A Low-Cost Modular Imaging System for Rapid, Multiplexed Immunofluorescence Detection in Clinical Tissues
Abstract
1. Introduction
2. Results
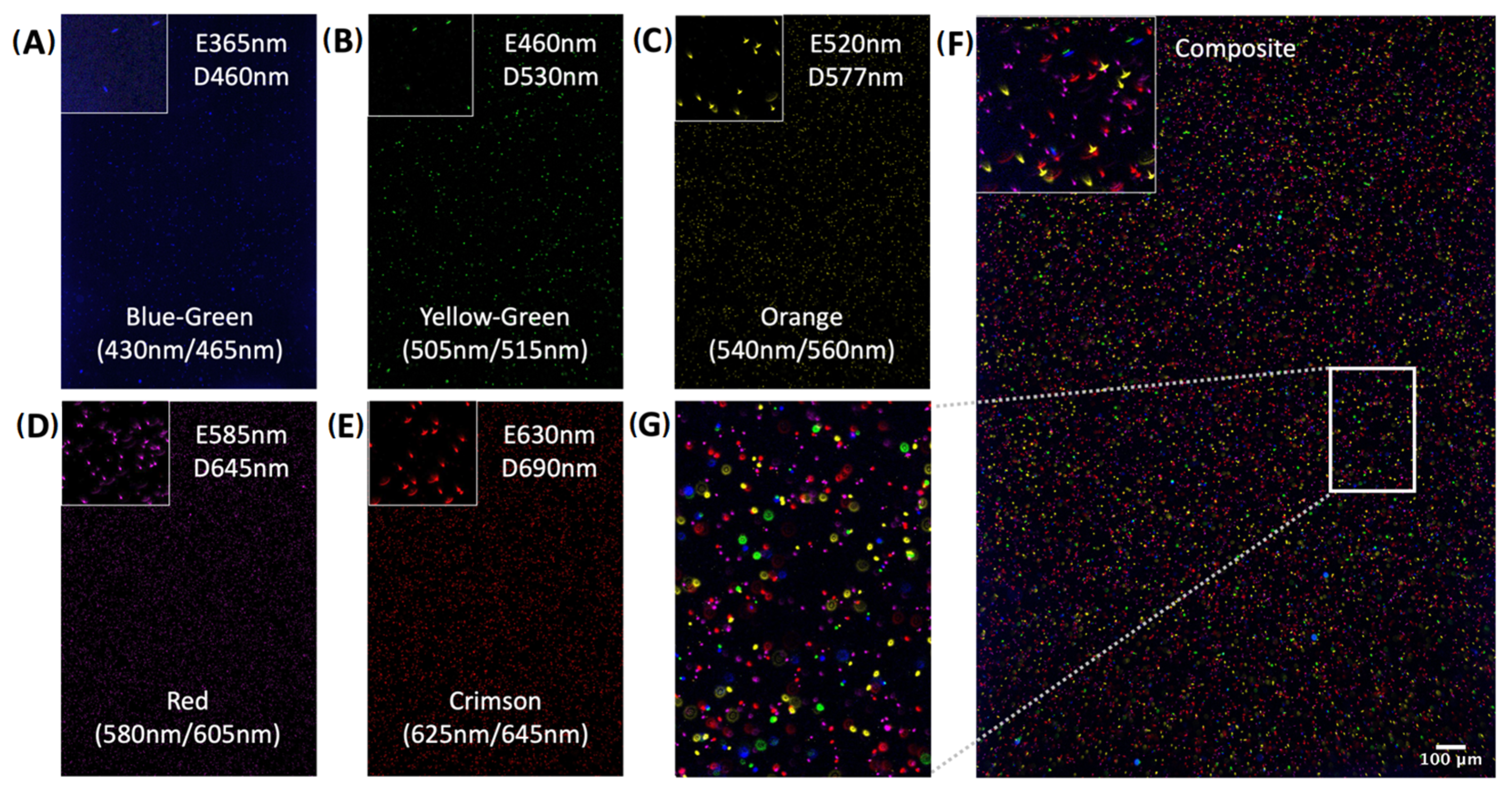
2.1. System Design
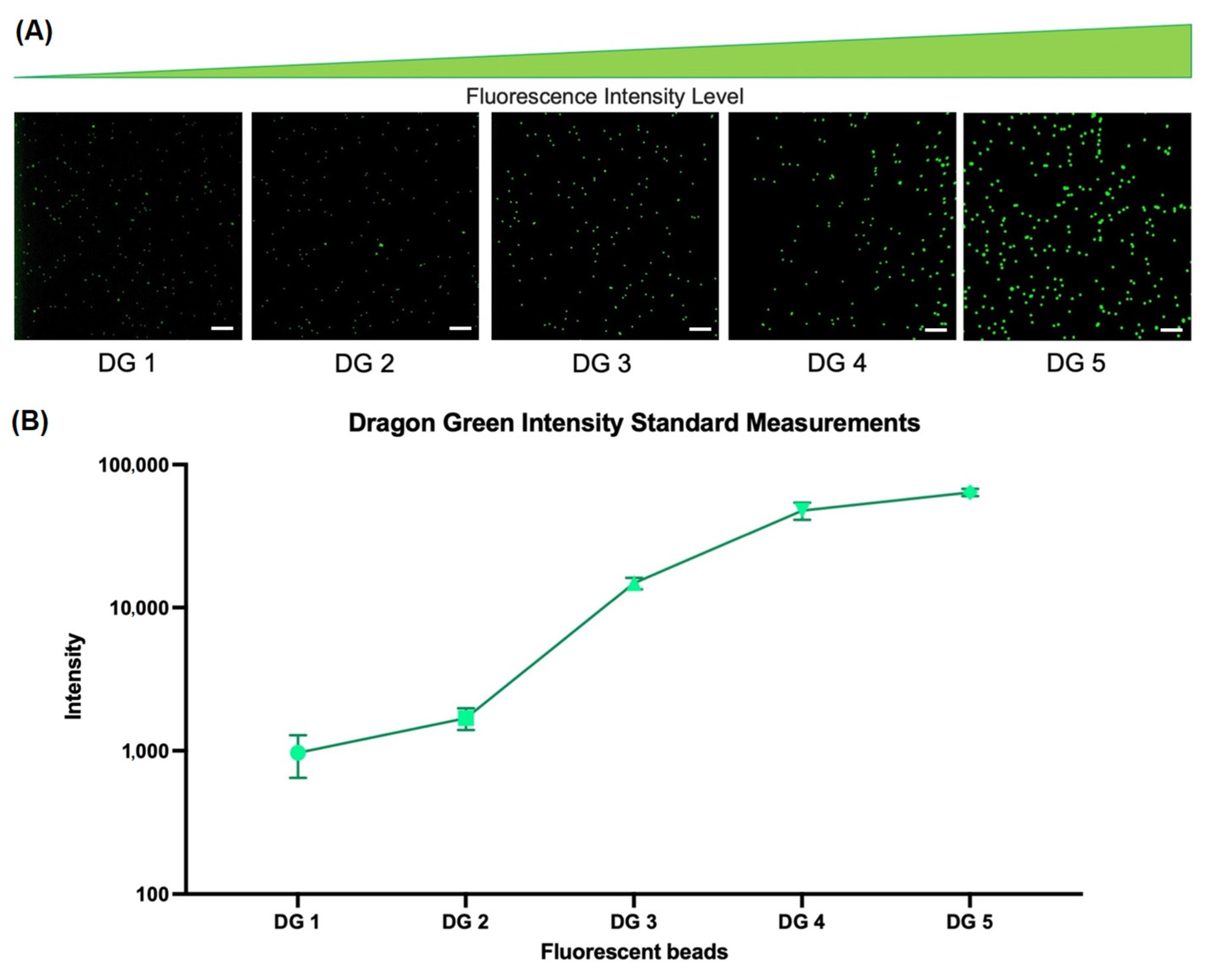
2.2. Evaluation of Specificity and Sensitivity
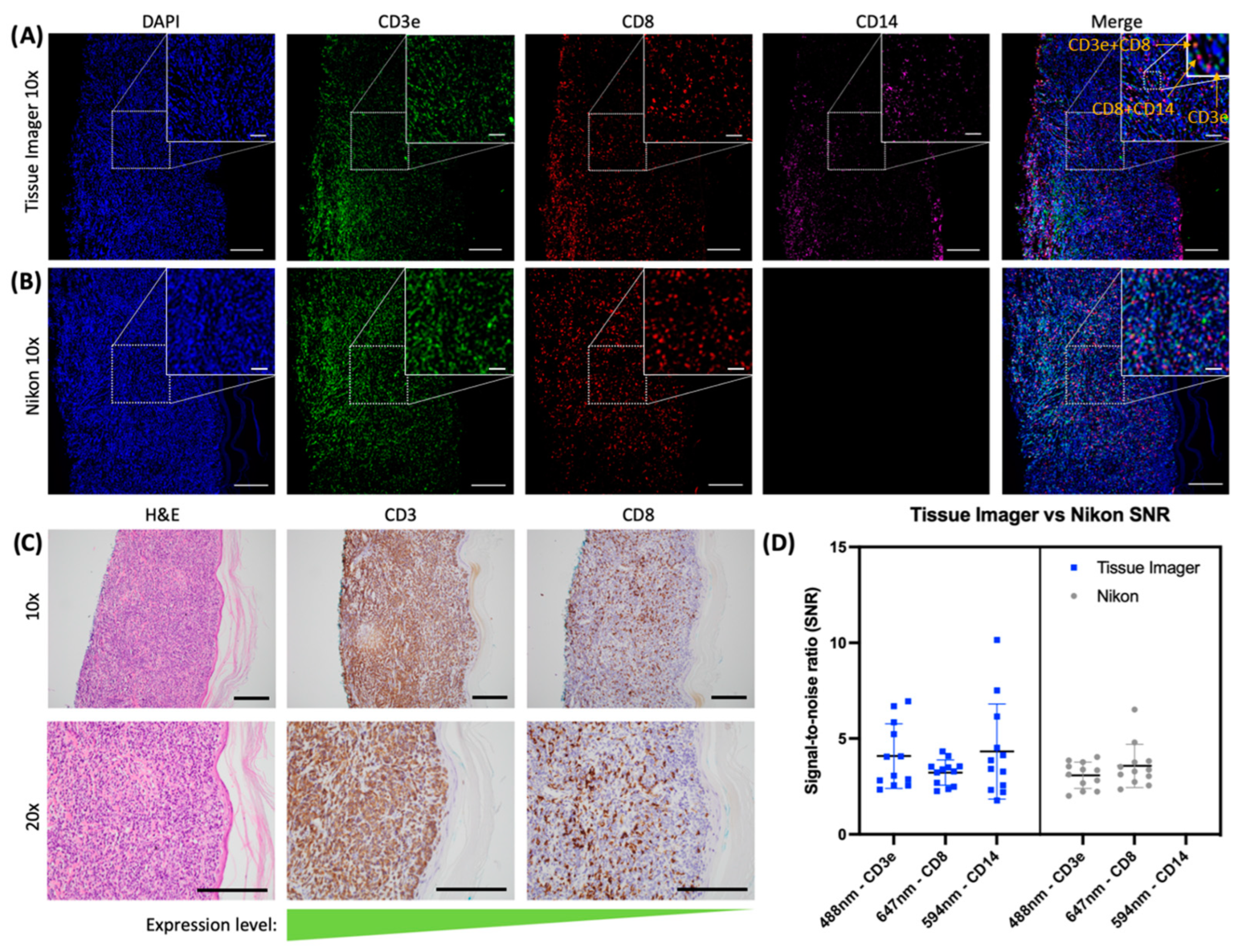
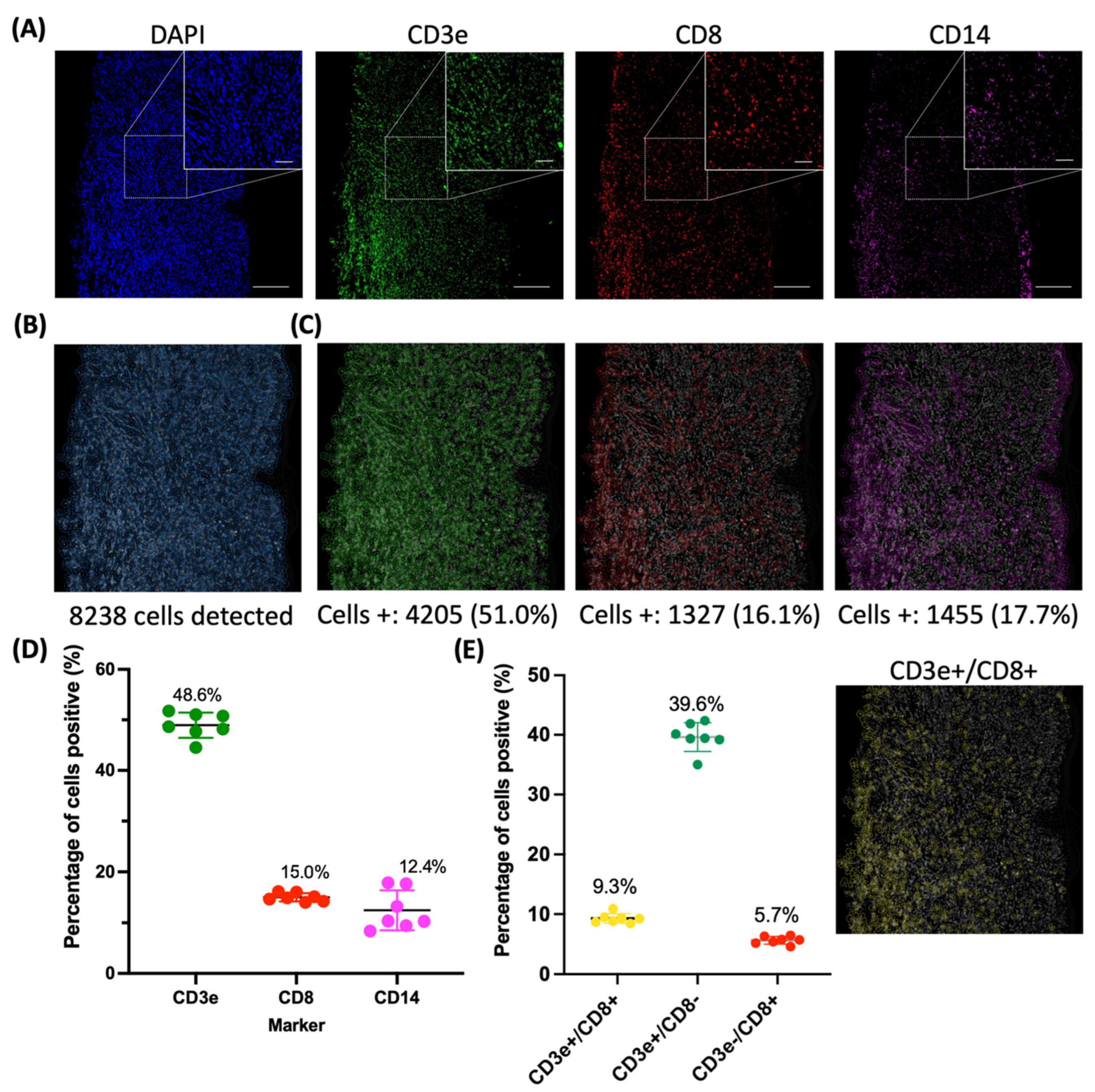
2.3. Evaluation of an Immune Panel on CTCL Tissue Samples
3. Discussion
4. Materials and Methods
4.1. Tissue Imager Design
4.2. Optical Resolution Characterization
4.3. Pixel Size Calibration Measurements
4.4. Fluorescent Beads
4.5. Preparation of FFPE Tissues
4.6. Antibody Staining
4.7. Image Acquisition and Data Transfer
4.8. ImageJ Image Processing
4.9. CellProfiler
4.10. Manual Counting
4.11. Statistical Analysis
4.12. Workflow Overview
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pollack, L.A.; Li, J.; Berkowitz, Z.; Weir, H.K.; Wu, X.-C.; Ajani, U.A.; Ekwueme, D.U.; Li, C.; Pollack, B.P. Melanoma survival in the United States, 1992 to 2005. J. Am. Acad. Dermatol. 2011, 65, S78–S86. [Google Scholar] [CrossRef] [PubMed]
- Bohndiek, S.E.; Brindle, K.M. Imaging and ‘omic’ methods for the molecular diagnosis of cancer. Expert Rev. Mol. Diagn 2010, 10, 417–434. [Google Scholar] [CrossRef] [PubMed]
