Biodiversity of rolB/C-like Natural Transgene in the Genus Vaccinium L. and Its Application for Phylogenetic Studies
Abstract
:1. Introduction
2. Results
2.1. Vaccinium rolB/C-like Gene: General Description
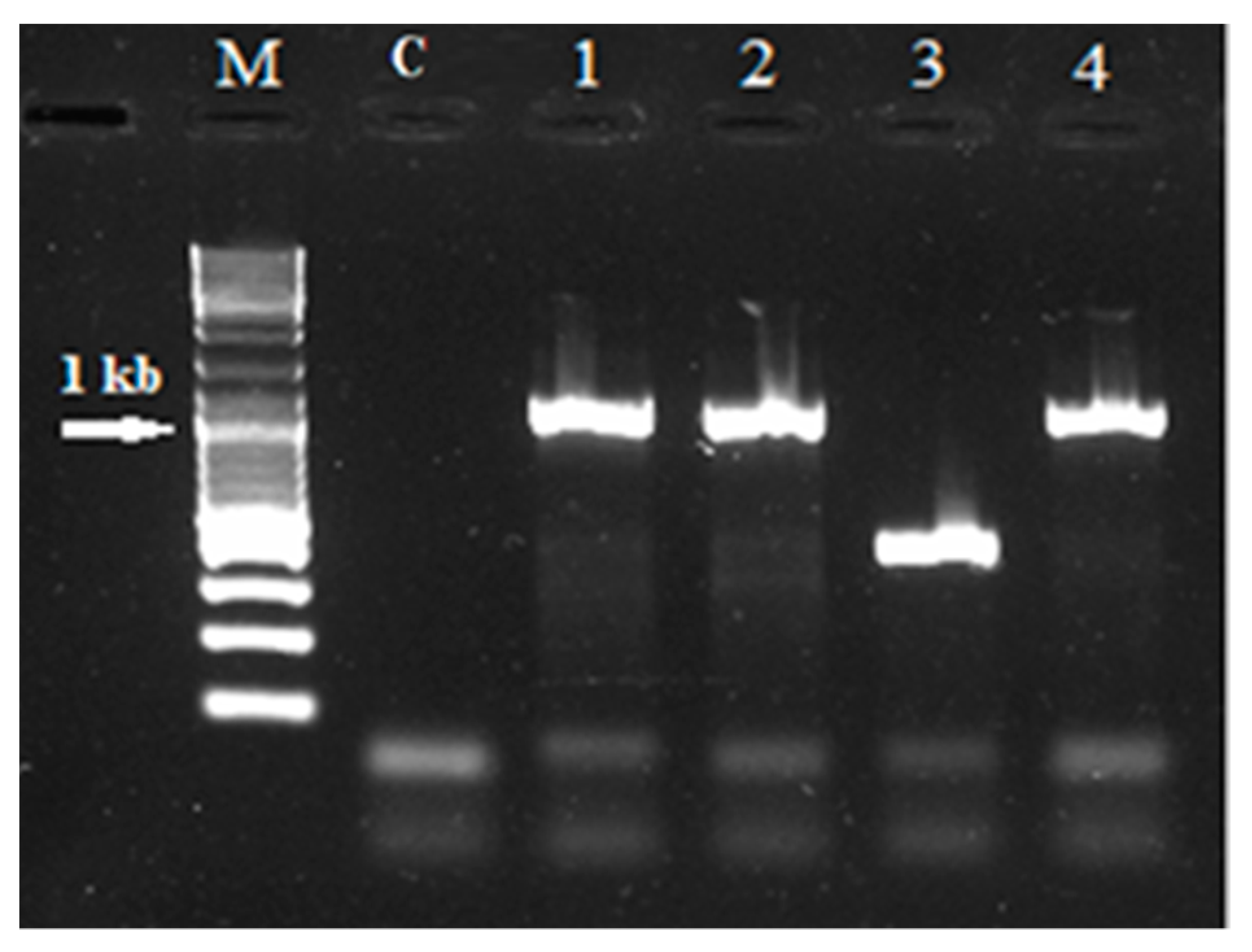
2.2. Intraspecific Variability of rolB/C-like Gene
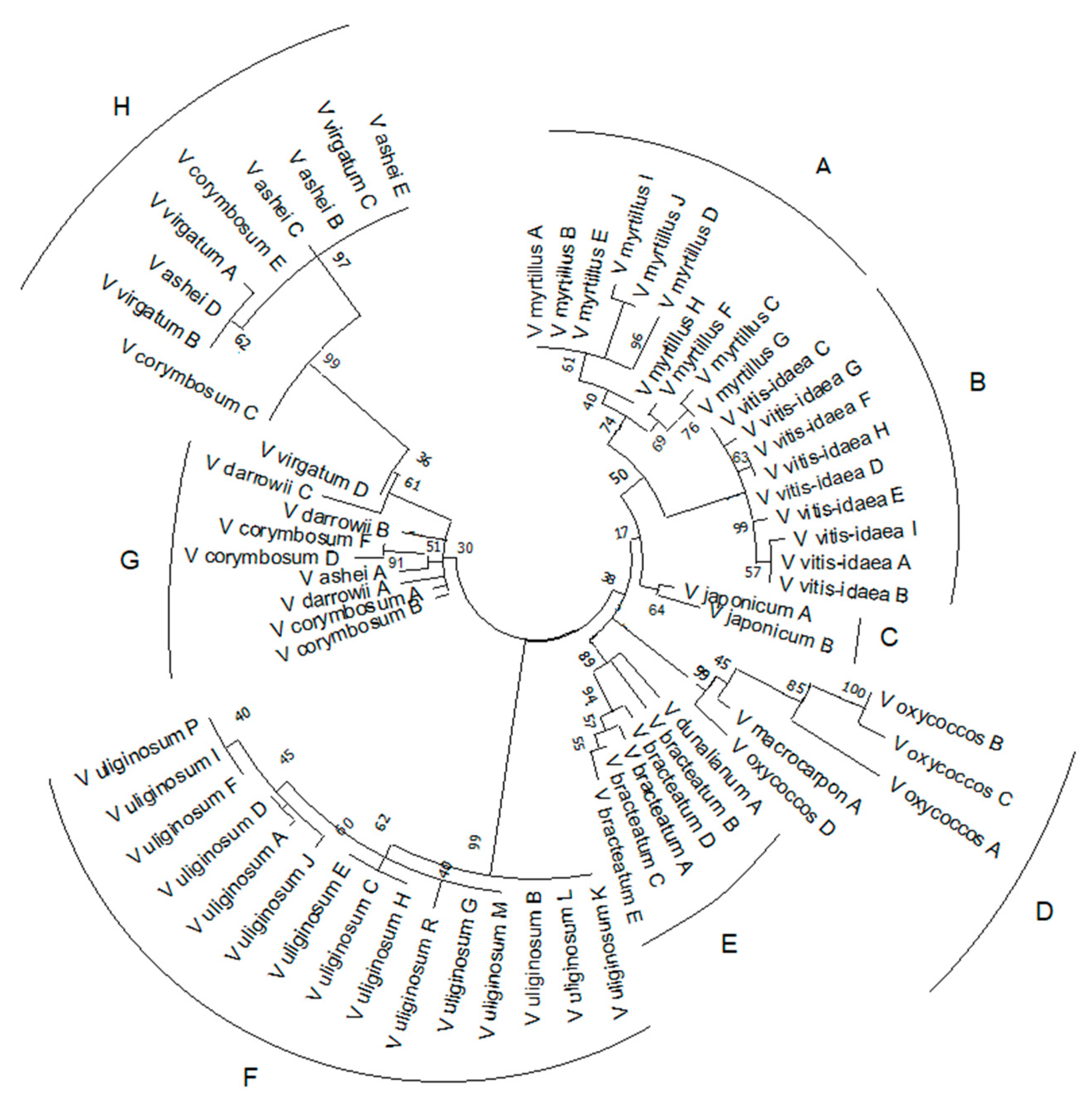
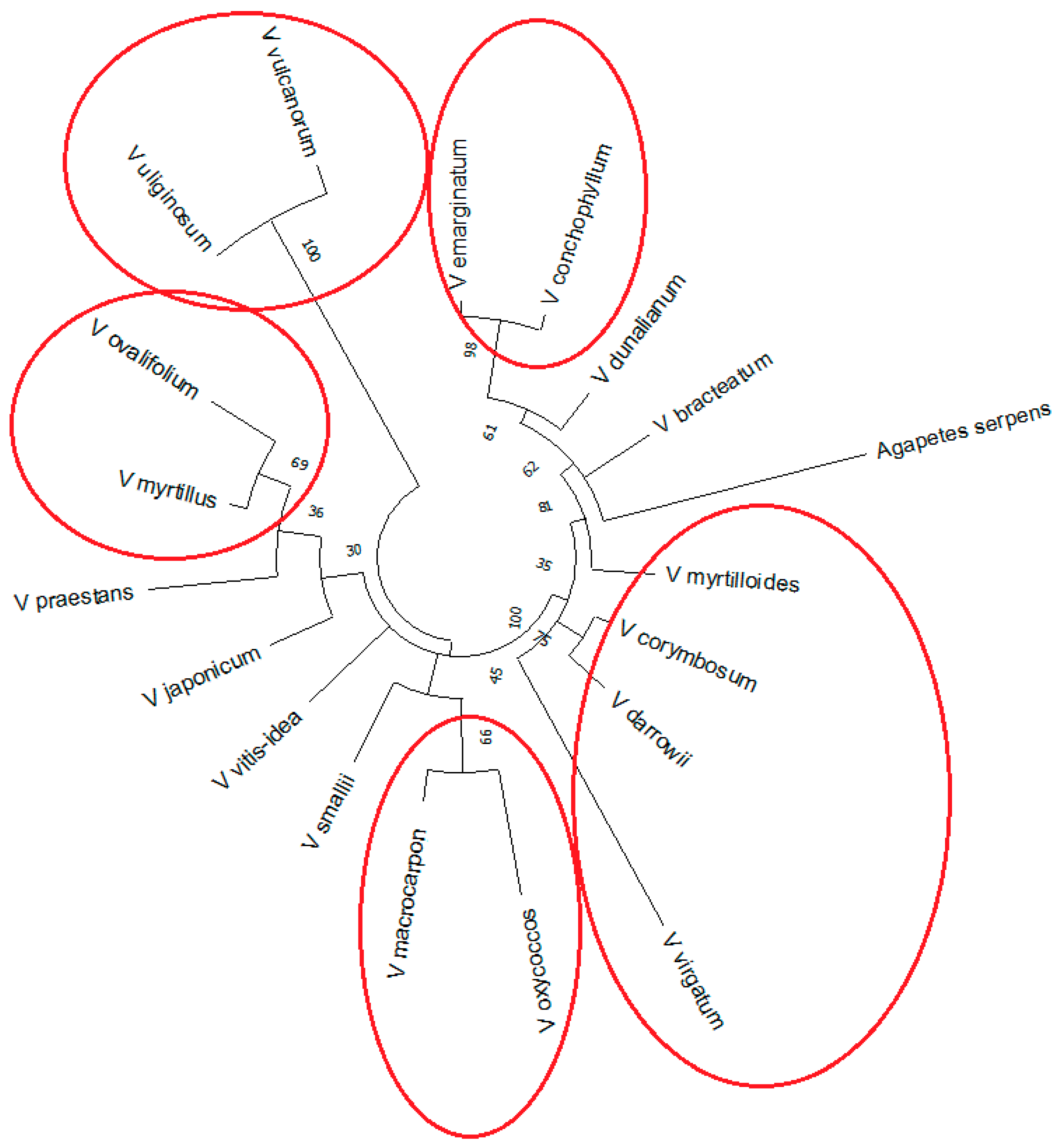
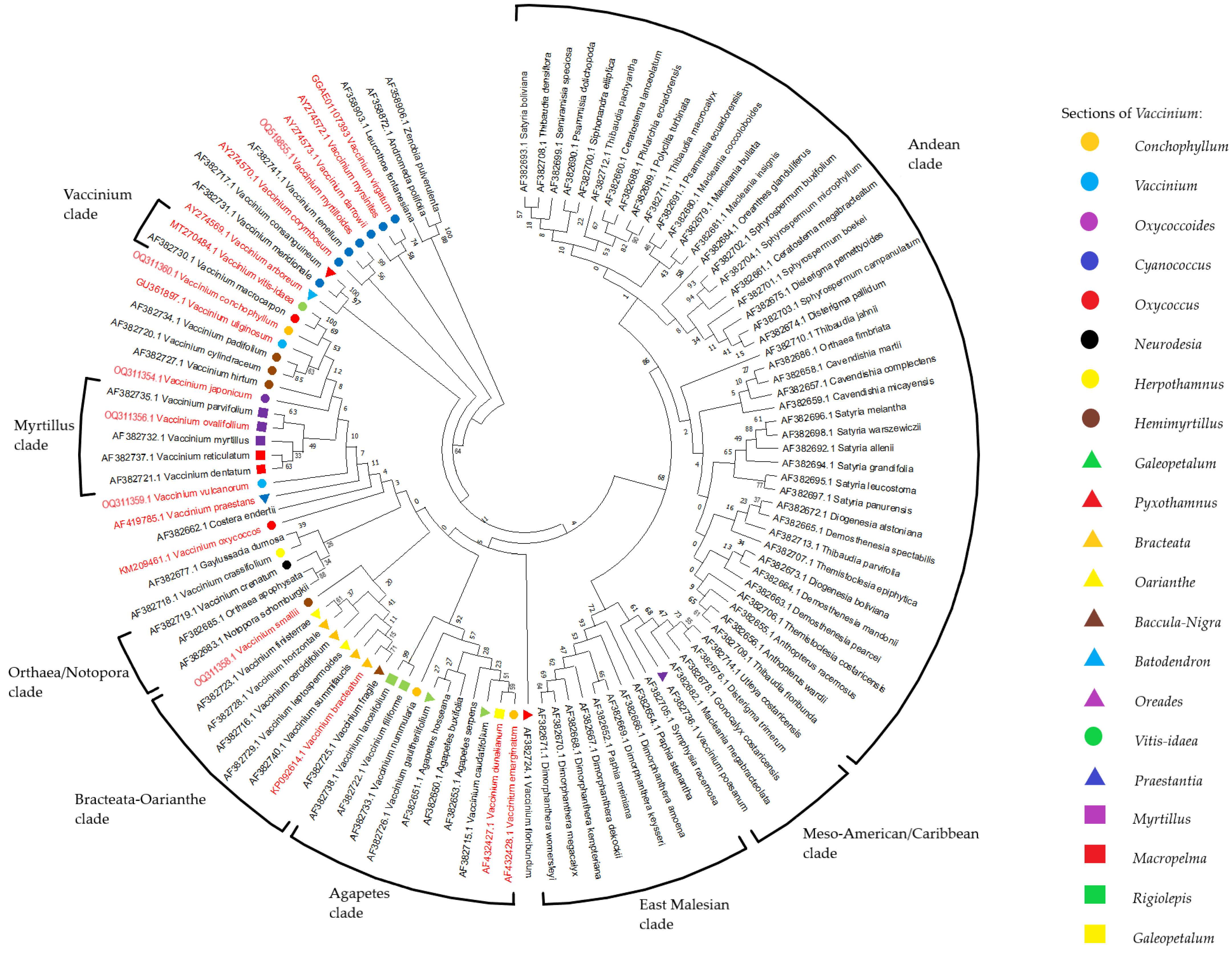
2.3. Reconstruction of Phylogenetic Relationship of Vaccinium Species Based on rolB/C-like Sequence
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. SRA Data
4.3. DNA Isolation
4.4. PCR
4.5. Molecular Cloning
4.6. DNA Sequencing
4.7. Allele Reconstruction from Sanger Sequencing Data
4.8. Allele Phasing
4.9. Phylogenetic Analysis
4.10. Prediction of Protein’s Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chilton, M.-D.; Drummond, M.H.; Merlo, D.J.; Sciaky, D.; Montoya, A.L.; Gordon, M.P.; Nester, E.W. Stable incorporation of plasmid DNA into higher plant cells: The molecular basis of crown gall tumorigenesis. Cell 1977, 11, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Oger, P.M.; Schrammeijer, B.; Hooykaas, P.J.J.; Farrand, S.K.; Winans, S.C. The bases of crown gall tumorigenesis. J. Bacteriol. 