Identification and Characterization of Development-Related microRNAs in the Red Flour Beetle, Tribolium castaneum
Abstract
1. Introduction
2. Results
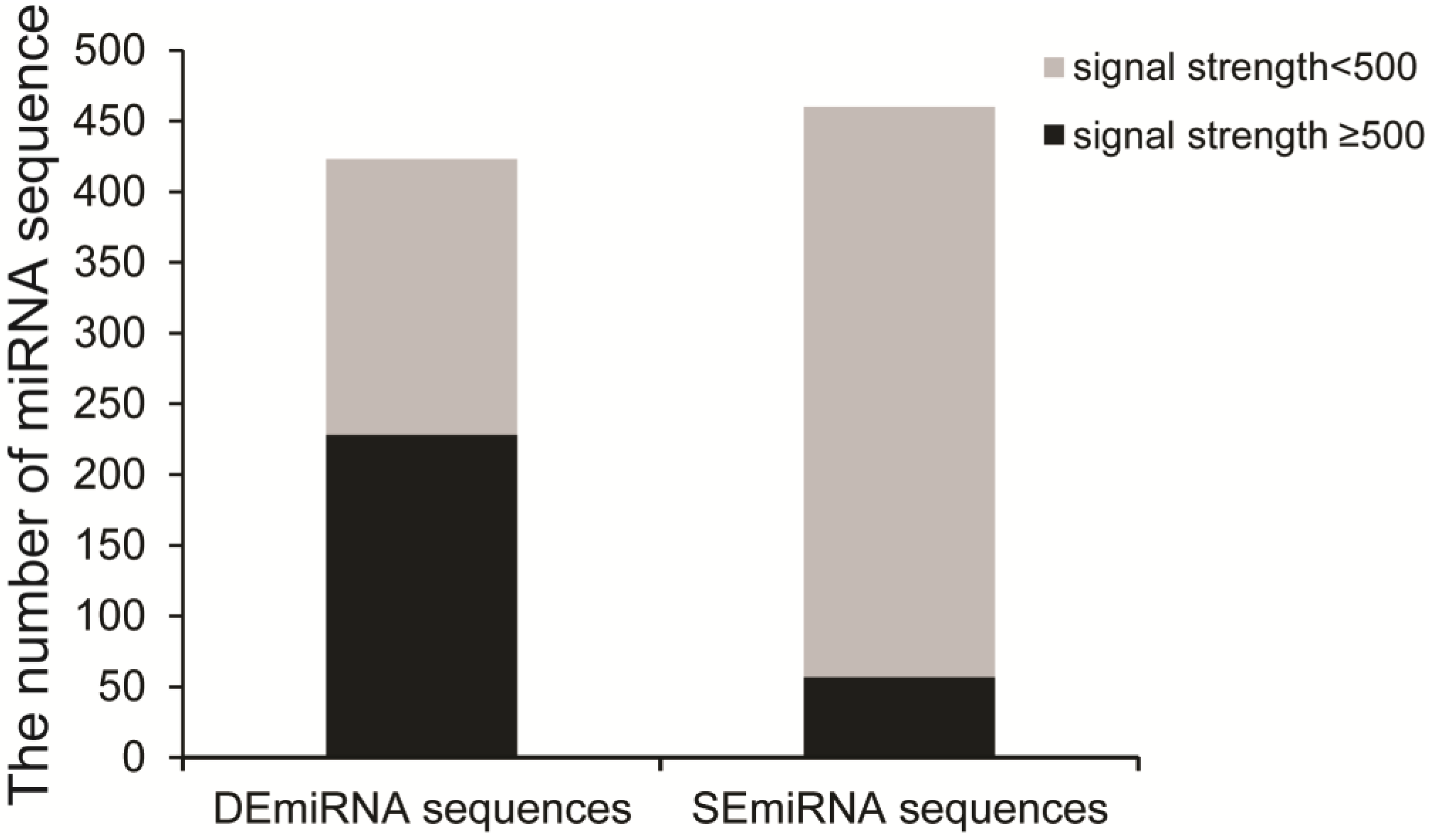
2.1. Identification of DEmiRNAs among Five Developmental Stages
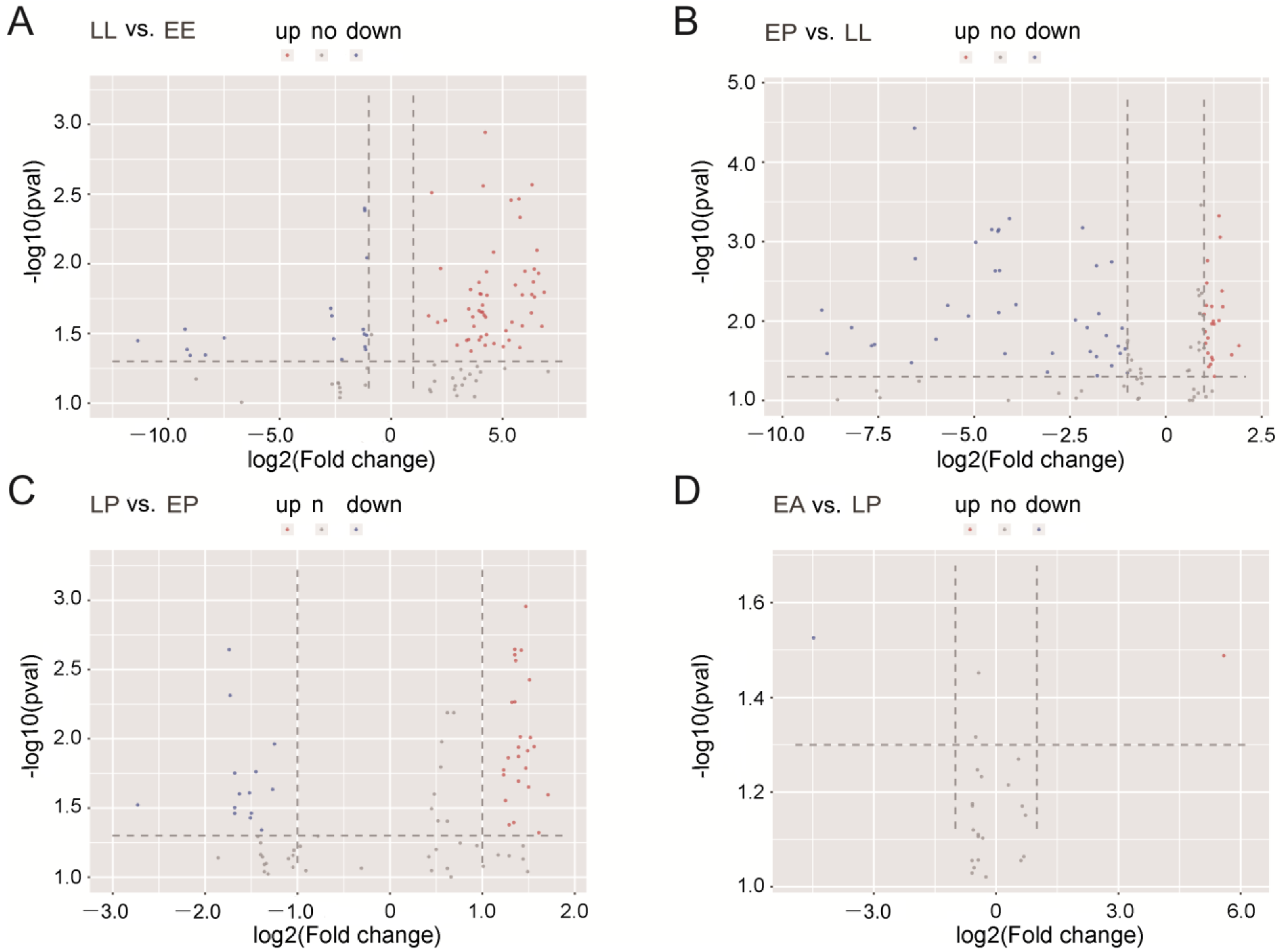
2.2. Identification of DEmiRNAs between Two Developmental Stages
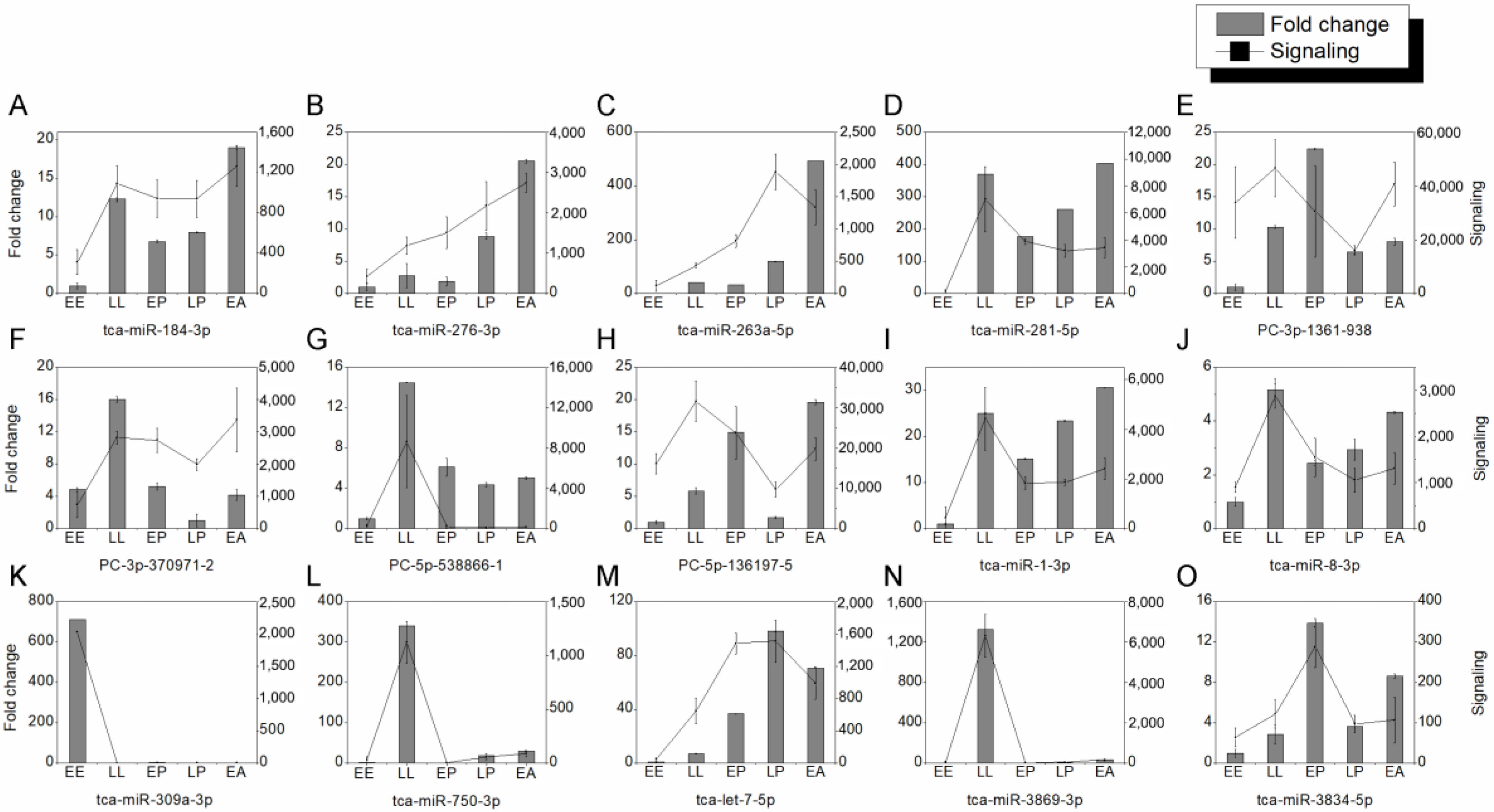
2.3. Analysis of Stage-Specific DEmiRNAs
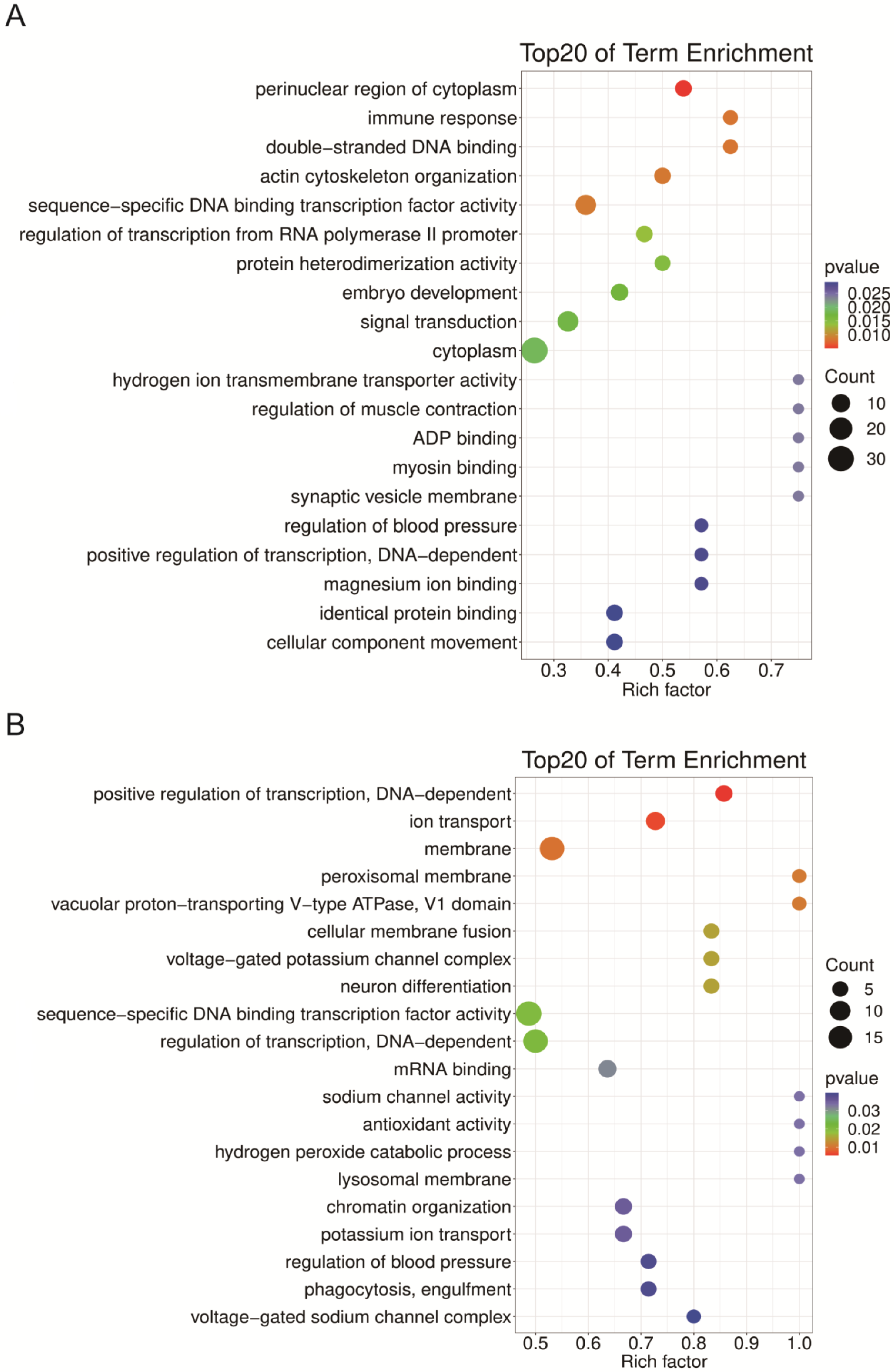
2.4. Target Prediction of DEmiRNAs
2.5. Functional Analysis of DEmiRNA Targets
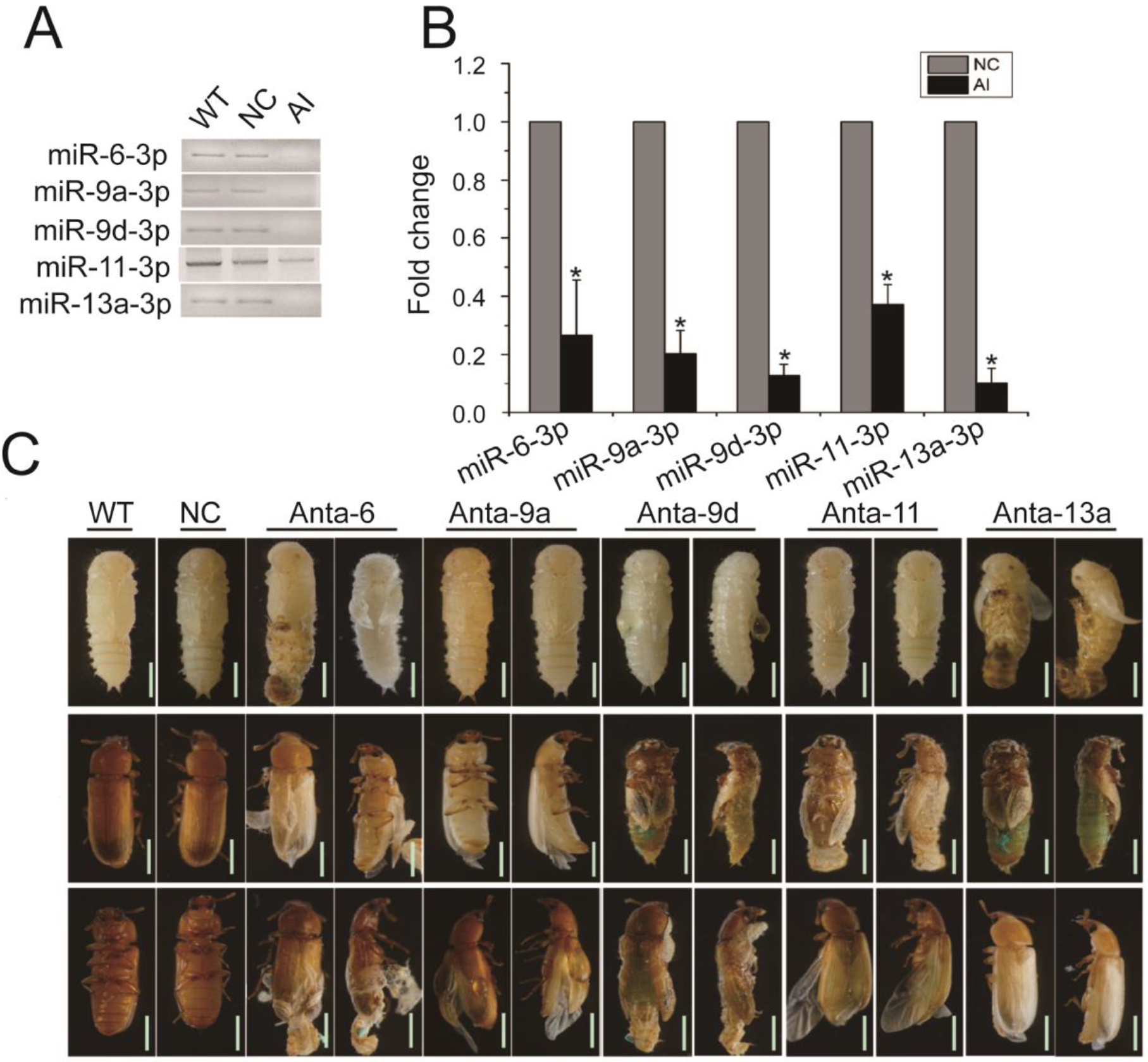
2.6. DEmiRNAs Are Required for Metamorphosis and Wing Development
3. Discussion
4. Materials and Methods
4.1. Insect Strains
4.2. RNA Isolation and cDNA Synthesis
4.3. Microarray Assay
4.4. Identification of Differentially Expressed miRNAs
4.5. Target Prediction for DEmiRNAs
4.6. Enrichment Analyses of Predicted DEmiRNA Targets
4.7. Quantitative Real-Time PCR
4.8. Antagomirs Synthesis and Injection
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Han, J.J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.J.; Zhang, B.T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Gene Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Weinberg, D.E.; Nakanishi, K.; Patel, D.J.; Bartel, D.P. The Inside-Out Mechanism of Dicers from Budding Yeasts. Cell 2011, 146, 262–276. [Google Scholar] [CrossRef]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Lakhotia, N.; Joshi, G.; Bhardwaj, A.R.; Katiyar-Agarwal, S.; Agarwal, M.; Jagannath, A.; Goel, S.; Kumar, A. Identification and characterization of miRNAome in root, stem, leaf and tuber developmental stages of potato (Solanum tuberosum L.) by high-throughput sequencing. BMC Plant Biol. 2014, 14, 6. [Google Scholar] [CrossRef]
- Hsin, J.P.; Lu, Y.; Loeb, G.B.; Leslie, C.S.; Rudensky, A.Y. The effect of cellular context on miR-155-mediated gene regulation in four major immune cell types. Nat. Immunol. 2018, 19, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Li, M.Y.; Li, P.F.; Cao, J.M. MicroRNAs in Cardiac Autophagy: Small Molecules and Big Role. Cells 2018, 7, 104. [Google Scholar] [CrossRef]
- Lim, D.H.; Lee, S.; Han, J.Y.; Choi, M.S.; Hong, J.S.; Seong, Y.; Kwon, Y.S.; Lee, Y.S. Ecdysone-responsive microRNA-252-5p controls the cell cycle by targeting Abi in Drosophila. FASEB J. 2018, 32, 4519–4533. [Google Scholar] [CrossRef]
