mol−1 and −54.3 kJ·mol−1, respectively.
- CB1954. The aziridine ring of this compound interacts with residue 70 in both enzymes, and despite minor differences in orientation it is the 4-nitro group rather than the 2-nitro group that is oriented toward the isoalloxazine in each case (Figure 8I,J). The distances between the oxygen of the reducible 4-nitro group of CB1954 and the H-N5 of isoalloxazine are equal to 5.5 Å (V. vulnificus NfsB) and 5.1 Å (NTR 2.0), whereas Ein are equal to −75.1 kJ·mol−1 and −56.4 kJ·mol−1, respectively. The 4-nitro group forms H-bonds with Lys14 and Lys74.
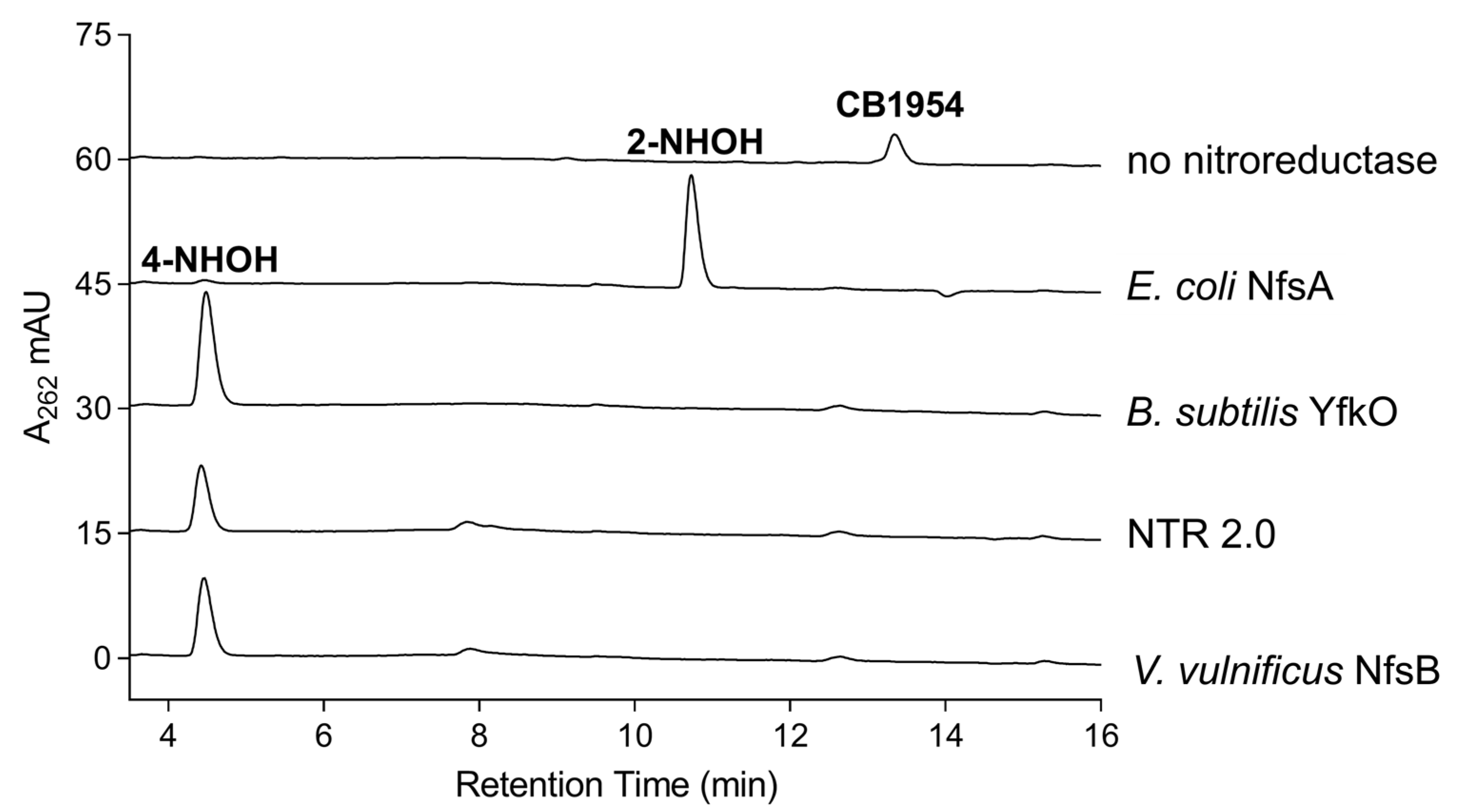
2.4. Analysis of CB1954 Reduction Products
3. Materials and Methods
3.1. Bacterial Strains, Media and Growth Conditions
3.2. Cloning, Expression and Purification of NTR 2.0
3.3. Crystallisation
3.4. X-ray Data Collection
3.5. Structure Solution and Refinement
3.6. Bacterial Cytotoxicity Assays
3.7. HPLC Identification of CB1954 Nitroreduction Products
3.8. Theoretical Modelling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ju, K.S.; Parales, R.E. Nitroaromatic compounds, from synthesis to biodegradation. Microbiol. Mol. Biol. Rev. 2010, 74, 250–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, E.M.; Little, R.F.; Mowday, A.M.; Rich, M.H.; Chan-Hyams, J.V.; Copp, J.N.; Smaill, J.B.; Patterson, A.V.; Ackerley, D.F. Nitroreductase gene-directed enzyme prodrug therapy: Insights and advances toward clinical utility. Biochem. J. 2015, 471, 131–153. [Google Scholar] [CrossRef] [PubMed]
- White, D.T.; Mumm, J.S. The nitroreductase system of inducible targeted ablation facilitates cell-specific regenerative studies in zebrafish. Methods 2013, 62, 232–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Chen, C.; Fu, J.; Lilley, B.; Berlinicke, C.; Hansen, B.; Ding, D.; Wang, G.; Wang, T.; Shou, D.; et al. Large-scale phenotypic drug screen identifies neuroprotectants in zebrafish and mouse models of retinitis pigmentosa. eLife 2021, 10, e57245. [Google Scholar] [CrossRef] [PubMed]
- Pisharath, H.; Parsons, M.J. Nitroreductase-mediated cell ablation in transgenic zebrafish embryos. Methods Mol. Biol. 2009, 546, 133–143. [Google Scholar]
- Sharrock, A.V.; Mulligan, T.S.; Hall, K.R.; Williams, E.M.; White, D.T.; Zhang, L.; Emmerich, K.; Matthews, F.; Nimmagadda, S.; Washington, S.; et al. NTR 2.0: A rationally engineered prodrug-converting enzyme with substantially enhanced efficacy for targeted cell ablation. Nat. Methods 2022, 19, 205–215. [Google Scholar] [CrossRef]
- Mathias, J.R.; Zhang, Z.; Saxena, M.T.; Mumm, J.S. Enhanced cell-specific ablation in zebrafish using a triple mutant of Escherichia coli nitroreductase. Zebrafish 2014, 11, 85–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Pitsawong, W.; Hoben, J.P.; Miller, A.F. Understanding the broad substrate repertoire of nitroreductase based on its kinetic mechanism. J. Biol. Chem. 2014, 289, 15203–15214. [Google Scholar] [CrossRef] [Green Version]
