The Immunological Profile of SARS-CoV-2 Infection in Children Is Linked to Clinical Severity and Age
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics of Patients and Controls
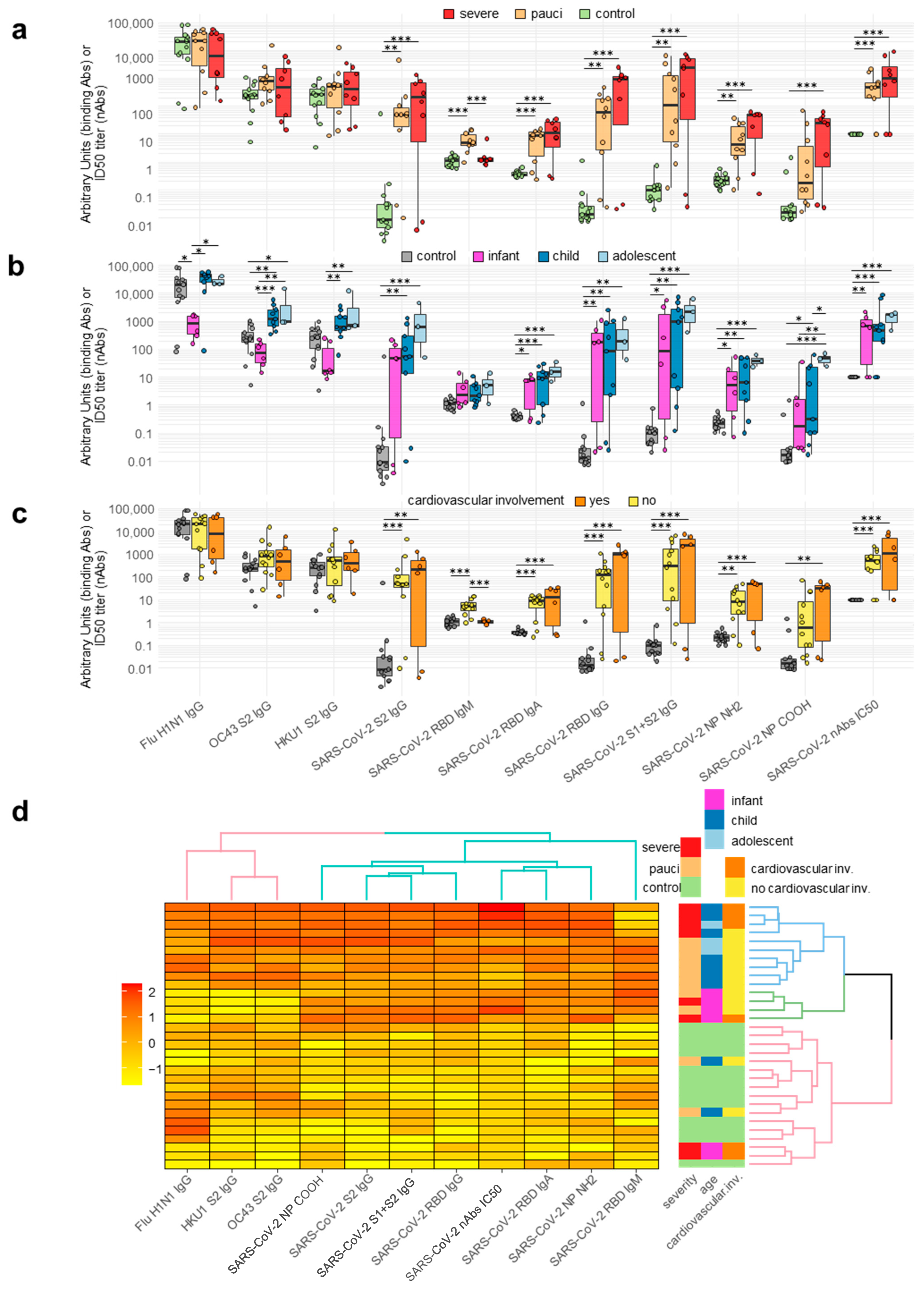
2.2. Antibody Responses of Patients and Controls
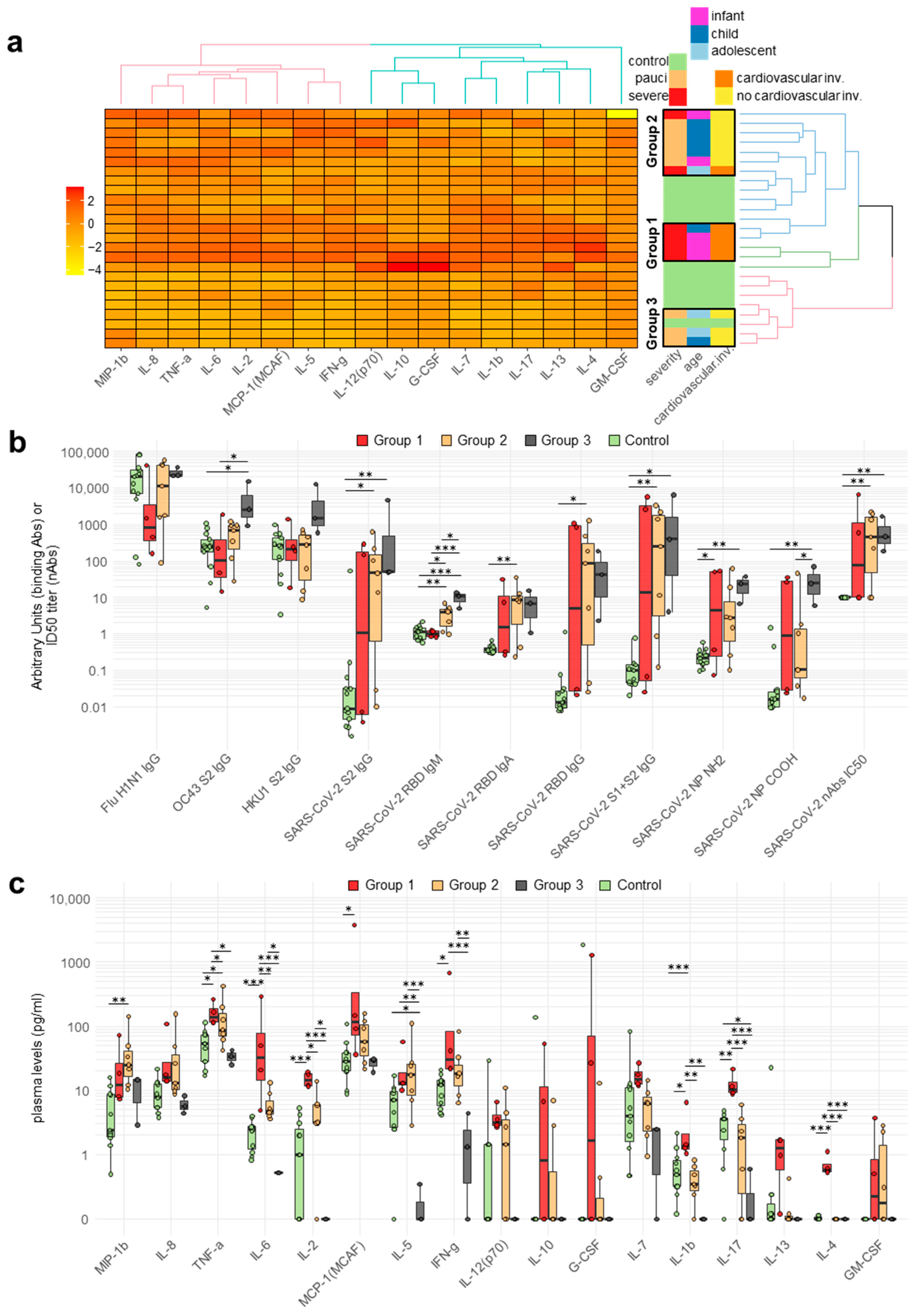
2.3. Cytokine and Chemokine Plasma Levels in Pediatric Patients
2.4. Correlation between Circulating Antibodies and Cytokine/Chemokine Responses of Patients
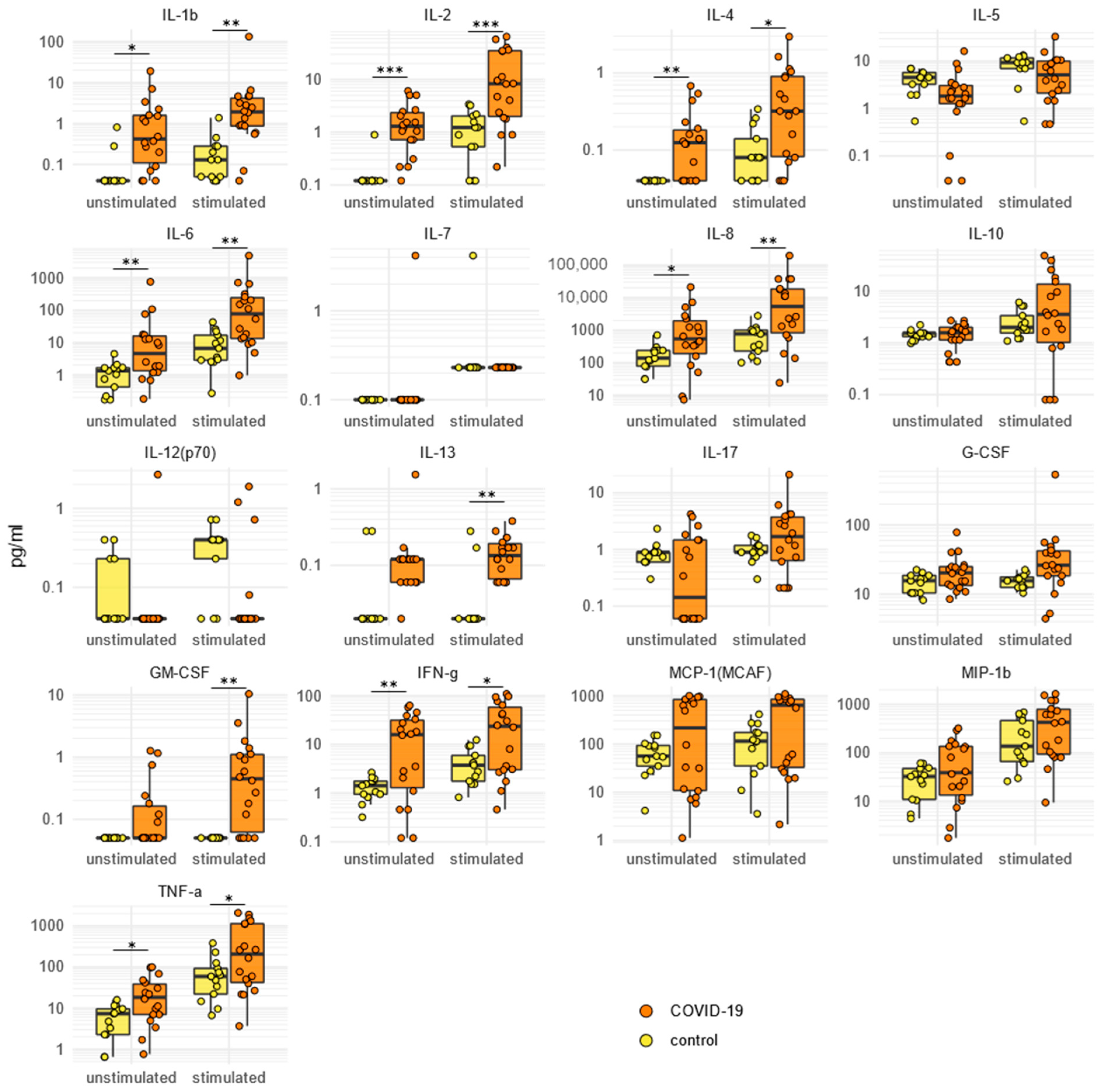
2.5. SARS-CoV-2-Specific Cytokine/Chemokine Secretion
2.6. SARS-CoV-2-Specific Immune Gene Expression
3. Discussion
4. Materials and Methods
4.1. Patient Population
4.2. SARS-CoV-2, Seasonal HCoVs, and H1N1 Flu Virus Antibody Assays
4.3. Isolation and Stimulation of Peripheral Blood Mononuclear Cells (PBMCs) with SARS-CoV-2-Specific Antigens
4.4. Quantigene Plex Gene Expression Assay
4.5. Multiplex Cytokine Analyses