- Hu, X.; Zhu, M.; Zhang, X.; Liu, B.; Liang, Z.; Huang, L.; Xu, J.; Yu, L.; Li, K.; Zar, M.S.; et al. Identification and characterization of circular RNAs in the silkworm midgut following Bombyx mori cytoplasmic polyhedrosis virus infection. RNA Biol. 2018, 15, 292–301. [Google Scholar] [CrossRef]
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyder, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The small RNA profile during Drosophila melanogaster development. Dev. Cell 2003, 5, 337–350. [Google Scholar] [CrossRef]
- Stark, A.; Kheradpour, P.; Parts, L.; Brennecke, J.; Hodges, E.; Hannon, G.J.; Kellis, M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007, 17, 1865–1879. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Vanlandingham, D.L.; Scholle, F.; Higgs, S.; Cullen, B.R. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genom. 2010, 11, 119. [Google Scholar] [CrossRef]
- He, P.A.; Nie, Z.; Chen, J.; Chen, J.; Lv, Z.; Sheng, Q.; Zhou, S.; Gao, X.; Kong, L.; Wu, X.; et al. Identification and characteristics of microRNAs from Bombyx mori. BMC Genom. 2008, 9, 248. [Google Scholar] [CrossRef]
- Wu, W.; Xiong, W.; Li, C.; Zhai, M.; Li, Y.; Ma, F.; Li, B. MicroRNA-dependent regulation of metamorphosis and identification of microRNAs in the red flour beetle, Tribolium castaneum. Genomics 2017, 109, 362–373. [Google Scholar] [CrossRef]
- Luo, Q.; Zhou, Q.; Yu, X.; Lin, H.; Hu, S.; Yu, J. Genome-wide mapping of conserved microRNAs and their host transcripts in Tribolium castaneum. J. Genet. Genom. 2008, 35, 349–355. [Google Scholar] [CrossRef]
- Singh, J.; Nagaraju, J. In silico prediction and characterization of microRNAs from red flour beetle (Tribolium castaneum). Insect Mol. Biol. 2008, 17, 427–436. [Google Scholar] [CrossRef]
- Ninova, M.; Ronshaugen, M.; Griffiths-Jones, S. MicroRNA evolution, expression, and function during short germband development in Tribolium castaneum. Genome Res. 2016, 26, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.H.; Wang, Z.L.; Tian, L.Q.; Gan, H.Y.; Zhang, S.W.; Zeng, Z.J. The integrative analysis of microRNA and mRNA expression in Apis mellifera following maze-based visual pattern learning. Insect Sci. 2014, 21, 619–636. [Google Scholar] [CrossRef] [PubMed]
- Zondag, L.; Dearden, P.K.; Wilson, M.J. Deep sequencing and expression of microRNAs from early honeybee (Apis mellifera) embryos reveals a role in regulating early embryonic patterning. BMC Evol. Biol. 2012, 12, 211. [Google Scholar] [CrossRef] [PubMed]