2000, 182, 3885–3895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nester, E.W. Agrobacterium: Nature’s genetic engineer. Front. Plant Sci. 2015, 5, 730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelvin, S.B. Integration of Agrobacterium T-DNA into the plant genome. Annu. Rev. Genet. 2017, 51, 195–217. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, B.; Citovsky, V. Pathways of DNA transfer to plants from Agrobacterium tumefaciens and related bacterial species. Ann. Rev. Phytopath. 2019, 57, 231–251. [Google Scholar] [CrossRef]
- Matveeva, T.V.; Otten, L. Widespread occurrence of natural transformation of plants by Agrobacterium. Plant Mol. Biol. 2019, 101, 415–437. [Google Scholar] [CrossRef]
- Tepfer, D. Genetic transformation using Agrobacterium rhizogenes. Physiol. Plant 1990, 79, 140–146. [Google Scholar] [CrossRef]
- Christey, M.C. Use of Ri-mediated transformation for production of transgenic plants. In Vitro Cell. Dev. Biol.-Plant 2001, 37, 687–700. [Google Scholar] [CrossRef]
- Lütken, H.; Clarke, J.L.; Müller, R. Genetic engineering and sustainable production of ornamentals: Current status and future directions. Plant Cell Rep. 2012, 31, 1141–1157. [Google Scholar] [CrossRef]
- White, F.F.; Garfinkel, D.J.; Huffman, G.A.; Gordon, M.P.; Nester, E.W. Sequence homologous to Agrobacterium rhizogenes T-DNA in the genomes of uninfected plants. Nature 1983, 301, 348–350. [Google Scholar] [CrossRef]
- Furner, I.J.; Huffman, G.A.; Amasino, R.M.; Garfinkel, D.J.; Gordon, M.P.; Nester, E.W. An Agrobacterium transformation in the evolution of the genus. Nicotiana. Nat. 1986, 319, 422–427. [Google Scholar] [CrossRef]
- Suzuki, K.; Yamashita, I.; Tanaka, N. Tobacco plants were transformed by Agrobacterium rhizogenes infection during their evolution. Plant J. 2002, 32, 775–787. [Google Scholar] [CrossRef]
- Matveeva, T.V.; Bogomaz, D.I.; Pavlova, O.A.; Nester, E.W.; Lutova, L.A. Horizontal gene transfer from genus Agrobacterium to the plant Linaria in nature. Mol. Plant Microbe Interact. 2012, 25, 1542–1551. [Google Scholar] [CrossRef] [Green Version]