- Parkinson, G.N.; Skelly, J.V.; Neidle, S. Crystal structure of FMN-dependent nitroreductase from Escherichia coli B: A prodrug-activating enzyme. J. Med. Chem. 2000, 43, 3624–3631. [Google Scholar] [CrossRef]
- Valiauga, B.; Williams, E.M.; Ackerley, D.F.; Čėnas, N. Reduction of quinones and nitroaromatic compounds by Escherichia coli nitroreductase A (NfsA): Characterization of kinetics and substrate specificity. Arch. Biochem. Biophys. 2017, 614, 14–22. [Google Scholar] [CrossRef]
- Williams, E.M. Development of Bacterial Nitroreductase Enzymes for Noninvasive Imaging in Cancer Gene Therapy. Ph.D. Thesis, School of Biological Sciences, Victoria University of Wellington, Wellington, New Zealand, 2013. [Google Scholar]
- Koder, R.L.; Haynes, C.A.; Rodgers, M.E.; Rodgers, D.W.; Miller, A.-F. Flavin thermodynamics explain the oxygen insensitivity of enteric nitroreductases. Biochemistry 2002, 41, 14197–14205. [Google Scholar] [CrossRef] [PubMed]
- Race, P.R.; Lovering, A.L.; Green, R.M.; Ossor, A.; White, S.A.; Searle, P.F.; Wrighton, C.J.; Hyde, E.I. Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone: Reversed binding orientations in different redox states of the enzyme. J. Biol. Chem. 2005, 280, 13256–13264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauzá, A.; Deyà, P.M.; Frontera, A. Anion-π Interactions in Supramolecular Chemistry and Catalysis. In Noncovalent Forces; Scheiner, S., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 471–500. [Google Scholar]
- Kokkonen, P.; Bednar, D.; Pinto, G.; Prokop, Z.; Damborsky, J. Engineering enzyme access tunnels. Biotechnol. Adv. 2019, 37, 107386. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [Green Version]
- Williams, E.M.; Sharrock, A.V.; Rylott, E.L.; Bruce, N.C.; MacKichan, J.K.; Ackerley, D.F. A cofactor consumption screen identifies promising NfsB family nitroreductases for dinitrotoluene remediation. Biotechnol. Lett. 2019, 41, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Valiauga, B.; Bagžiūnas, G.; Sharrock, A.V.; Ackerley, D.F.; Čėnas, N. E. coli nitroreductase-A as a candidate for gene-directed prodrug therapy: Potentiometric data and modes of substrate binding. In Proceedings of the SCT 29th Young Research Fellows Meeting, Nantes, France, 4–6 July 2022. [Google Scholar]
- Day, M.A.; Jarrom, D.; Christofferson, A.J.; Graziano, A.E.; Anderson, J.L.R.; Searle, P.F.; Hyde, E.I.; White, S.A. The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN. Biochem. J. 2021, 478, 2601–2617. [Google Scholar] [CrossRef]
- Helsby, N.A.; Ferry, D.M.; Patterson, A.V.; Pullen, S.M.; Wilson, W.R. 2-Amino metabolites are key mediators of CB1954 and SN23862 bystander effects in nitroreductase GDEPT. Br. J. Cancer 2004, 90, 1084–1092. [Google Scholar] [CrossRef] [Green Version]