- Burlingame, E.A.; McDonnell, M.; Schau, G.F.; Thibault, G.; Lanciault, C.; Morgan, T.; Johnson, B.E.; Corless, C.; Gray, J.W.; Chang, Y.H. SHIFT: Speedy histological-to-immunofluorescent translation of a tumor signature enabled by deep learning. Sci. Rep. 2020, 10, 17507. [Google Scholar] [CrossRef] [PubMed]
- Blom, S.; Paavolainen, L.; Bychkov, D.; Turkki, R.; Mäki-Teeri, P.; Hemmes, A.; Välimäki, K.; Lundin, J.; Kallioniemi, O.; Pellinen, T. Systems pathology by multiplexed immunohistochemistry and whole-slide digital image analysis. Sci. Rep. 2017, 7, 15580. [Google Scholar] [CrossRef] [PubMed]
- Kalra, J.; Baker, J. Multiplex Immunohistochemistry for Mapping the Tumor Microenvironment. In Signal Transduction Immunohistochemistry; Springer: Berlin/Heidelberg, Germany, 2017; pp. 237–251. [Google Scholar]
- Duraiyan, J.; Govindarajan, R.; Kaliyappan, K.; Palanisamy, M. Applications of immunohistochemistry. J. Pharm. Bioallied. Sci. 2012, 4, S307–S309. [Google Scholar]
- Hester, C.A.; Augustine, M.M.; Choti, M.A.; Mansour, J.C.; Minter, R.M.; Polanco, P.M.; Porembka, M.; Wang, S.; Yopp, A.C. Comparative outcomes of adenosquamous carcinoma of the pancreas: An analysis of the National Cancer Database. J. Surg. Oncol. 2018, 118, 21–30. [Google Scholar] [CrossRef]
- Parra, E.R.; Ferrufino-Schmidt, M.C.; Tamegnon, A.; Zhang, J.; Solis, L.; Jiang, M.; Ibarguen, H.; Haymaker, C.; Lee, J.J.; Bernatchez, C.; et al. Immuno-profiling and cellular spatial analysis using five immune oncology multiplex immunofluorescence panels for paraffin tumor tissue. Sci. Rep. 2021, 11, 8511. [Google Scholar] [CrossRef]