- Kyndt, T.; Quispe, D.; Zhai, H.; Jarret, R.; Ghislain, M.; Liu, Q.; Gheysen, G.; Kreuze, J.F. The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: An example of a naturally transgenic food crop. Proc. Natl. Acad. Sci. USA 2015, 112, 5844–5849. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, D.; Wang, Y.; Dong, H.; Yuan, Y.; Yang, W.; Lai, D.; Zhang, M.; Jiang, L.; Li, Z. Parasitic plant dodder (Cuscuta spp.): A new natural Agrobacterium-to-plant horizontal gene transfer species. Sci. China Life Sci. 2020, 63, 312–316. [Google Scholar] [CrossRef]
- Matveeva, T.V. New naturally transgenic plants: 2020 up-date. Biol. Commun. 2021, 66, 36–46. [Google Scholar] [CrossRef]
- Bogomaz, F.D.; Matveeva, T.V. Expression sequences of opine synthase genes in natural GMOs based on analysis of their transcriptomes. Plant Biotechnol. Breed. 2022, 5, 15–24. [Google Scholar] [CrossRef]
- Lipatov, P.Y.; Bogomaz, F.D.; Gosudarev, K.D.; Kondrashova, S.A.; Kuchevsky, M.V.; Malyuga, N.L.; Myagkiy, E.V.; Sergeenkova, M.V.; Tverdokhlebova, V.R.; Shtina, A.D.; et al. New cellular T-DNAs in naturally transgenic plants. Ecol. Genet. 2022, 20, 40–41. [Google Scholar] [CrossRef]
- Matveeva, T.V. Why do plants need agrobacterial genes? Ecol. Genet. 2021, 19, 365–375. [Google Scholar] [CrossRef]
