Genome-Wide Identification and Chilling Stress Analysis of the NF-Y Gene Family in Melon
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of Melon NF-Y Genes
2.2. Gene Structure of CmNF-Ys
2.3. Motif Composition and Multiple Alignments of the CmNF-Ys
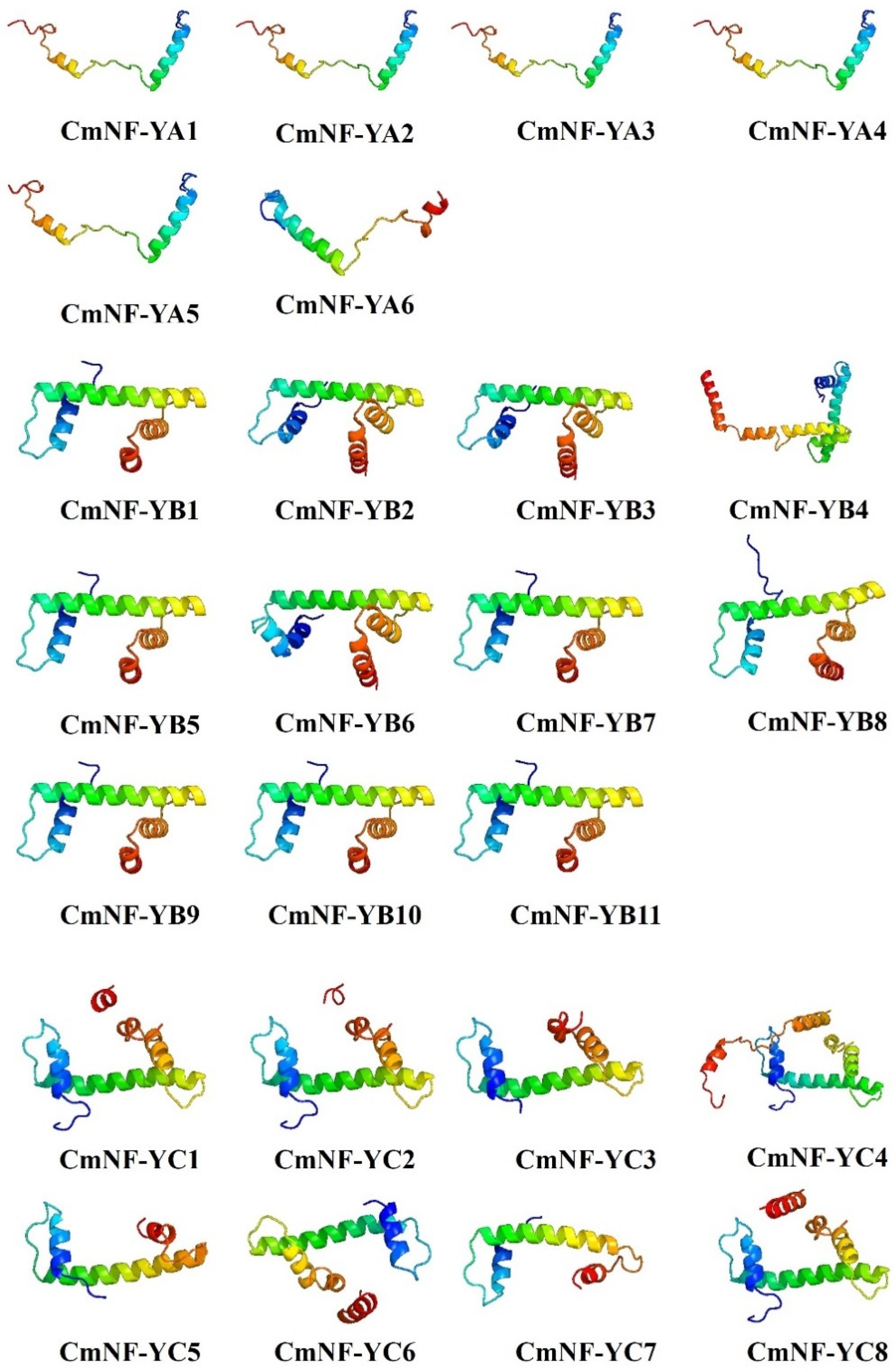
2.4. Secondary and Tertiary Structure Prediction of CmNF-Ys
2.5. Phylogenetic Analysis of CmNF-Ys
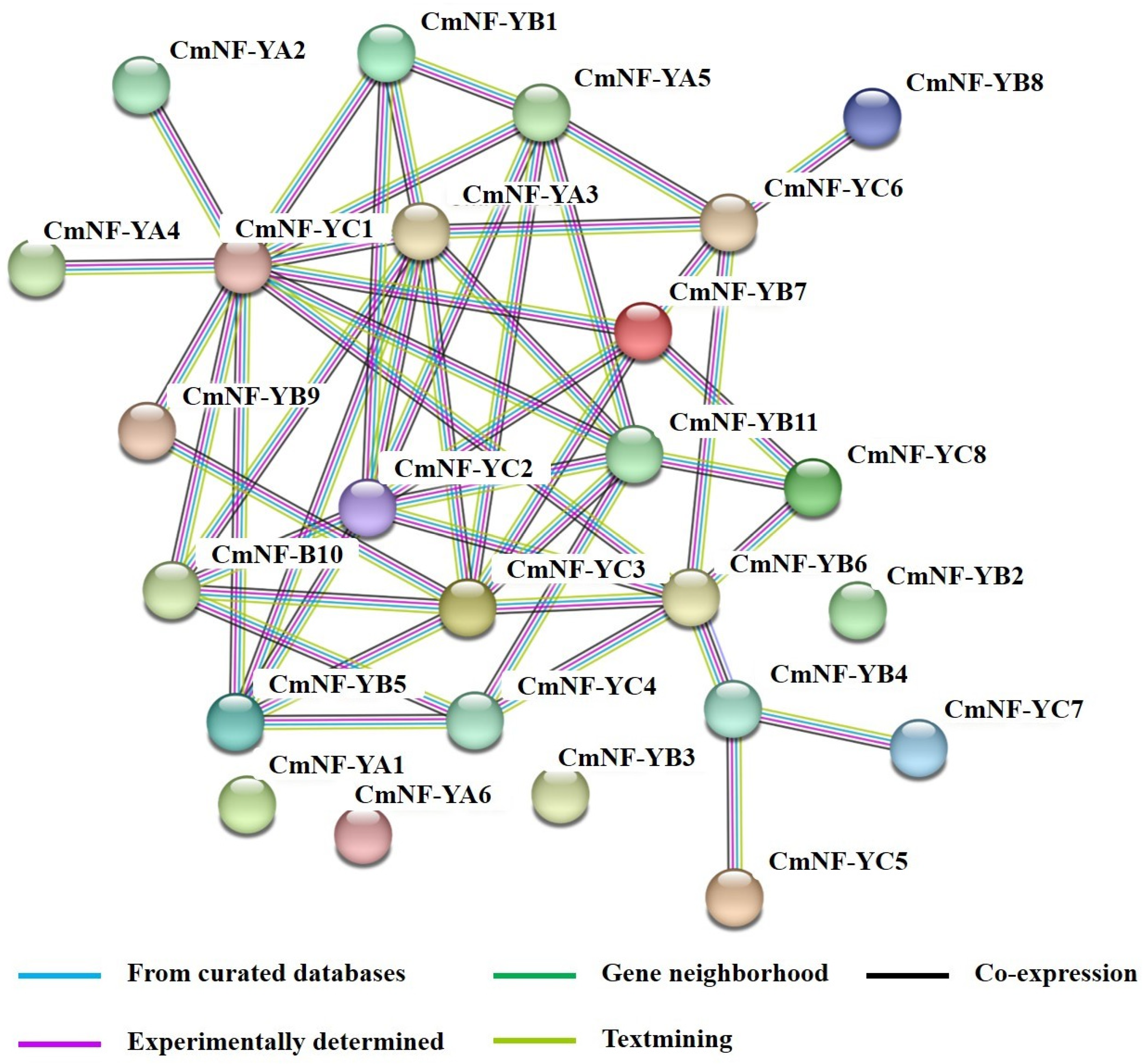
2.6. Protein–Protein Association Network Analysis of CmNF-Ys
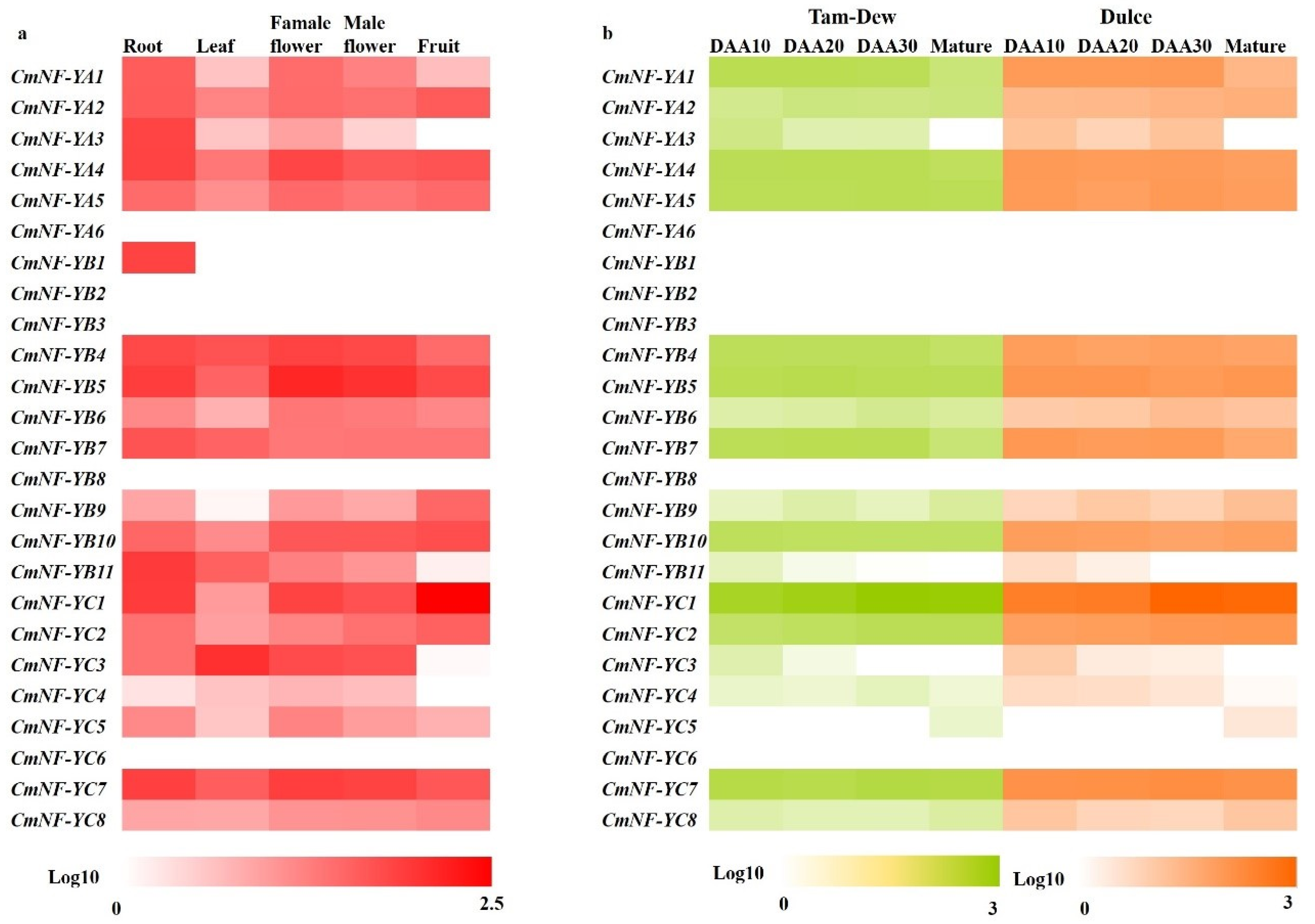
2.7. Expression Profiles of CmNF-Ys in Various Tissues and Fruits during Development
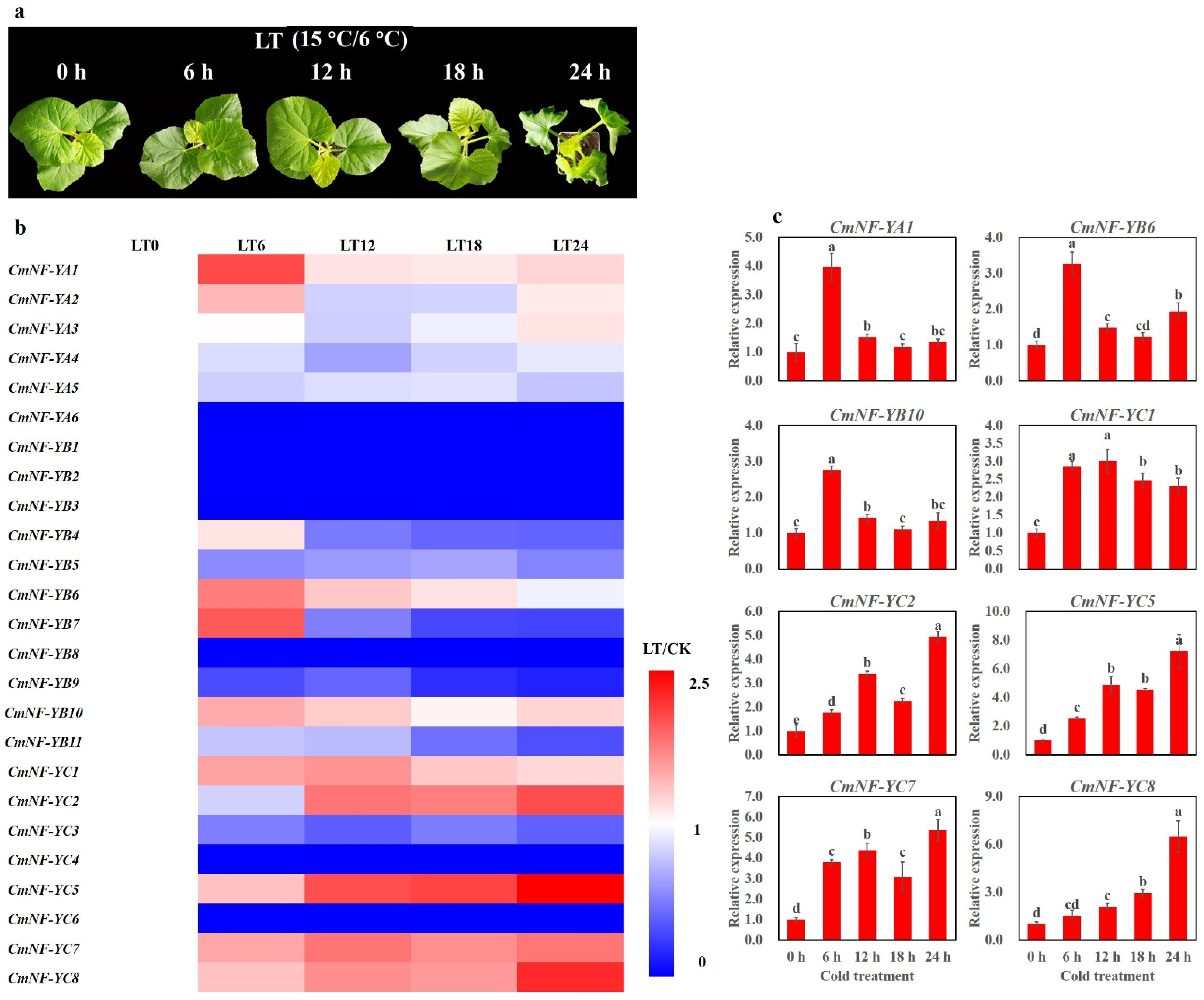
2.8. Expression Profiles of CmNF-Ys under Cold Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Cold Treatment
4.2. Identification of NF-Y Family Members in Melon
4.3. Analysis of Basic Characteristics of CmNF-Y Members
4.4. Multiple Sequence Alignment and Phylogenetic Analysis
4.5. Genetic Structure and Conserved Motif Analyses
4.6. Protein Secondary, Tertiary Structure, and Association Network Analysis
4.7. RNA Isolation and Analysis of CmNF-Ys Expression Patterns
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Myers, Z.A.; Holt, B.F. NUCLEAR FACTOR-Y: Still complex after all these years? Curr. Opin. Plant Biol. 2018, 45, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, D.; Wu, Y.; Voigt, A.; Adams, R.; Schramm, P.; Grimm, B. Studies on Differential Nuclear Translocation Mechanism and Assembly of the Three Subunits of the Arabidopsis thaliana Transcription Factor NF-Y. Mol. Plant 2012, 5, 876–888. [Google Scholar] [CrossRef] [PubMed]
- Gnesutta, N.; Kumimoto, R.W.; Swain, S.; Chiara, M.; Siriwardana, C.; Horner, D.S.; Holt, B.F.; Mantovani, R. CONSTANS Imparts DNA Sequence Specificity to the Histone Fold NF-YB/NF-YC Dimer. Plant Cell 2017, 29, 1516–1532. [Google Scholar] [CrossRef] [PubMed]
- Baudin, M.; Laloum, T.; Lepage, A.; Rípodas, C.; Ariel, F.; Frances, L.; Crespi, M.; Gamas, P.; Blanco, F.A.; Zanetti, M.E.; et al. A Phylogenetically Conserved Group of Nuclear Factor-Y Transcription Factors Interact to Control Nodulation in Legumes. Plant Physiol. 2015, 169, 2761–2773. [Google Scholar]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis thaliana Nuclear Factor Y Transcription Factors. Front. Plant Sci. 2017, 7, 2045. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, Y.; Lai, W.; Hu, L.; Jiang, L.; Liu, S. In Silico Identification and Expression Analysis of Nuclear Factor Y (Nf-Y) Transcription Factors in Cucumber. Agronomy 2020, 10, 236. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Zhu, J.; Ma, W.; Li, Z.; Bi, Z.; Sun, C.; Bai, J.; Zhang, J.; Liu, Y. Genome-Wide Identification and Analysis of the NF-Y Gene Family in Potato (Solanum tuberosum L.). Front. Genet. 2021, 12, 739989. [Google Scholar] [CrossRef]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef]
- Bai, A.N.; Lu, X.D.; Li, D.Q.; Liu, J.X.; Liu, C.M. NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 2016, 26, 384–388. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Wang, X.; Han, X.; An, Y.; Lin, S.; Shen, C.; Wen, J.; Liu, C.; Yin, W.; et al. Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef]
- Sorin, C.; Declerck, M.; Christ, A.; Blein, T.; Ma, L.; Lelandais-Briere, C.; Njo, M.F.; Beeckman, T.; Crespi, M.; Hartmann, C. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 2014, 202, 1197–1211. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Li, T.; Xu, P.B.; Li, L.; Du, S.S.; Lian, H.L.; Yang, H.Q. DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett. 2016, 590, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Ma, C.; Xu, Y.; Wang, T.; Chen, Y.; Lu, J.; Zhang, L.; Jiang, C.Z.; Hong, B.; Gao, J. Control of chrysanthemum flowering through integration with an aging pathway. Nat. Commun. 2017, 8, 829. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Cao, Y.; Ku, L.; Yao, W.; Cao, Y.; Ren, Z.; Dou, D.; Wang, H.; Ren, Z.; Liu, H.; et al. Dual functions of ZmNF-YA3 in photoperiod-dependent flowering and abiotic stress responses in maize. J. Exp. Bot. 2018, 69, 5177–5189. [Google Scholar] [CrossRef]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.Y.; Ahn, J.H. Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant 2019, 12, 489–505. [Google Scholar] [CrossRef]
- Li, S.; Li, K.; Ju, Z.; Cao, D.; Fu, D.; Zhu, H.; Zhu, B.; Luo, Y. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genom. 2016, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Wu, F.; Jiang, G.; Xiao, L.; Li, Z.; Duan, X.; Jiang, Y. Genome-wide identification, characterization and expression analysis of NF-Y gene family in relation to fruit ripening in banana. Postharvest Bio Tec. 2019, 151, 98–110. [Google Scholar] [CrossRef]