- Stack, E.C.; Wang, C.; Roman, K.A.; Hoyt, C.C. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 2014, 70, 46–58. [Google Scholar] [CrossRef]
- Feng, Z.; Jensen, S.M.; Messenheimer, D.J.; Farhad, M.; Neuberger, M.; Bifulco, C.B.; Fox, B.A. Multispectral imaging of T and B cells in murine spleen and tumor. J. Immunol. 2016, 196, 3943–3950. [Google Scholar] [CrossRef]
- Sanchez, K.; Kim, I.; Chun, B.; Pucilowska, J.; Redmond, W.L.; Urba, W.J.; Martel, M.; Wu, Y.; Campbell, M.; Sun, Z.; et al. Multiplex immunofluorescence to measure dynamic changes in tumor-infiltrating lymphocytes and PD-L1 in early-stage breast cancer. Breast Cancer Res. 2021, 23, 2. [Google Scholar] [CrossRef]
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J.; et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020, 38, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Manesse, M.; Patel, K.K.; Bobrow, M.; Downing, S.R. The InSituPlex® Staining Method for Multiplexed Immunofluorescence Cell Phenotyping and Spatial Profiling of Tumor FFPE Samples. In Biomarkers for Immunotherapy of Cancer; Springer: Berlin/Heidelberg, Germany, 2020; pp. 585–592. [Google Scholar]
- Pedroza-Gonzalez, A.; Zhou, G.; Vargas-Mendez, E.; Boor, P.P.; Mancham, S.; Verhoef, C.; Polak, W.G.; Grünhagen, D.; Pan, Q.; La Janssen, H.; et al. Tumor-infiltrating plasmacytoid dendritic cells promote immunosuppression by Tr1 cells in human liver tumors. Oncoimmunology 2015, 4, e1008355. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef]