- Matveeva, T.V. Agrobacterium-mediated transformation in the evolution of plants. Curr. Top. Microbiol. Immun. 2018, 418, 421–441. [Google Scholar] [CrossRef]
- Quispe-Huamanquispe, D.G.; Gheysen, G.; Yang, J.; Jarret, R.; Rossel, G.; Kreuze, J.F. The horizontal gene transfer of Agrobacterium T-DNAs into the series batatas (Genus Ipomoea) genome is not confined to hexaploid sweetpotato. Sci. Rep. 2019, 9, 12584. [Google Scholar] [CrossRef] [Green Version]
- Khafizova, G.V.; Matveeva, T.V. Agrobacterium-mediated transformation of Nicotiana glauca and Nicotiana sylvestris. Vavilovskii Zhurnal Genetiki I Selektsii 2022, 26, 697–703. [Google Scholar] [CrossRef]
- Chen, K.; Zhurbenko, P.; Danilov, L.; Matveeva, T.; Otten, L. Conservation of an Agrobacterium cT-DNA insert in Camellia section Thea reveals the ancient origin of tea plants from a genetically modified ancestor. Front. Plant Sci. 2022, 13, 997762. [Google Scholar] [CrossRef]
- Zhurbenko, P.M.; Klimenko, F.N. PhaseAll: A simple tool for read-based allele phasing. Ecol. Genet. 2022, 20, 32. [Google Scholar] [CrossRef]
- Index Nominum Genericorum. Available online: https://naturalhistory2.si.edu/botany/ing/ (accessed on 30 December 2022).