- Shi, H.; Ye, T.; Zhong, B.; Liu, X.; Jin, R.; Chan, Z. AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol. 2014, 203, 554–567. [Google Scholar] [CrossRef]
- Sato, H.; Suzuki, T.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NF-YB2 and NF-YB3 Have Functionally Diverged and Differentially Induce Drought and Heat Stress-Specific Genes. Plant Physiol. 2019, 180, 1677–1690. [Google Scholar] [CrossRef]
- Lee, D.K.; Kim, H.I.; Jang, G.; Chung, P.J.; Jeong, J.S.; Kim, Y.S.; Bang, S.W.; Jung, H.; Choi, Y.D.; Kim, J.K. The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci. 2015, 241, 199–210. [Google Scholar] [CrossRef]
- Wang, B.; Li, Z.; Ran, Q.; Li, P.; Peng, Z.; Zhang, J. ZmNF-YB16 Overexpression Improves Drought Resistance and Yield by Enhancing Photosynthesis and the Antioxidant Capacity of Maize Plants. Front. Plant Sci. 2018, 9, 709. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-J.; Fang, Y.; Fu, Y.-R.; Huang, J.-G.; Wu, C.-A.; Zheng, C.-C. NFYA1 Is Involved in Regulation of Postgermination Growth Arrest Under Salt Stress in Arabidopsis. PLoS ONE 2013, 8, e61289. [Google Scholar] [CrossRef] [PubMed]
- Selvam, M.P.; Somu, V.; Moin, M.; Reddy, M.; Yugandhar, P.; Balachandran, S.; Kirti, P. Activation-tagging in indica rice identifies a novel transcription factor subunit, NF-YC13 associated with salt tolerance. Sci. Rep. 2017, 7, 9341. [Google Scholar]
- Panahi, B.; Mohammadi, S.; Ruzicka, K.; Abbasi Holaso, H.; Mehrjerdi, M. Genome-wide identification and co-expression network analysis of nuclear factor-Y in barley revealed potential functions in salt stress. Physiol. Mol. Biol. Plants 2019, 25, 485–495. [Google Scholar] [CrossRef]
- Ren, C.; Zhang, Z.; Wang, Y.; Li, S.; Liang, Z. Genome-wide identification and characterization of the NF-Y gene family in grape (Vitis vinifera L.). BMC Genom. 2016, 17, 605. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zheng, Y.; Guo, Y.; Chen, X.; Sun, Y.; Yang, J.; Ye, N. Identification, expression, and putative target gene analysis of nuclear factor-Y (NF-Y) transcription factors in tea plant (Camellia sinensis). Planta 2019, 250, 1671–1686. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Wen, S.; Lan, C.; Yu, Y.; Chen, G. Genome-Wide Identification and Expression Profile Analysis of the NF-Y Transcription Factor Gene Family in Petunia hybrida. Plants 2020, 9, 336. [Google Scholar] [CrossRef]