- Sade-Feldman, M.; Yizhak, K.; Bjorgaard, S.L.; Ray, J.P.; de Boer, C.G.; Jenkins, R.W.; Lieb, D.J.; Chen, J.H.; Frederick, D.T.; Barzily-Rokni, M.; et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 2018, 175, 998–1013.e20. [Google Scholar] [CrossRef] [PubMed]
- Amaria, R.N.; Reddy, S.; Tawbi, H.A.; Davies, M.A.; Ross, M.I.; Glitza, I.C.; Cormier, J.N.; Lewis, C.; Hwu, W.-J.; Hanna, E.; et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat. Med. 2018, 24, 1649–1654. [Google Scholar] [CrossRef] [PubMed]
- Toki, M.I.; Merritt, C.R.; Wong, P.F.; Smithy, J.W.; Kluger, H.M.; Syrigos, K.N.; Ong, G.T.; Warren, S.E.; Beechem, J.M.; Rimm, D.L. High-plex predictive marker discovery for melanoma immunotherapy–treated patients using digital spatial profiling. Clin. Cancer Res. 2019, 25, 5503–5512. [Google Scholar] [CrossRef] [PubMed]
- Brede, C.; Friedrich, M.; Jordán-Garrote, A.-L.; Riedel, S.S.; Bäuerlein, C.A.; Heinze, K.G.; Bopp, T.; Schulz, S.; Mottok, A.; Kiesel, C.; et al. Mapping immune processes in intact tissues at cellular resolution. J. Clin. Investig. 2012, 122, 4439–4446. [Google Scholar] [CrossRef]
- Bagherani, N.; Smoller, B.R. An overview of cutaneous T cell lymphomas. F1000Research 2016, 5, 1882. [Google Scholar] [CrossRef]