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Viner, R.M.; Mytton, O.T.; Bonell, C.; Melendez-Torres, G.J.; Ward, J.; Hudson, L.; Waddington, C.; Thomas, J.; Russell, S.; van der Klis, F.; et al. Susceptibility to SARS-CoV-2 Infection Among Children and Adolescents Compared with Adults: A Systematic Review and Meta-Analysis. JAMA Pediatr. 2021, 175, 143–156. [Google Scholar] [CrossRef]
- Sharma, C.; Ganigara, M.; Galeotti, C.; Burns, J.; Berganza, F.M.; Hayes, D.A.; Singh-Grewal, D.; Bharath, S.; Sajjan, S.; Bayry, J. Multisystem Inflammatory Syndrome in Children and Kawasaki Disease: A Critical Comparison. Nat. Rev. Rheumatol. 2021, 17, 731–748. [Google Scholar] [CrossRef]
- Patel, J.M. Multisystem Inflammatory Syndrome in Children (MIS-C). Curr. Allergy Asthma Rep. 2022, 22, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. Why Is COVID-19 Less Severe in Children? A Review of the Proposed Mechanisms Underlying the Age-Related Difference in Severity of SARS-CoV-2 Infections. Arch. Dis. Child. 2021, 106, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Brodin, P. SARS-CoV-2 Infections in Children: Understanding Diverse Outcomes. Immunity 2022, 55, 201–209. [Google Scholar] [CrossRef]
- Dispinseri, S.; Secchi, M.; Pirillo, M.F.; Tolazzi, M.; Borghi, M.; Brigatti, C.; De Angelis, M.L.; Baratella, M.; Bazzigaluppi, E.; Venturi, G.; et al. Neutralizing Antibody Responses to SARS-CoV-2 in Symptomatic COVID-19 Is Persistent and Critical for Survival. Nat. Commun. 2021, 12, 2670. [Google Scholar] [CrossRef] [PubMed]
- Petrara, M.R.; Bonfante, F.; Costenaro, P.; Cantarutti, A.; Carmona, F.; Ruffoni, E.; Di Chiara, C.; Zanchetta, M.; Barzon, L.; Donà, D.; et al. Asymptomatic and Mild SARS-CoV-2 Infections Elicit Lower Immune Activation and Higher Specific Neutralizing Antibodies in Children Than in Adults. Front. Immunol. 2021, 12, 741796. [Google Scholar] [CrossRef]
- Karron, R.A.; Garcia Quesada, M.; Schappell, E.A.; Schmidt, S.D.; Deloria Knoll, M.; Hetrich, M.K.; Veguilla, V.; Doria-Rose, N.; Dawood, F.S. Binding and Neutralizing Antibody Responses to SARS-CoV-2 in Very Young Children Exceed Those in Adults. JCI Insight 2022, 7, e157963. [Google Scholar] [CrossRef]
- Dong, Y.; Mo, X.; Hu, Y.; Qi, X.; Jiang, F.; Jiang, Z.; Tong, S. Epidemiology of COVID-19 Among Children in China. Pediatrics 2020, 145, e20200702. [Google Scholar] [CrossRef]
- CDC Multisystem Inflammatory Syndrome (MIS). Available online: https://www.cdc.gov/mis/index.html (accessed on 25 July 2022).
- Multisystem Inflammatory Syndrome in Children and Adolescents with COVID-19. Available online: https://www.who.int/publications-detail-redirect/multisystem-inflammatory-syndrome-in-children-and-adolescents-with-covid-19 (accessed on 25 July 2022).