- Liu, R.; Wu, M.; Liu, H.L.; Gao, Y.M.; Chen, J.; Yan, H.W.; Xiang, Y. Genome-wide identification and expression analysis of the NF-Y transcription factor family in Populus. Physiol. Plant 2021, 171, 309–327. [Google Scholar] [CrossRef]
- Li, M.; Duan, X.; Liu, T.; Qi, H. Short-term suboptimal low temperature has short- and long-term effects on melon seedlings. Sci. Hortic 2022, 297, 110967. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; González, V.; Henaff, E.; Camara, F.; Cozzuto, L.; Lowy, E.; Alioto, T.; et al. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef]
- Liu, P.; Wang, S.; Wang, X.; Yang, X.; Li, Q.; Wang, C.; Chen, C.; Shi, Q.; Ren, Z.; Wang, L. Genome-wide characterization of two-component system (TCS) genes in melon (Cucumis melo L.). Plant Physiol. Biochem. 2020, 151, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Gao, L.; Zhang, Y.; Zhang, F.; Yang, X.; Huang, D. Genome-wide investigation of the NAC transcription factor family in melon (Cucumis melo L.) and their expression analysis under salt stress. Plant Cell Rep. 2016, 35, 1827–1839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, Z.; Wang, F.; Zhong, C.; Liu, Y.; Li, Z.; Wang-Pruski, G.; Zhang, Z. Genome-wide Identification and Characteristics Analysis of Melon (Cucumis melo L.) MYB Transcription Factors and Their Responses to Autotoxicity and Saline-alkali Stress. Trop. Plant Biol. 2022, 15, 93–109. [Google Scholar] [CrossRef]
- Tian, Z.; Han, J.; Che, G.; Hasi, A. Genome-wide characterization and expression analysis of SAUR gene family in Melon (Cucumis melo L.). Planta 2022, 255, 123. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, K.; Zhang, J.; Wang, J.; Li, H.; Yang, X.; Shi, Q. Genome-wide characterization of MATE family members in Cucumis melo L. and their expression profiles in response to abiotic and biotic stress. Hortic. Plant J. 2022, 8, 474–488. [Google Scholar] [CrossRef]
- Dolfini, D.; Gatta, R.; Mantovani, R. NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. 2012, 47, 29–49. [Google Scholar] [CrossRef]
- Chayut, N.; Yuan, H.; Ohali, S.; Meir, A.; Yeselson, Y.; Portnoy, V.; Zheng, Y.; Fei, Z.; Lewinsohn, E.; Katzir, N.; et al. A bulk segregant transcriptome analysis reveals metabolic and cellular processes associated with Orange allelic variation and fruit β-carotene accumulation in melon fruit. BMC Plant Biol. 2015, 15, 274. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhu, J.; Yang, Y. Genome-Wide Identification and Expression Analysis of NF-Y Transcription Factor Families in Watermelon (Citrullus lanatus). J. Plant Growth Reg. 2017, 36, 590–607. [Google Scholar] [CrossRef]