- Tarabadkar, E.S.; Shinohara, M.M. Skin directed therapy in cutaneous T-cell lymphoma. Front. Oncol. 2019, 9, 260. [Google Scholar] [CrossRef]
- Berti, E.; Tomasini, D.; Vermeer, M.H.; Meijer, C.J.; Alessi, E.; Willemze, R. Primary cutaneous CD8-positive epidermotropic cytotoxic T cell lymphomas: A distinct clinicopathological entity with an aggressive clinical behavior. Am. J. Pathol. 1999, 155, 483–492. [Google Scholar] [CrossRef]
- Breslauer, D.N.; Maamari, R.N.; Switz, N.A.; Lam, W.A.; Fletcher, D.A. Mobile phone based clinical microscopy for global health applications. PLoS ONE 2009, 4, e6320. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.T.; Knapper, J.; Stirling, J.; Mduda, J.; Mkindi, C.; Mayagaya, V.; Mwakajinga, G.A.; Nyakyi, P.T.; Sanga, V.L.; Carbery, D.; et al. Robotic microscopy for everyone: The OpenFlexure microscope. Biomed. Opt. Express 2020, 11, 2447–2460. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Qi, H.; Luo, W.; Tseng, D.; Ki, S.J.; Wan, Z.; Göröcs, Z.; Bentolila, L.A.; Wu, T.-T.; Sun, R.; et al. Fluorescent imaging of single nanoparticles and viruses on a smart phone. ACS Nano 2013, 7, 9147–9155. [Google Scholar] [CrossRef] [PubMed]
- Yelleswarapu, V.R.; Jeong, H.-H.; Yadavali, S.; Issadore, D. Ultra-high throughput detection (1 million droplets per second) of fluorescent droplets using a cell phone camera and time domain encoded optofluidics. Lab Chip 2017, 17, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.; Campa, F.; Shih, W.