- Knox, R.J.; Friedlos, F.; Marchbank, T.; Roberts, J.J. Bioactivation of CB 1954: Reaction of the active 4-hydroxylamino derivative with thioesters to form the ultimate DNA-DNA interstrand crosslinking species. Biochem. Pharmacol. 1991, 42, 1691–1697. [Google Scholar] [CrossRef] [PubMed]
- Helsby, N.A.; Wheeler, S.J.; Pruijn, F.B.; Palmer, B.D.; Yang, S.; Denny, W.A.; Wilson, W.R. Effect of nitroreduction on the alkylating reactivity and cytotoxicity of the 2,4-dinitrobenzamide-5-aziridine CB1954 and the corresponding nitrogen mustard SN23862: Distinct mechanisms of bioreductive activation. Chem. Res. Toxicol. 2003, 16, 469–478. [Google Scholar] [CrossRef]
- Knox, R.J.; Friedlos, F.; Jarman, M.; Roberts, J.J. A new cytotoxic, DNA interstrand crosslinking agent, 5-(aziridin-1-yl)-4-hydroxylamino-2-nitrobenzamide, is formed from 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB 1954) by a nitroreductase enzyme in Walker carcinoma cells. Biochem. Pharmacol. 1988, 37, 4661–4669. [Google Scholar] [CrossRef]
- Osswald, A.; Sun, Z.; Grimm, V.; Ampem, G.; Riegel, K.; Westendorf, A.M.; Sommergruber, W.; Otte, K.; Dürre, P.; Riedel, C.U. Three-dimensional tumor spheroids for in vitro analysis of bacteria as gene delivery vectors in tumor therapy. Microb. Cell Factories 2015, 14, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anlezark, G.M.; Vaughan, T.; Fashola-Stone, E.; Michael, N.P.; Murdoch, H.; Sims, M.A.; Stubbs, S.; Wigley, S.; Minton, N.P. Bacillus amyloliquefaciens orthologue of Bacillus subtilis ywrO encodes a nitroreductase enzyme which activates the prodrug CB1954. Microbiology 2002, 148, 297–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vass, S.O.; Jarrom, D.; Wilson, W.R.; Hyde, E.I.; Searle, P.F. E. coli NfsA: An alternative nitroreductase for prodrug activation gene therapy in combination with CB1954. Br. J. Cancer 2009, 100, 1903–1911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prosser, G.A.; Copp, J.N.; Mowday, A.M.; Guise, C.P.; Syddall, S.P.; Williams, E.M.; Horvat, C.N.; Swe, P.M.; Ashoorzadeh, A.; Denny, W.A.; et al. Creation and screening of a multi-family bacterial oxidoreductase library to discover novel nitroreductases that efficiently activate the bioreductive prodrugs CB1954 and PR-104A. Biochem. Pharmacol. 2013, 85, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Prosser, G.A.; Patterson, A.V.; Ackerley, D.F. uvrB gene deletion enhances SOS chromotest sensitivity for nitroreductases that preferentially generate the 4-hydroxylamine metabolite of the anti-cancer prodrug CB1954. J. Biotechnol. 2010, 150, 190–194. [Google Scholar] [CrossRef]