- An, Y.; Suo, X.; Niu, Q.; Yin, S.; Chen, L. Genome-Wide Identification and Analysis of the NF-Y Transcription Factor Family Reveal Its Potential Roles in Salt Stress in Alfalfa (Medicago sativa L.). Int. J. Mol. Sci. 2022, 23, 6426. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Blanc, G.; Hokamp, K.; Wolfe, K. A Recent Polyploidy Superimposed on Older Large-Scale Duplications in the. Genome Res. 2003, 13, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Carriero, N.; Gerstein, M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet 2004, 20, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Fischer, R.; Goldberg, R.; Harada, J. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA 2003, 100, 2152–2156. [Google Scholar] [CrossRef]
- Kumimoto, R.; Zhang, Y.; Siefers, N.; Holt, B. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010, 63, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Kumimoto, R.; Adam, L.; Hymus, G.; Repetti, P.; Reuber, T.; Marion, C.; Hempel, F.; Ratcliffe, O. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar] [CrossRef]
- Wang, R.; Zhu, L.; Zhang, Y.; Fan, J.; Li, L. Genome-wide analysis of poplar NF-YB gene family and identified PtNF-YB1 important in regulate flowering timing in transgenic plants. BMC Plant Biol. 2019, 19, 251. [Google Scholar] [CrossRef]
- Su, H.; Chen, Z.; Dong, Y.; Ku, L.; Abou-Elwafa, S.F.; Ren, Z.; Cao, Y.; Dou, D.; Liu, Z.; Liu, H.; et al. Identification of ZmNF-YC2 and its regulatory network for maize flowering time. J. Exp. Bot. 2021, 72, 7792–7807. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, Z.; Tao, Q.; Chen, X.; Shui, C.; Ren, X.; Yu, L.; Liang, M. Brassica napus miR169 regulates BnaNF-YA in salinity, drought and ABA responses. Environ. Exp. Bot. 2022, 199, 104882. [Google Scholar] [CrossRef]
- He, X.; Liu, G.; Li, B.; Xie, Y.; Wei, Y.; Shang, S.; Tian, L.; Shi, H. Functional analysis of the heterotrimeric NF-Y transcription factor complex in cassava disease resistance. Ann. Bot. 2020, 124, 1185–1198. [Google Scholar] [CrossRef]