-C. Open-source do-it-yourself multi-color fluorescence smartphone microscopy. Biomed. Opt. Express 2017, 8, 5075–5086. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Tokura, Y.; Yagi, H.; Seo, N.; Takagi, T.; Takigawa, M. Nonerythrodermic, leukemic variant of cutaneous T-cell lymphoma with indolent clinical course: Th2-type tumor cells lacking T-cell receptor/CD3 expression and coinfiltrating tumoricidal CD8+ T cells. J. Am. Acad. Dermatol. 2000, 43, 946–954. [Google Scholar] [CrossRef]
- Pujol, B.F.; Lucibello, F.C.; Gehling, U.M.; Lindemann, K.; Weidner, N.; Zuzarte, M.-L.; Adamkiewicz, J.; Elsässer, H.-P.; Müller, R.; Havemann, K. Endothelial-like cells derived from human CD14 positive monocytes. Differentiation 2000, 65, 287–300. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef]
- Lamprecht, M.R.; Sabatini, D.M.; Carpenter, A.E. CellProfiler™: Free, versatile software for automated biological image analysis. Biotechniques 2007, 42, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Finotello, F.; Rieder, D.; Hackl, H.; Trajanoski, Z. Next-generation computational tools for interrogating cancer immunity. Nat. Rev. Genet. 2019, 20, 724–746. [Google Scholar] [CrossRef] [PubMed]