- Legeai, F.; Rizk, G.; Walsh, T.; Edwards, O.; Gordon, K.; Lavenier, D.; Leterme, N.; Mereau, A.; Nicolas, J.; Tagu, D.; et al. Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acyrthosiphon pisum. BMC Genom. 2010, 11, 281. [Google Scholar] [CrossRef] [PubMed]
- Kirkness, E.F.; Haas, B.J.; Sun, W.; Braig, H.R.; Perotti, M.A.; Clark, J.M.; Lee, S.H.; Robertson, H.M.; Kennedy, R.C.; Elhaik, E.; et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Natl. Acad. Sci. USA 2010, 107, 12168–12173. [Google Scholar] [CrossRef] [PubMed]
- Sohail, S.; Tariq, K.; Sajid, M.; Ali, M.W.; Peng, W.; Zhang, H. miR-125-3p and miR-276b-3p Regulate the Spermatogenesis of Bactrocera dorsalis by Targeting the orb2 Gene. Genes 2022, 13, 1861. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, S.; Li, T.; Li, H.; Zhang, H.; Zheng, W. RNA sequencing identifies an ovary-enriched microRNA, miR-311-3p, involved in ovarian development and fecundity by targeting Endophilin B1 in Bactrocera dorsalis. Pest Manag. Sci. 2022, 79, 688–700. [Google Scholar] [CrossRef]
- Zhang, Q.; Dou, W.; Taning, C.N.T.; Yu, S.S.; Yuan, G.R.; Shang, F.; Smagghe, G.; Wang, J.J. miR-309a is a regulator of ovarian development in the oriental fruit fly Bactrocera dorsalis. PLoS Genet. 2022, 18, e1010411. [Google Scholar] [CrossRef] [PubMed]
- He, Y.Z.; Aksoy, E.; Ding, Y.; Raikhel, A.S. Hormone-dependent activation and repression of microRNAs by the ecdysone receptor in the dengue vector mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2021, 118, e2102417118. [Google Scholar] [CrossRef]
- Aksoy, E.; Raikhel, A.S. Juvenile hormone regulation of microRNAs is mediated by E75 in the Dengue vector mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2021, 118, e2102851118. [Google Scholar] [CrossRef]
- Wu, W.; Zhai, M.; Li, C.; Yu, X.; Song, X.; Gao, S.; Li, B. Multiple functions of miR-8-3p in the development and metamorphosis of the red flour beetle, Tribolium castaneum. Insect Mol. Biol. 2019, 28, 208–221. [Google Scholar] [CrossRef]
- Gomez-Orte, E.; Belles, X. MicroRNA-dependent metamorphosis in hemimetabolan insects. Proc. Natl. Acad. Sci. USA 2009, 106, 21678–21682. [Google Scholar] [CrossRef]
- Lozano, J.; Montanez, R.; Belles, X. MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway. Proc. Natl. Acad. Sci. USA 2015, 112, 3740–3745. [Google Scholar] [CrossRef]