- Garcia-Beltran, W.F.; Lam, E.C.; Astudillo, M.G.; Yang, D.; Miller, T.E.; Feldman, J.; Hauser, B.M.; Caradonna, T.M.; Clayton, K.L.; Nitido, A.D.; et al. COVID-19-Neutralizing Antibodies Predict Disease Severity and Survival. Cell 2021, 184, 476–488.e11. [Google Scholar] [CrossRef]
- Weisberg, S.P.; Connors, T.J.; Zhu, Y.; Baldwin, M.R.; Lin, W.-H.; Wontakal, S.; Szabo, P.A.; Wells, S.B.; Dogra, P.; Gray, J.; et al. Distinct Antibody Responses to SARS-CoV-2 in Children and Adults across the COVID-19 Clinical Spectrum. Nat. Immunol. 2021, 22, 25–31. [Google Scholar] [CrossRef]
- Bahar, B.; Jacquot, C.; Mo, Y.D.; DeBiasi, R.L.; Campos, J.; Delaney, M. Kinetics of Viral Clearance and Antibody Production Across Age Groups in Children with Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J. Pediatr. 2020, 227, 31–37.e1. [Google Scholar] [CrossRef] [PubMed]
- Cotugno, N.; Ruggiero, A.; Bonfante, F.; Petrara, M.R.; Zicari, S.; Pascucci, G.R.; Zangari, P.; De Ioris, M.A.; Santilli, V.; Manno, E.C.; et al. Virological and Immunological Features of SARS-CoV-2-Infected Children Who Develop Neutralizing Antibodies. Cell. Rep. 2021, 34, 108852. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Hurst, J.H.; Lorang, C.G.; Aquino, J.N.; Rodriguez, J.; Pfeiffer, T.S.; Singh, T.; Semmes, E.C.; Lugo, D.J.; Rotta, A.T.; et al. Asymptomatic or Mild Symptomatic SARS-CoV-2 Infection Elicits Durable Neutralizing Antibody Responses in Children and Adolescents. JCI Insight 2021, 6, e150909. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.; Thomas, P.G.; Randolph, A.G. Immunology of SARS-CoV-2 Infection in Children. Nat. Immunol. 2022, 23, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Aydillo, T.; Rombauts, A.; Stadlbauer, D.; Aslam, S.; Abelenda-Alonso, G.; Escalera, A.; Amanat, F.; Jiang, K.; Krammer, F.; Carratala, J.; et al. Immunological Imprinting of the Antibody Response in COVID-19 Patients. Nat. Commun. 2021, 12, 3781. [Google Scholar] [CrossRef]
- Shrock, E.; Fujimura, E.; Kula, T.; Timms, R.T.; Lee, I.-H.; Leng, Y.; Robinson, M.L.; Sie, B.M.; Li, M.Z.; Chen, Y.; et al. Viral Epitope Profiling of COVID-19 Patients Reveals Cross-Reactivity and Correlates of Severity. Science 2020, 370, eabd4250. [Google Scholar] [CrossRef]
- Sasson, J.M.; Campo, J.J.; Carpenter, R.M.; Young, M.K.; Randall, A.Z.; Trappl-Kimmons, K.; Oberai, A.; Hung, C.; Edgar, J.; Teng, A.A.; et al. Diverse Humoral Immune Responses in Younger and Older Adult COVID-19 Patients. mBio 2021, 12, e01229-21. [Google Scholar] [CrossRef]
- Dowell, A.C.; Butler, M.S.; Jinks, E.; Tut, G.; Lancaster, T.; Sylla, P.; Begum, J.; Bruton, R.; Pearce, H.; Verma, K.; et al. Children Develop Robust and Sustained Cross-Reactive Spike-Specific Immune Responses to SARS-CoV-2 Infection. Nat. Immunol. 2022, 23, 40–49. [Google Scholar] [CrossRef]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.-E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 2020, 27, 992–1000.e3. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients with COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Acharya, D.; Liu, G.; Gack, M.U. Dysregulation of Type I Interferon Responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 397–398. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired Type I Interferon Activity and Inflammatory Responses in Severe COVID-19 Patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Graff, K.; Smith, C.; Silveira, L.; Jung, S.; Curran-Hays, S.; Jarjour, J.; Carpenter, L.; Pickard, K.; Mattiucci, M.; Fresia, J.; et al. Risk Factors for Severe COVID-19 in Children. Pediatr. Infect. Dis. J. 2021, 40, e137–e145. [Google Scholar] [CrossRef]
- DeBiasi, R.L.; Song, X.; Delaney, M.; Bell, M.; Smith, K.; Pershad, J.; Ansusinha, E.; Hahn, A.; Hamdy, R.; Harik, N.; et al. Severe Coronavirus Disease-2019 in Children and Young Adults in the Washington, DC, Metropolitan Region. J. Pediatr. 2020, 223, 199–203.e1. [Google Scholar] [CrossRef]
- Sobolewska-Pilarczyk, M.; Pokorska-Śpiewak, M.; Stachowiak, A.; Marczyńska, M.; Talarek, E.; Ołdakowska, A.; Kucharek, I.; Sybilski, A.; Mania, A.; Figlerowicz, M.; et al. COVID-19 Infections in Infants. Sci. Rep. 2022, 12, 7765. [Google Scholar] [CrossRef]
- Soraya, G.V.; Ulhaq, Z.S. Interleukin-6 Levels in Children Developing SARS-CoV-2 Infection. Pediatr. Neonatol. 2020, 61, 253–254. [Google Scholar] [CrossRef]
- Curatola, A.; Chiaretti, A.; Ferretti, S.; Bersani, G.; Lucchetti, D.; Capossela, L.; Sgambato, A.; Gatto, A. Cytokine Response to SARS-CoV-2 Infection in Children. Viruses 2021, 13, 1868. [Google Scholar] [CrossRef]
- Ozsurekci, Y.; Aykac, K.; Er, A.G.; Halacli, B.; Arasli, M.; Oygar, P.D.; Gürlevik, S.; Cura Yayla, B.C.; Karakaya, J.; Alp, A.; et al. Predictive Value of Cytokine/Chemokine Responses for the Disease Severity and Management in Children and Adult Cases with COVID-19. J. Med. Virol. 2021, 93, 2828–2837. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Wang, X.; Liu, P.; Liang, X.; Ge, Y.; Tian, H.; Chang, H.; Zhou, H.; Zeng, M.; Xu, J. Mild Cytokine Elevation, Moderate CD4+ T Cell Response and Abundant Antibody Production in Children with COVID-19. Virol. Sin. 2020, 35, 734–743. [Google Scholar] [CrossRef]
- Müller, T.M.; Becker, E.; Wiendl, M.; Schulze, L.L.; Voskens, C.; Völkl, S.; Kremer, A.E.; Neurath, M.F.; Zundler, S. Circulating Adaptive Immune Cells Expressing the Gut Homing Marker A4β7 Integrin Are Decreased in COVID-19. Front. Immunol. 2021, 12, 639329. [Google Scholar] [CrossRef] [PubMed]
- Secchi, M.; Bazzigaluppi, E.; Brigatti, C.; Marzinotto, I.; Tresoldi, C.; Rovere-Querini, P.; Poli, A.; Castagna, A.; Scarlatti, G.; Zangrillo, A.; et al. COVID-19 Survival Associates with the Immunoglobulin Response to the SARS-CoV-2 Spike Receptor Binding Domain. J. Clin. Investig. 2020, 130, 6366–6378. [Google Scholar] [CrossRef] [PubMed]
- Fenyö, E.M.; Heath, A.; Dispinseri, S.; Holmes, H.; Lusso, P.; Zolla-Pazner, S.; Donners, H.; Heyndrickx, L.; Alcami, J.; Bongertz, V.; et al. International Network for Comparison of HIV Neutralization Assays: The NeutNet Report. PLoS ONE 2009, 4, e4505. [Google Scholar] [CrossRef] [PubMed]