- Engbjerg, J.S.; Costanzo, V.; Sardella, D.; Bordoni, L.; Jakobsen, S.; D’Apolito, L.; Frøkiær, J.; Trepiccione, F.; Capasso, G.; Frische, S. The Probe for Renal Organic Cation Secretion (4-Dimethylaminostyryl)-N-Methylpyridinium (ASP+)) Shows Amplified Fluorescence by Binding to Albumin and Is Accumulated In Vivo. Mol. Imaging 2022, 2022, 7908357. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, J.; Jian, H.; Wei, C.; Shiu, J.; Ganesan, A.; Zhao, W.; Hedde, P.N. A Low-Cost Modular Imaging System for Rapid, Multiplexed Immunofluorescence Detection in Clinical Tissues. Int. J. Mol. Sci. 2023, 24, 7008. https://doi.org/10.3390/ijms24087008
Gu J, Jian H, Wei C, Shiu J, Ganesan A, Zhao W, Hedde PN. A Low-Cost Modular Imaging System for Rapid, Multiplexed Immunofluorescence Detection in Clinical Tissues. International Journal of Molecular Sciences. 2023; 24(8):7008. https://doi.org/10.3390/ijms24087008
Chicago/Turabian StyleGu, Joshua, Hannah Jian, Christine Wei, Jessica Shiu, Anand Ganesan, Weian Zhao, and Per Niklas Hedde. 2023. "A Low-Cost Modular Imaging System for Rapid, Multiplexed Immunofluorescence Detection in Clinical Tissues" International Journal of Molecular Sciences 24, no. 8: 7008. https://doi.org/10.3390/ijms24087008
APA StyleGu, J., Jian, H., Wei, C., Shiu, J., Ganesan, A., Zhao, W., & Hedde, P. N. (2023). A Low-Cost Modular Imaging System for Rapid, Multiplexed Immunofluorescence Detection in Clinical Tissues. International Journal of Molecular Sciences, 24(8), 7008. https://doi.org/10.3390/ijms24087008