- He, Q.; Zhang, Y.; Dong, W. MicroRNA miR-927 targets the juvenile hormone primary response gene Kruppel homolog1 to control Drosophila developmental growth. Insect Mol. Biol. 2020, 29, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fu, Y.; Cheng, H.; Zhao, C.; Huang, Q.; Chang, M.; Qiu, W.; Shen, Y.; Li, D. lncR26319/miR-2834/EndophilinA axis regulates oogenesis of the silkworm, Bombyx mori. Insect Sci. 2023, 30, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.J.; Zhu, F.; Liu, Y.J.; Li, Z.; Moural, T.W.; Liu, X.M.; Liu, X. MicroRNAs miR-14 and miR-2766 regulate tyrosine hydroxylase to control larval-pupal metamorphosis in Helicoverpa armigera. Pest Manag. Sci. 2022, 78, 3540–3550. [Google Scholar] [CrossRef] [PubMed]
- Inui, T.; Sezutsu, H.; Daimon, T. MicroRNA let-7 is required for hormonal regulation of metamorphosis in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2022, 145, 103784. [Google Scholar] [CrossRef]
- Duan, T.F.; Gao, S.J.; Wang, H.C.; Li, L.; Li, Y.Y.; Tan, Y.; Pang, B.P. MicroRNA let-7-5p targets the juvenile hormone primary response gene Kruppel homolog 1 and regulates reproductive diapause in Galeruca daurica. Insect Biochem. Mol. Biol. 2022, 142, 103727. [Google Scholar] [CrossRef]
- Wang, Y.; Zou, H.; Lai, J.; Zhang, Z.; Sun, W. The miR-282-5p regulates larval moulting process by targeting chitinase 5 in Bombyx mori. Insect Mol. Biol. 2022, 31, 190–201. [Google Scholar] [CrossRef]
- Zhang, J.; Wen, D.; Li, E.Y.; Palli, S.R.; Li, S.; Wang, J.; Liu, S. MicroRNA miR-8 promotes cell growth of corpus allatum and juvenile hormone biosynthesis independent of insulin/IGF signaling in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2021, 136, 103611. [Google Scholar] [CrossRef]
- Shang, F.; Niu, J.; Ding, B.Y.; Zhang, W.; Wei, D.D.; Wei, D.; Jiang, H.B.; Wang, J.J. The miR-9b microRNA mediates dimorphism and development of wing in aphids. Proc. Natl. Acad. Sci. USA 2020, 117, 8404–8409. [Google Scholar] [CrossRef]
- Mukherjee, S.; Paricio, N.; Sokol, N.S. A stress-responsive miRNA regulates BMP signaling to maintain tissue homeostasis. Proc. Natl. Acad. Sci. USA 2021, 118, e2022583118. [Google Scholar] [CrossRef]
- Pandey, M.; Bansal, S.; Bar, S.; Yadav, A.K.; Sokol, N.S.; Tennessen, J.M.; Kapahi, P.; Chawla, G. miR-125-chinmo pathway regulates dietary restriction-dependent enhancement of lifespan in Drosophila. Elife 2021, 10, e62621. [Google Scholar] [CrossRef]