- Qi, M.; Zheng, W.; Zhao, X.; Hohenstein, J.D.; Kandel, Y.; O’Conner, S.; Wang, Y.; Du, C.; Nettleton, D.; MacIntosh, G.C.; et al. QQS orphan gene and its interactor NF-YC4 reduce susceptibility to pathogens and pests. Plant Biotechnol. J. 2019, 17, 252–263. [Google Scholar] [CrossRef]
- Alam, M.; Tanaka, T.; Nakamura, H.; Ichikawa, H.; Kobayashi, K.; Yaeno, T.; Yamaoka, N.; Shimomoto, K.; Takayama, K.; Nishina, H.; et al. Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol. J. 2014, 13, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Ding, Z.; Zhu, Z.; Zhang, H.; Gao, P.; Luan, F. Genetic Mapping and Nucleotide Diversity of Two Powdery Mildew Resistance Loci in Melon (Cucumis melo). Phytopathology 2020, 110, 1970–1979. [Google Scholar]
- Oumouloud, A.; El-Otmani, M.; Chikh-Rouhou, H.; Garcés-Claver, A.; Gonzalez Torres, R.; Perl-Treves, R.; Álvarez, J. Breeding melon for resistance to Fusarium wilt: Recent developments. Euphytica 2013, 15, 155–169. [Google Scholar] [CrossRef]
- Li, M.; Duan, X.; Wang, Q.; Chen, W.; Qi, H. A new morphological method to identify cold tolerance of melon at seedling stage. Funct. Plant Biol. 2020, 47, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, W.; Du, Q.; Xiao, H.; Li, J.; Wang, J.; Shang, F. Abscisic acid and hydrogen peroxide regulate proline homeostasis in melon seedlings under cold stress by forming a bidirectional closed loop. Environ. Exp. Bot. 2023, 205, 105102. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010, 2, 1090–1103. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.; Nastou, K.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020, 49, D605–D612. [Google Scholar] [CrossRef]
- Latrasse, D.; Rodriguez-Granados, N.Y.; Veluchamy, A.; Mariappan, K.G.; Bevilacqua, C.; Crapart, N.; Camps, C.; Sommard, V.; Raynaud, C.; Dogimont, C.; et al. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenet Chromatin 2017, 10, 22. [Google Scholar] [CrossRef] [PubMed]





| Gene | Gene ID | Gene Locus | Protein | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Size (aa) | MW (kDa) | pI | Instability Index (%) | Aliphatic Index | GRAVY | Subcellular Localization | |||
| CmNF-YA1 | MELO3C023554 | Chr01: 33657884 … 33661524 (−) | 318 | 35.64 | 7.90 | 42.85 | 66.86 | −0.784 | Nucleus |
| CmNF-YA2 | MELO3C015196 | Chr02: 6195415 .. 6200409 (−) | 322 | 35.43 | 7.15 | 57.36 | 54.84 | −0.792 | Nucleus |
| CmNF-YA3 | MELO3C009551 | Chr04: 31271731 .. 31275259 (+) | 325 | 35.70 | 9.40 | 57.01 | 71.14 | −0.473 | Nucleus |