- Copp, J.N.; Hanson-Manful, P.; Ackerley, D.F.; Patrick, W.M. Error-prone PCR and effective generation of gene variant libraries for directed evolution. Methods Mol. Biol. 2014, 1179, 3–22. [Google Scholar]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Leslie, A.G.; Powell, H.R. Processing diffraction data with mosflm. In Evolving Methods for Macromolecular Crystallography; Springer: Berlin/Heidelberg, Germany, 2007; pp. 41–51. [Google Scholar]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radveikienė, I.; Palinauskas, D.; Ragauskaitė, E.; Bagdžiūnas, G. Self-assembled cyclodextrins-based nanostructures on indium-tin-oxide for a detection of catecholamine neurotransmitters. Appl. Surf. Sci. 2022, 600, 154170. [Google Scholar] [CrossRef]



| Wavelength (Å) | 0.9537 |
|---|---|
| Space group | C 2 2 21 |
| Unit-cell parameters (Å, °) | a = 68.43, b = 137.36, c = 107.04, α/β/γ = 90/90/90 |
| Resolution range (Å) | 68.68–1.85 (1.89–1.85) |
| Measured reflections | 624,970 |
| Unique reflections | 43,344 |
| Multiplicity | 14.4 |
| Temperature (K) | 100 |
| Matthews coefficient (Å3 Da−1) | 2.32 |
| Solvent content (%) | 47 |
| No. of molecules in ASU | 2 |
| Completeness (%) | 99.9 (99.7) |
| Mean I/σ(I) | 18.3 (3.7) |
| Rmerge† (%) | 12 (79.2) |
| CC(1/2) | 0.999 |
| Wilson B factor (Å2) | 10.275 |
| Rwork | 0.165 |
| Rfree | 0.196 |
| Protein atoms | 3342 |
| Other ions/molecules | 7 |
| Number of waters | 184 |
| B factors (proteins) | 18.5 (Chain A)/19.8 (Chain B) |
| B factors (water) | 23.9 |
| RMSD | |
| Bond angles (°) | 1.6771 |
| Bond lengths (Å) | 0.0120 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharrock, A.V.; Mumm, J.S.; Bagdžiūnas, G.; Čėnas, N.; Arcus, V.L.; Ackerley, D.F. The Crystal Structure of Engineered Nitroreductase NTR 2.0 and Impact of F70A and F108Y Substitutions on Substrate Specificity. Int. J. Mol. Sci. 2023, 24, 6633. https://doi.org/10.3390/ijms24076633
Sharrock AV, Mumm JS, Bagdžiūnas G, Čėnas N, Arcus VL, Ackerley DF. The Crystal Structure of Engineered Nitroreductase NTR 2.0 and Impact of F70A and F108Y Substitutions on Substrate Specificity. International Journal of Molecular Sciences. 2023; 24(7):6633. https://doi.org/10.3390/ijms24076633
Chicago/Turabian StyleSharrock, Abigail V., Jeff S. Mumm, Gintautas Bagdžiūnas, Narimantas Čėnas, Vickery L. Arcus, and David F. Ackerley. 2023. "The Crystal Structure of Engineered Nitroreductase NTR 2.0 and Impact of F70A and F108Y Substitutions on Substrate Specificity" International Journal of Molecular Sciences 24, no. 7: 6633. https://doi.org/10.3390/ijms24076633
APA StyleSharrock, A. V., Mumm, J. S., Bagdžiūnas, G., Čėnas, N., Arcus, V. L., & Ackerley, D. F. (2023). The Crystal Structure of Engineered Nitroreductase NTR 2.0 and Impact of F70A and F108Y Substitutions on Substrate Specificity. International Journal of Molecular Sciences, 24(7), 6633. https://doi.org/10.3390/ijms24076633