- Fernandes, J.; Varghese, J. Sexually dimorphic microRNA miR-190 regulates lifespan in male Drosophila. RNA Biol. 2022, 19, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Calvi, B.R.; Hundley, H.A.; Sokol, N.S. MicroRNA mediated regulation of the onset of enteroblast differentiation in the Drosophila adult intestine. Cell Rep. 2022, 41, 111495. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xia, X.; Li, J.; Igaki, T. Tumor elimination by clustered microRNAs miR-306 and miR-79 via noncanonical activation of JNK signaling. Elife 2022, 11, e77340. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Smith, L.; Poulton, J.; Deng, W.M. The microRNA miR-7 regulates Tramtrack69 in a developmental switch in Drosophila follicle cells. Development 2013, 140, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Yatsenko, A.S.; Shcherbata, H.R. Drosophila miR-9a targets the ECM receptor Dystroglycan to canalize myotendinous junction formation. Dev. Cell 2014, 28, 335–348. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, B.; Roy, S.; Saha, T.T.; Kokoza, V.A.; Li, M.; Raikhel, A.S. microRNA-309 targets the Homeobox gene SIX4 and controls ovarian development in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2016, 113, E4828–E4836. [Google Scholar] [CrossRef]
- Song, J.; Li, W.; Zhao, H.; Gao, L.; Fan, Y.; Zhou, S. The microRNAs let-7 and miR-278 regulate insect metamorphosis and oogenesis by targeting the juvenile hormone early-response gene Kruppel-homolog 1. Development 2018, 145, dev170670. [Google Scholar] [CrossRef]
- Peng, W.; Yu, S.; Handler, A.M.; Tu, Z.; Saccone, G.; Xi, Z.; Zhang, H. miRNA-1-3p is an early embryonic male sex-determining factor in the Oriental fruit fly Bactrocera dorsalis. Nat. Commun. 2020, 11, 932. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Fu, J. The microRNA miR-14 Regulates Egg-Laying by Targeting EcR in Honeybees (Apis mellifera). Insects 2021, 12, 351. [Google Scholar] [CrossRef]
- Iovino, N.; Pane, A.; Gaul, U. miR-184 has multiple roles in Drosophila female germline development. Dev. Cell 2009, 17, 123–133. [Google Scholar] [CrossRef]