- Vorsa, N.; Zalapa, J. Domestication, Genetics, and Genomics of the American Cranberry. Plant Breed. Rev. 2019, 40, 279–315. [Google Scholar] [CrossRef]
- Camp, W.H. On the Structure of Populations in the Genus Vaccinium. Brittonia 1942, 4, 189–204. [Google Scholar] [CrossRef]
- Camp, W. The North American blueberries with notes on other groups of Vacciniaceae. Brittonia 1945, 5, 203–275. [Google Scholar] [CrossRef] [Green Version]
- Hancock, J.; Lyrene, P.; Finn, C.; Vorsa, N.; Lobos, G. Blueberries and Cranberries. Temp. Fruit Crop Breed. 2008, 115–150. [Google Scholar] [CrossRef]
- Vander Kloet, S.P. The Genus Vaccinium in North America; Agriculture Canada: Ottawa, ON, Canada, 1988. [Google Scholar]
- Kron, K.; Powell, E.; Luteyn, J. Phylogenetic relationships within the blueberry tribe (Vaccinieae, Ericaceae) based on sequence data from matK and nuclear ribosomal ITS regions, with comments on the placement of Satyria. Am. J. Bot. 2002, 89, 327–336. [Google Scholar] [CrossRef]
- Powell, E.; Kron, K. Molecular Systematics of the Northern Andean Blueberries (Vaccinieae, Vaccinioideae, Ericaceae). Int. J. Plant Sci. 2003, 164, 987–995. [Google Scholar] [CrossRef]
- Soltis, D.; Mavrodiev, E.; Doyle, J.; Rauscher, J.; Soltis, P. ITS and ETS Sequence Data and Phylogeny Reconstruction in Allopolyploids and Hybrids. Syst. Bot. 2008, 33, 7–20. [Google Scholar] [CrossRef]
- Diaz-Garcia, L.; Garcia-Ortega, L.; González-Rodríguez, M.; Delaye, L.; Iorizzo, M.; Zalapa, J. Chromosome-Level Genome Assembly of the American Cranberry (Vaccinium macrocarpon Ait.) and Its Wild Relative Vaccinium microcarpum. Front. Plant Sci. 2021, 12, 137. [Google Scholar] [CrossRef] [PubMed]
- Kawash, J.; Colt, K.; Hartwick, N.T.; Abramson, B.W.; Vorsa, N.; Polashock, J.J.; Michael, T.P. Contrasting a reference cranberry genome to a crop wild relative provides insights into adaptation, domestication, and breeding. PLoS ONE 2022, 17, e0264966. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, S.; Fujikawa, M.; Yamane, H.; Shirasawa, K.; Babiker, E.; Tao, R. Genomic insight into the developmental history of southern highbush blueberry populations. Heredity 2020, 126, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J. Blueberry Plant Denominated ‘Draper’. U.S. Patent No. USPP15103P3, 23 January 2003. [Google Scholar]
- Otten, L. The Agrobacterium Phenotypic Plasticity (Plast) Genes. Agrobacterium Biol. 2018, 418, 375–419. [Google Scholar] [CrossRef]
- Levesque, H.; Delepelaire, P.; Rouzé, P.; Slightom, J.; Tepfer, D. Common evolutionary origin of the central portions of the Ri TL-DNA of Agrobacterium rhizogenes and the Ti T-DNAs of Agrobacterium tumefaciens. Plant Mol. Biol. 1988, 11, 731–744. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Amosova, A.V.; Belyakov, E.A.; Zhurbenko, P.M.; Mikhailova, Y.V.; Punina, E.O.; Shneyer, V.S.; Loskutov, I.G. Genetic Con-sequences of Interspecific Hybridization, Its Role in Speciation and Phenotypic Diversity of Plants. Russ. J. Genet. 2019, 55, 278–294. [Google Scholar] [CrossRef]
- Zhidkin, R.R.; Matveeva, T.V. Phylogeny problems of the genus Vaccinium L. and ways to solve them. Ecol. Genet. 2022, 20, 151–164. [Google Scholar] [CrossRef]
- Schlautman, B.; Covarrubias-Pazaran, G.; Fajardo, D.; Steffan, S.; Zalapa, J. Dis-criminating power of microsatellites in cranberry organelles for taxonomic studies in Vaccinium and Ericaceae. Genet. Resour. Crop Evol. 2016, 64, 451–466. [Google Scholar] [CrossRef]
- Draper, J.; Scott, R.; Armitage, P.; Walden, R. Plant Genetic Transformation and Gene Expression—A Laboratory Manual; Blackwell Science: Oxford, UK, 1988. [Google Scholar]
- Inoue, H.; Nojima, H.; Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef]