| CmNF-YA4 | MELO3C014590 | Chr05: 1114809 .. 1119924 (+) | 341 | 37.07 | 6.53 | 54.09 | 49.62 | −0.926 | Nucleus |
| CmNF-YA5 | MELO3C007077 | Chr08: 594883 .. 611363 (+) | 169 | 18.95 | 9.32 | 66.59 | 57.16 | −0.193 | Nucleus |
| CmNF-YA6 | MELO3C023161 | Chr08: 16875004 .. 16880763 (−) | 206 | 23.47 | 8.94 | 68.50 | 55.44 | −0.94 | Nucleus |
| CmNF-YB1 | MELO3C016042 | Chr01: 32765510 .. 32766166 (−) | 161 | 17.49 | 5.16 | 41.97 | 55.09 | −0.816 | Nucleus |
| CmNF-YB2 | MELO3C015318 | Chr02: 737865 .. 738293 (+) | 142 | 16.50 | 5.17 | 39.28 | 65.99 | −0.962 | Nucleus |
| CmNF-YB3 | MELO3C015320 | Chr02: 743726 .. 744109 (+) | 127 | 14.69 | 6.15 | 25.58 | 70.79 | −0.754 | Nucleus |
| CmNF-YB4 | MELO3C017276 | Chr02: 24713746 .. 24717503 (−) | 210 | 23.58 | 5.05 | 65.10 | 74.81 | −0.391 | Nucleus |
| CmNF-YB5 | MELO3C009309 | Chr04: 32949453 .. 32953167 (−) | 117 | 12.97 | 5.86 | 51.56 | 66.84 | −0.669 | Nucleus |
| CmNF-YB6 | MELO3C014503 | Chr05: 1870042 .. 1872775 (+) | 155 | 17.48 | 5.01 | 48.06 | 79.29 | −0.842 | Nucleus |
| CmNF-YB7 | MELO3C014103 | Chr06: 36946833 .. 36947294 (+) | 153 | 16.42 | 5.23 | 41.61 | 60.07 | −0.614 | Nucleus |
| CmNF-YB8 | MELO3C014120 | Chr06: 37108875 .. 37109569 (+) | 210 | 22.86 | 5.58 | 45.97 | 66.90 | −0.535 | Nucleus |
| CmNF-YB9 | MELO3C017568 | Chr07: 25210242 .. 25211051 (−) | 225 | 24.65 | 7.76 | 50.93 | 64.62 | −0.633 | Nucleus |
| CmNF-YB10 | MELO3C011726 | Chr10: 5430171 .. 5434588 (−) | 174 | 19.00 | 5.11 | 44.08 | 64.02 | −0.638 | Nucleus |
| CmNF-YB11 | MELO3C025951 | Chr11: 14106203 .. 14107884 (+) | 200 | 20.56 | 6.31 | 31.98 | 48.35 | −0.648 | Nucleus |
| CmNF-YC1 | MELO3C018689 | Chr01: 2153793 .. 2156247 (−) | 266 | 29.96 | 5.96 | 71.76 | 63.53 | −0.705 | Nucleus |
| CmNF-YC2 | MELO3C015332 | Chr02: 804011 .. 809860 (−) | 258 | 28.61 | 5.89 | 59.94 | 70.35 | −0.517 | Nucleus/Cytoplasm |
| CmNF-YC3 | MELO3C026204 | Chr02: 26758439 .. 26761732 (−) | 228 | 24.99 | 5.07 | 65.10 | 71.58 | −0.433 | Nucleus/Cytoplasm |
| CmNF-YC4 | MELO3C011458 | Chr03: 25878114 .. 25879023 (+) | 140 | 15.71 | 9.04 | 51.67 | 81.57 | −0.57 | Nucleus |
| CmNF-YC5 | MELO3C014530 | Chr05: 1604408 .. 1608430 (−) | 279 | 31.13 | 4.93 | 42.67 | 71.29 | −0.846 | Nucleus |
| CmNF-YC6 | MELO3C016347 | Chr07: 23806907 .. 23807260 (+) | 117 | 12.89 | 7.76 | 51.19 | 87.61 | 0.035 | Nucleus |