- He, J.; Chen, Q.; Wei, Y.; Jiang, F.; Yang, M.; Hao, S.; Guo, X.; Chen, D.; Kang, L. MicroRNA-276 promotes egg-hatching synchrony by up-regulating brm in locusts. Proc. Natl. Acad. Sci. USA 2016, 113, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.H.; Lee, S.; Choi, M.S.; Han, J.Y.; Seong, Y.; Na, D.; Kwon, Y.S.; Lee, Y.S. The conserved microRNA miR-8-3p coordinates the expression of V-ATPase subunits to regulate ecdysone biosynthesis for Drosophila metamorphosis. FASEB J. 2020, 34, 6449–6465. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, J.; Ling, L.; Luo, X.; Yang, D.; Yang, X.; Zhang, X.; Huang, Y. miR-34 regulates larval growth and wing morphogenesis by directly modulating ecdysone signalling and cuticle protein in Bombyx mori. RNA Biol. 2020, 17, 1342–1351. [Google Scholar] [CrossRef]
- Jiang, J.; Ge, X.; Li, Z.; Wang, Y.; Song, Q.; Stanley, D.W.; Tan, A.; Huang, Y. MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform B in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2013, 43, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, F.; Lee, J.A.; Gao, F.B. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006, 20, 2793–2805. [Google Scholar] [CrossRef]
- Bejarano, F.; Smibert, P.; Lai, E.C. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev. Biol. 2010, 338, 63–73. [Google Scholar] [CrossRef]
- Hyun, S.; Lee, J.H.; Jin, H.; Nam, J.; Namkoong, B.; Lee, G.; Chung, J.; Kim, V.N. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 2009, 139, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, D.M.; Caparros, E.; Dominguez, M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO J. 2011, 30, 756–769. [Google Scholar] [CrossRef]
- Kennell, J.A.; Gerin, I.; MacDougald, O.A.; Cadigan, K.M. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 15417–15422. [Google Scholar] [CrossRef]
- Lim, D.H.; Lee, S.; Han, J.Y.; Choi, M.S.; Hong, J.S.; Lee, Y.S. MicroRNA miR-252 targets mbt to control the developmental growth of Drosophila. Insect Mol. Biol. 2019, 28, 444–454. [Google Scholar] [CrossRef]
- Rubio, M.; de Horna, A.; Belles, X. MicroRNAs in metamorphic and non-metamorphic transitions in hemimetabolan insect metamorphosis. BMC Genom. 2012, 13, 386. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhai, M.; Yu, R.; Song, X.; Feng, F.; Gao, H.; Li, B. MiR-3017b contributes to metamorphosis by targeting sarco/endoplasmic reticulum Ca(2+) ATPase in Tribolium castaneum. Insect Mol. Biol. 2022, 31, 286–296. [Google Scholar] [CrossRef]
- Bushati, N.; Stark, A.; Brennecke, J.; Cohen, S.M. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 2008, 18, 501–506. [Google Scholar] [CrossRef]