- Saulle, I.; Vanetti, C.; Goglia, S.; Vicentini, C.; Tombetti, E.; Garziano, M.; Clerici, M.; Biasin, M. A New ERAP2/Iso3 Isoform Expression Is Triggered by Different Microbial Stimuli in Human Cells. Could It Play a Role in the Modulation of SARS-CoV-2 Infection? Cells 2020, 9, 1951. [Google Scholar] [CrossRef] [PubMed]

| Patients | Controls | |||
|---|---|---|---|---|
| # | % | # | % | |
| Median age (range) in years | 6.1 (0.1–14.7) | - | 9.4 (1.0–16.6) | - |
| Female | 12 | 33 | 5 | 38.5 |
| Comorbidities | 0 | 0 | 0 | 0 |
| White cell count (average ± SD) | 8071 ± 3506 | - | - | |
| Neutrophils (average ± SD) | 2962 ± 2234 | 35 ± 15.4 | - | |
| Lymphocytes (average ± SD) | 4215 ± 1976 | 53.2 ± 14.8 | - | - |
| Positive RT-PCR based on respiratory specimen | 13/19 | 68.4 | - | - |
| IgG Anti-SARS-CoV-2 | 11/13 | 84.6 | - | - |
| Paucisymptomatic/moderate (PM) | 10 | 55% | - | - |
| Severe/critical (SC) | 8 | 45% | - | - |
| IFNA2 | CASP1 | CD209 | TAP1 |
| IL2 | ACTB | IL1B | MPO |
| IL28A | TLR4 | IL10 | NLRP3 |
| IL17A | IL18 | IL1RN | IL6R |
| CCL2 | IL7 | ABCA1 | HAVCR2 |
| TNFRSF4 | PTGS2 | ELOVL6 | CXCL10 |
| IFNB1 | MX1 | CD38 | IFNG |
| PPARG | IFITM3 | IL12B | NOD2 |
| CRP | CD44 | NOS2 | CCL3 |
| PPIB | CLEC4M | CH25H | CD69 |
| COVID19 | IFITM1 | IFI16 | AGTR1 |
| CCL5 | CD274 | ACAT1 | ERAP1 |
| GAPDH | IL1A | HPRT1 | TBP |
| PDCD1 | IL12A | NOD1 | ERAP2 |
| HMGCS1 | ITGA4 | CSF3 | |
| TLR8 | NR1H3 | AGTR2 | |
| IL22 | IL8 | TLR3 | |
| CSF2 | ITGB7 | IL6 | |
| PYCARD | TLR7 | ACE | |
| TNF | ACE2 | TMPRSS2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanetti, C.; Lampasona, V.; Stracuzzi, M.; Fenizia, C.; Biasin, M.; Saulle, I.; Limanaqi, F.; Abdelsalam, A.; Loretelli, C.; Paradiso, L.; et al. The Immunological Profile of SARS-CoV-2 Infection in Children Is Linked to Clinical Severity and Age. Int. J. Mol. Sci. 2023, 24, 6779. https://doi.org/10.3390/ijms24076779
Vanetti C, Lampasona V, Stracuzzi M, Fenizia C, Biasin M, Saulle I, Limanaqi F, Abdelsalam A, Loretelli C, Paradiso L, et al. The Immunological Profile of SARS-CoV-2 Infection in Children Is Linked to Clinical Severity and Age. International Journal of Molecular Sciences. 2023; 24(7):6779. https://doi.org/10.3390/ijms24076779
Chicago/Turabian StyleVanetti, Claudia, Vito Lampasona, Marta Stracuzzi, Claudio Fenizia, Mara Biasin, Irma Saulle, Fiona Limanaqi, Ahmed Abdelsalam, Cristian Loretelli, Laura Paradiso, and et al. 2023. "The Immunological Profile of SARS-CoV-2 Infection in Children Is Linked to Clinical Severity and Age" International Journal of Molecular Sciences 24, no. 7: 6779. https://doi.org/10.3390/ijms24076779
APA StyleVanetti, C., Lampasona, V., Stracuzzi, M., Fenizia, C., Biasin, M., Saulle, I., Limanaqi, F., Abdelsalam, A., Loretelli, C., Paradiso, L., Longoni, E., Barcellini, L., Piemonti, L., Marzinotto, I., Dispinseri, S., Amendola, A., Fappani, C., Tanzi, E., Clerici, M. S., ... Trabattoni, D. (2023). The Immunological Profile of SARS-CoV-2 Infection in Children Is Linked to Clinical Severity and Age. International Journal of Molecular Sciences, 24(7), 6779. https://doi.org/10.3390/ijms24076779