- Zhidkin, R.R.; Zhurbenko, P.M.; Gorodilova, E.Y.; Matveeva, T.V. Molecular genetic and bioinformatic approaches for the allele reconstruction of the rolB/C-like gene in representatives of the genus Vaccinium L. Ecol. Genet. 2022, 22, 36–37. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, 10–14. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Van der Auwera, G.A.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra, 1st ed.; O’Reilly Media: Sebastopol, CA, USA, 2020. [Google Scholar]
- Martin, M.; Patterson, M.; Garg, S.; OFischer, S.; Pisanti, N.; WKlau, G.; Schöenhuth, A.; Marschall, T. WhatsHap: Fast and accurate read-based phasing. bioRxiv 2016. [Google Scholar] [CrossRef] [Green Version]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]


| Section | Species | NGS Data | SRR Accession Number(s) |
|---|---|---|---|
| Oxycoccoides (Hooker f.) Sleumer | V. japonicum Miq. | rolB/C-like gene is present in the genome, and coverage is sufficient to assemble a full-length gene | SRR13349629 |
| Oxycoccus (Hill) Koch | V. macrocarpon Aiton | rolB/C-like gene described earlier, in new SRA data coverage is sufficient to assemble a full-length gene | SRR13376387 SRR13376383 SRR13376388 |
| V. oxycoccos L. | rolB/C-like gene is present in the genome, and coverage is sufficient to assemble a full-length gene | SRR14876344 | |
| Cyanococcus A. Gray | V. virgatum Aiton/V. ashei Reade | rolB/C-like gene is present in the genome, and coverage is sufficient to assemble a full-length gene | SRR12686860 SRR12686864 |
| V. darrowii Camp | rolB/C-like gene is present in the genome, and coverage is sufficient to assemble a full-length gene | SRR13865261 SRR15673719 | |
| V. corymbosum L. | rolB/C-like gene was described earlier, in new SRA data coverage is sufficient to assemble a full-length gene | SRR8298002 SRR8298006 | |
| V. stenophyllum Steud. | rolB/C-like gene is present in the genome, but coverage is not sufficient to assemble a full-length gene | SRR19401505 | |
| V. tenellum Aiton | rolB/C-like gene is present in the genome, but coverage is not sufficient to assemble a full-length gene | SRR19908712 | |
| Hemimyrtillus Sleumer | V. smallii A. Gray | rolB/C-like gene is present in the genome, but coverage is not sufficient to assemble a full-length gene | SRR19400793 |
| Myrtillus Dumortier | V. myrtillus L. | rolB/C-like gene is present in the genome, and coverage is sufficient to assemble a full-length gene | SRR7523927 SRR7523930 SRR7523926 |
| V. scoparium Leiberg ex Coville | rolB/C-like gene is present in the genome, but coverage is not sufficient to assemble a full-length gene | SRR19405468 | |
| V. yatabe Makino | rolB/C-like gene is present in the genome, but coverage is not sufficient to assemble a full-length gene | SRR19400792 | |
| Polycodium (Rafinesque) Rehder | V. stamineum L. | rolB/C-like gene is present in the genome, but coverage is not sufficient to assemble a full-length gene | SRR19401493 |
| Pseudocephalanthos C.Y.Wu and R.C.Fang. | V. dunalianum Wight | rolB/C-like gene is present in the genome, and coverage is sufficient to assemble a full-length gene | SRR7768528 SRR2057022 |
| Bracteata J.J. Smith | V. bracteatum Thunb. | rolB/C-like gene is present in the genome, and coverage is sufficient to assemble a full-length gene | SRR17459400 SRR17459403 SRR17459408 |
| V. wrightii A.Gray | rolB/C-like gene is present in the genome, but coverage is not sufficient to assemble a full-length gene | SRR19401672 | |
| Vaccinium L. | V. uliginosum L. | rolB/C-like gene is present in the genome, and coverage is sufficient to assemble a full-length gene | SRR7686610 SRR7686609 |
| Vitis-idaea (Moench) Koch | V. vitis-idaea L. | rolB/C-like gene is present in the genome, and coverage is sufficient to assemble a full-length gene | SRR5799277 SRR5799278 SRR5799279 |