- Li, X.; Zhang, F.; Coates, B.; Wei, C.; Zhu, X.; Zhang, Y.; Zhou, X. Temporal analysis of microRNAs associated with wing development in the English grain aphid, Sitobion avenae (F.) (Homoptera: Aphidiae). Insect Biochem. Mol Biol. 2022, 142, 103579. [Google Scholar] [CrossRef] [PubMed]
- Katti, P.; Thimmaya, D.; Madan, A.; Nongthomba, U. Overexpression of miRNA-9 Generates Muscle Hypercontraction Through Translational Repression of Troponin-T in Drosophila melanogaster Indirect Flight Muscles. G3 2017, 7, 3521–3531. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Li, X.; Li, Y. Epithelial microRNA-9a regulates dendrite growth through Fmi-Gq signaling in Drosophila sensory neurons. Dev. Neurobiol. 2016, 76, 225–237. [Google Scholar] [CrossRef]
- Biryukova, I.; Asmar, J.; Abdesselem, H.; Heitzler, P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev. Biol. 2009, 327, 487–496. [Google Scholar] [CrossRef]
- Rahman, S.; Modak, C.; Akter, M.; Alam, M.S. Role of MicroRNA Genes miR-1000 and miR-375 in Forming Olfactory Conditional Memory in Drosophila melanogaster. MicroRNA 2020, 9, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Fu, X.; Du, J.; Wu, B.; Zhao, X.; Zhu, J.; Zhao, Z. Regulation of circadian rhythm and sleep by miR-375-timeless interaction in Drosophila. FASEB J. 2020, 34, 16536–16551. [Google Scholar] [CrossRef]
- Beeman, R.W.; Stuart, J.J.; Brown, S.J.; Denell, R.E. Structure and function of the homeotic gene complex (HOM-C) in the beetle, Tribolium castaneum. Bioessays 1993, 15, 439–444. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, Q.; Eicken, C.; Sheng, N.; Zhang, X.; Yang, L.; Gao, X. MicroRNA profiling using microParaflo microfluidic array technology. Methods Mol. Biol. 2012, 822, 153–182. [Google Scholar]
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Horwich, M.D.; Zamore, P.D. Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nat. Protoc. 2008, 3, 1537–1549. [Google Scholar] [CrossRef]
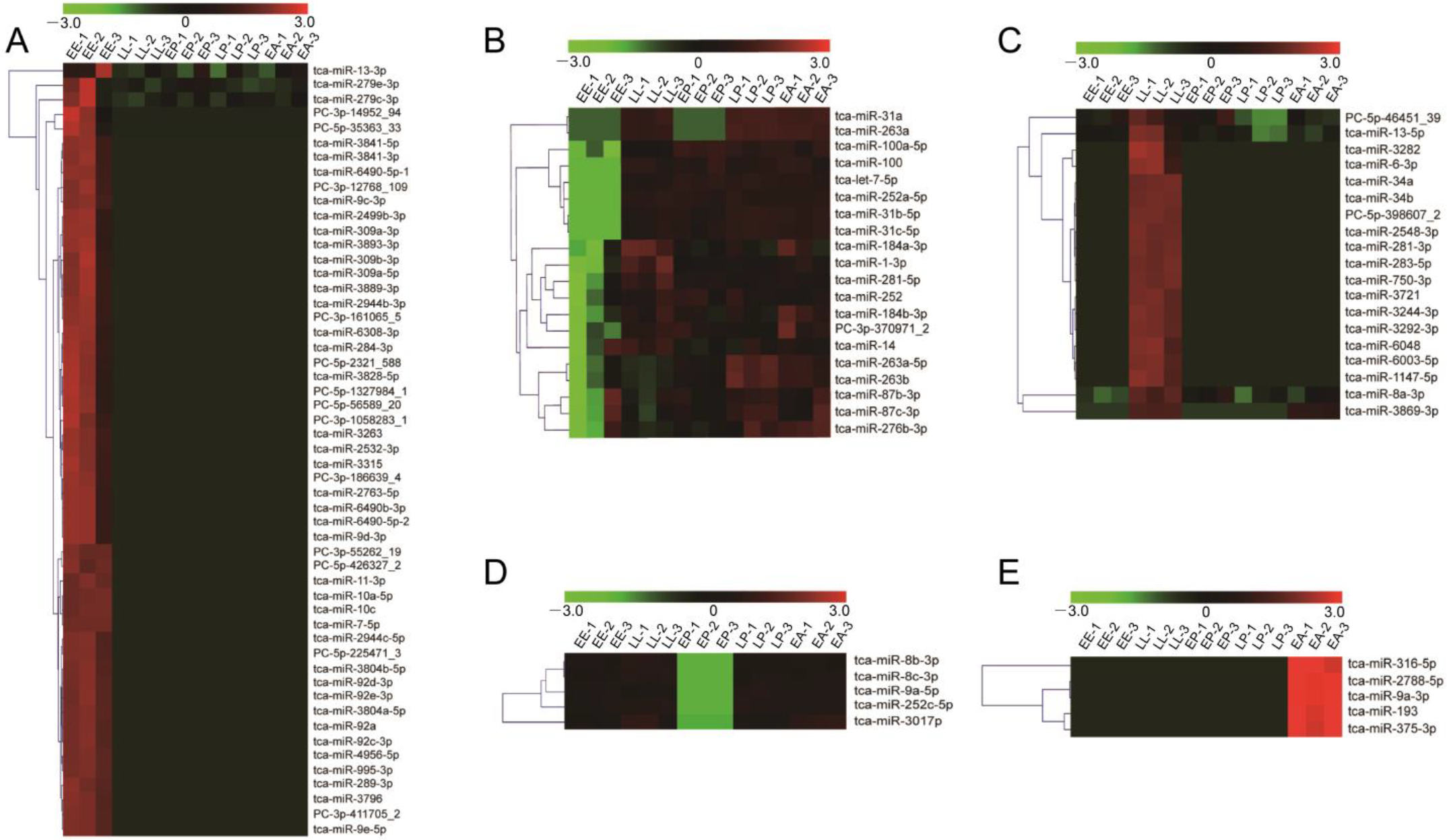


| Cluster | DEmiRNA No. | DEmiRNA | Target Sites | Target Genes |
|---|---|---|---|---|
| A | 53 | tca-miR-13-3p, tca-miR-279e-3p, tca-miR-279c-3p, PC-3p-14952_94, PC-5p-35363_33, tca-miR-3841-5p, tca-miR-3841-3p, tca-miR-6490-5p-1, PC-3p-12768_109, tca-miR-9c-3p, tca-miR-2499b-3p, tca-miR-309a-3p, tca-miR-3893-3p, tca-miR-309b-3p, tca-miR-309a-5p, tca-miR-3889-3p, tca-miR-2944b-3p, PC-3p-161065_5, tca-miR-6308-3p, tca-miR-284-3p, PC-5p-2321_588, tca-miR-3828-5p, PC-5p-1327984_1, PC-5p-56589_20, PC-3p-1058283_1, tca-miR