| CmNF-YC7 | MELO3C011797 | Chr10: 4748948 .. 4753271 (−) | 282 | 31.56 | 5.20 | 41.42 | 63.30 | −0.931 | Nucleus |
| CmNF-YC8 | MELO3C025765 | Chr11: 28166692 .. 28169738 (−) | 272 | 30.78 | 9.52 | 48.03 | 56.03 | −1.129 | Nucleus |
| Proteins | α-Helix % | Extended Strand % | β-Turn % | Random Coil % |
|---|---|---|---|---|
| CmNF-YA1 | 18.55 | 7.55 | 4.72 | 69.18 |
| CmNF-YA2 | 21.74 | 9.94 | 3.42 | 64.90 |
| CmNF-YA3 | 14.46 | 16.92 | 4.62 | 64.00 |
| CmNF-YA4 | 19.35 | 7.04 | 2.64 | 70.97 |
| CmNF-YA5 | 33.14 | 10.65 | 6.51 | 49.70 |
| CmNF-YA6 | 20.39 | 16.99 | 2.91 | 59.71 |
| CmNF-YB1 | 46.58 | 6.21 | 3.73 | 43.48 |
| CmNF-YB2 | 59.15 | 0.70 | 3.53 | 36.62 |
| CmNF-YB3 | 56.69 | 6.30 | 3.94 | 33.07 |
| CmNF-YB4 | 59.04 | 6.67 | 6.67 | 27.62 |
| CmNF-YB5 | 58.12 | 0.85 | 5.13 | 35.90 |
| CmNF-YB6 | 65.16 | 6.45 | 3.23 | 25.16 |
| CmNF-YB7 | 47.06 | 5.23 | 9.80 | 37.91 |
| CmNF-YB8 | 40.00 | 11.90 | 4.29 | 43.81 |
| CmNF-YB9 | 39.56 | 3.11 | 3.56 | 53.77 |
| CmNF-YB10 | 51.15 | 1.72 | 4.60 | 42.53 |
| CmNF-YB11 | 40.00 | 8.50 | 6.50 | 45.00 |
| CmNF-YC1 | 47.06 | 5.23 | 9.80 | 37.91 |
| CmNF-YC2 | 47.67 | 5.02 | 2.15 | 45.16 |
| CmNF-YC3 | 40.79 | 11.84 | 5.26 | 42.11 |
| CmNF-YC4 | 36.43 | 8.57 | 3.57 | 51.43 |
| CmNF-YC5 | 46.32 | 8.46 | 7.72 | 37.50 |
| CmNF-YC6 | 47.01 | 14.53 | 5.98 | 32.48 |
| CmNF-YC7 | 45.75 | 1.77 | 1.77 | 50.71 |
| CmNF-YC8 | 33.08 | 8.27 | 4.51 | 54.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Du, Q.; Li, J.; Wang, H.; Xiao, H.; Wang, J. Genome-Wide Identification and Chilling Stress Analysis of the NF-Y Gene Family in Melon. Int. J. Mol. Sci. 2023, 24, 6934. https://doi.org/10.3390/ijms24086934
Li M, Du Q, Li J, Wang H, Xiao H, Wang J. Genome-Wide Identification and Chilling Stress Analysis of the NF-Y Gene Family in Melon. International Journal of Molecular Sciences. 2023; 24(8):6934. https://doi.org/10.3390/ijms24086934
Chicago/Turabian StyleLi, Meng, Qingjie Du, Juanqi Li, Hu Wang, Huaijuan Xiao, and Jiqing Wang. 2023. "Genome-Wide Identification and Chilling Stress Analysis of the NF-Y Gene Family in Melon" International Journal of Molecular Sciences 24, no. 8: 6934. https://doi.org/10.3390/ijms24086934
APA StyleLi, M., Du, Q., Li, J., Wang, H., Xiao, H., & Wang, J. (2023). Genome-Wide Identification and Chilling Stress Analysis of the NF-Y Gene Family in Melon. International Journal of Molecular Sciences, 24(8), 6934. https://doi.org/10.3390/ijms24086934