| Relation to any section is not defined | V. talamancense (Wilbur and Luteyn) Luteyn | rolB/C-like gene is present in the genome, but coverage is not sufficient to assemble a full-length gene | SRR19401435 |
| V. schoddei Sleumer | rolB/C-like gene is present in the genome, but coverage is not sufficient to assemble a full-length gene | SRR19401400 | |
| V. stanleyi Schweinf | rolB/C-like gene is present in the genome, and coverage is sufficient to assemble a full-length gene | SRR19354672 | |
| V. wollastonii Wernham | rolB/C-like gene is present in the genome, but coverage is not sufficient to assemble a full-length gene | SRR19401324 |
| Species | Number of Genotypes | Number of Unique Alleles | Number of Homozygotes | Number of Heterozygotes | Ploidy of Studied Species | Max. Alleles per Plant |
|---|---|---|---|---|---|---|
| V. myrtillus | 13 | 10 | 10 | 3 | Diploids | 2 |
| V. corymbosum | 2 | 6 | 0 | 2 | Tetraploids | 4 |
| V. macrocarpon | 3 | 1 | 3 | 0 | Diploids | 1 |
| V. oxycoccos | 4 | 4 | 3 | 1 | Diploids, tetraploids and hexaploids | 2 |
| V. virgatum/ashei | 5 | 9 | 3 | 2 | Hexaploids | 3 |
| V. japonicum | 2 | 2 | 2 | 0 | Diploids | 1 |
| V. vitis-idaea | 18 | 9 | 5 | 13 | Diploids | 2 |
| V. uliginosum | 14 | 18 | 0 | 14 | Diploids, tetraploids and hexaploids | 3 |
| V. dunalianum | 2 | 1 | 2 | 0 | Diploids | 1 |
| V. bracteatum | 3 | 5 | 1 | 2 | Diploids | 2 |
| V. darrowii | 2 | 3 | 0 | 2 | Diploids | 2 |
| Primers | Tm | Amplicon Size | Purpose of Use | |
|---|---|---|---|---|
| Name | Sequence | |||
| VaccFvn | tcctaaccctaaccctgacc | 50 | 1503 | Amplification of the entire gene and flanking sequences |
| VaccRvn | aactcgtgattgtacctcgtt | |||
| VaccR | ttactggccggtcctcatca | 55 | 1002 | Amplification and sequencing of the entire gene |
| VaccF | cacgtgtaaagcccgtgatgtt | |||
| Vacc_ts_F | taaagcctgccactgcgatt | 55 | 838 | Efficient amplification of 838 out of 876 bp of coding sequence in any of studied Vaccinium genotype |
| Vacc_ts_pR | ccaatcgccacgagtaactaac | |||
| Vacc_reR | ccgaagttcgccgtcctg | n/a * | n/a | Primers for sequencing of gene fragments |
| Vacc_reF | tccaagccatcgacctactc | |||
| pJET1.2 Forward Sequencing Primer | cgactcactatagggagagcggc | 58 | n/a | Real-time PCR from colonies and sequencing |
| pJET1.2 Reverse Sequencing Primer | aagaacatcgattttccatggcag | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhidkin, R.; Zhurbenko, P.; Bogomaz, O.; Gorodilova, E.; Katsapov, I.; Antropov, D.; Matveeva, T. Biodiversity of rolB/C-like Natural Transgene in the Genus Vaccinium L. and Its Application for Phylogenetic Studies. Int. J. Mol. Sci. 2023, 24, 6932. https://doi.org/10.3390/ijms24086932
Zhidkin R, Zhurbenko P, Bogomaz O, Gorodilova E, Katsapov I, Antropov D, Matveeva T. Biodiversity of rolB/C-like Natural Transgene in the Genus Vaccinium L. and Its Application for Phylogenetic Studies. International Journal of Molecular Sciences. 2023; 24(8):6932. https://doi.org/10.3390/ijms24086932
Chicago/Turabian StyleZhidkin, Roman, Peter Zhurbenko, Olesya Bogomaz, Elizaveta Gorodilova, Ivan Katsapov, Dmitry Antropov, and Tatiana Matveeva. 2023. "Biodiversity of rolB/C-like Natural Transgene in the Genus Vaccinium L. and Its Application for Phylogenetic Studies" International Journal of Molecular Sciences 24, no. 8: 6932. https://doi.org/10.3390/ijms24086932
APA StyleZhidkin, R., Zhurbenko, P., Bogomaz, O., Gorodilova, E., Katsapov, I., Antropov, D., & Matveeva, T. (2023). Biodiversity of rolB/C-like Natural Transgene in the Genus Vaccinium L. and Its Application for Phylogenetic Studies. International Journal of Molecular Sciences, 24(8), 6932. https://doi.org/10.3390/ijms24086932