-3263, tca-miR-2532-3p, tca-miR-3315, PC-3p-186639_4, tca-miR-2763-5p, tca-miR-6490b-3p, tca-miR-6490-5p-2, tca-miR-9d-3p, PC-3p-55262_19, PC-5p-426327_2, tca-miR-11-3p, tca-miR-10a-5p, tca-miR-10c, tca-miR-7-5p, tca-miR-2944c-5p, PC-5p-225471_3, tca-miR-3804b-5p, tca-miR-92d-3p, tca-miR-92e-3p, tca-miR-3804a-5p, tca-miR-92a, tca-miR-92c-3p, tca-miR-4956-5p, tca-miR-995-3p, tca-miR-289-3p, tca-miR-3796, PC-3p-411705_2, tca-miR-9e-5p | 1792 | 748 |
| B | 20 | tca-miR-31a, tca-miR-263a, tca-miR-100a-5p, tca-miR-100, tca-let-7-5p, tca-miR-252a-5p, tca-miR-31b-5p, tca-miR-31c-5p, tca-miR-184a-3p, tca-miR-1-3p, tca-miR-281-5p, tca-miR-252, tca-miR-184b-3p, PC-3p-370971_2, tca-miR-14, tca-miR-263a-5p, tca-miR-263b, tca-miR-87b-3p, tca-miR-87c-3p, tca-miR-276b-3p | 728 | 379 |
| C | 19 | PC-5p-46451_39, tca-miR-13-5p, tca-miR-3282, tca-miR-6-3p, tca-miR-34a, tca-miR-34b, PC-5p-398607_2, tca-miR-2548-3p, tca-miR-281-3p, tca-miR-283-5p, tca-miR-750-3p, tca-miR-3721, tca-miR-3244-3p, tca-miR-3292-3p, tca-miR-6048, tca-miR-6003-5p, tca-miR-1147-5p, tca-miR-8a-3p, tca-miR-3869-3p | 665 | 460 |
| D | 5 | tca-miR-8b-3p, tca-miR-8c-3p, tca-miR-9a-5p, tca-miR-252c-5p, tca-miR-3017b | 389 | 234 |
| E | 5 | tca-miR-316-5p, tca-miR-2788-5p, tca-miR-9a-3p, tca-miR-193, tca-miR-375-3p | 148 | 132 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Wu, W.; Tang, J.; Feng, F.; Chen, P.; Li, B. Identification and Characterization of Development-Related microRNAs in the Red Flour Beetle, Tribolium castaneum. Int. J. Mol. Sci. 2023, 24, 6685. https://doi.org/10.3390/ijms24076685
Li C, Wu W, Tang J, Feng F, Chen P, Li B. Identification and Characterization of Development-Related microRNAs in the Red Flour Beetle, Tribolium castaneum. International Journal of Molecular Sciences. 2023; 24(7):6685. https://doi.org/10.3390/ijms24076685
Chicago/Turabian StyleLi, Chengjun, Wei Wu, Jing Tang, Fan Feng, Peng Chen, and Bin Li. 2023. "Identification and Characterization of Development-Related microRNAs in the Red Flour Beetle, Tribolium castaneum" International Journal of Molecular Sciences 24, no. 7: 6685. https://doi.org/10.3390/ijms24076685
APA StyleLi, C., Wu, W., Tang, J., Feng, F., Chen, P., & Li, B. (2023). Identification and Characterization of Development-Related microRNAs in the Red Flour Beetle, Tribolium castaneum. International Journal of Molecular Sciences, 24(7), 6685. https://doi.org/10.3390/ijms24076685






