Genome-Wide Meta-Analysis Identifies Multiple Novel Rare Variants to Predict Common Human Infectious Diseases Risk
Abstract
1. Introduction
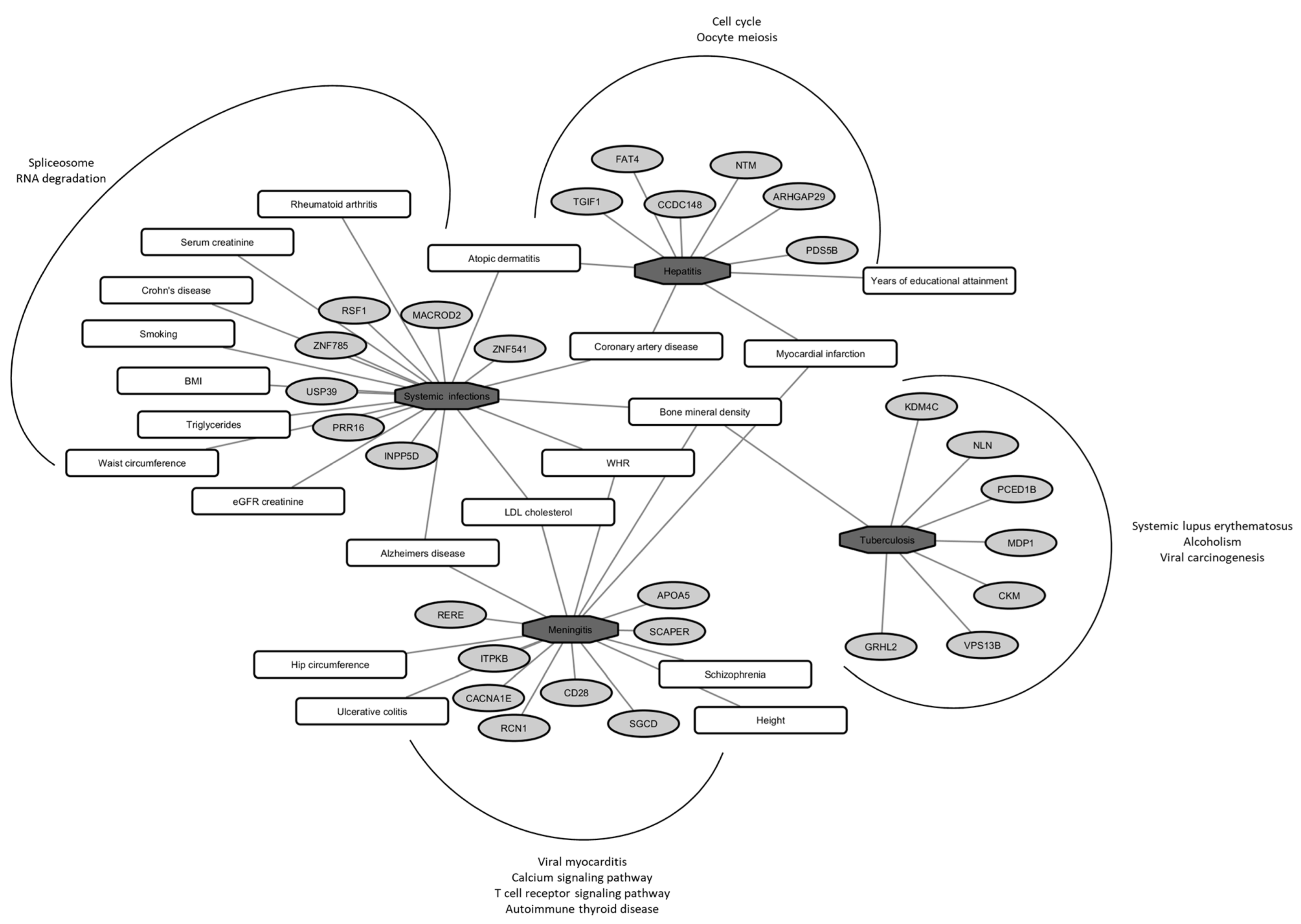
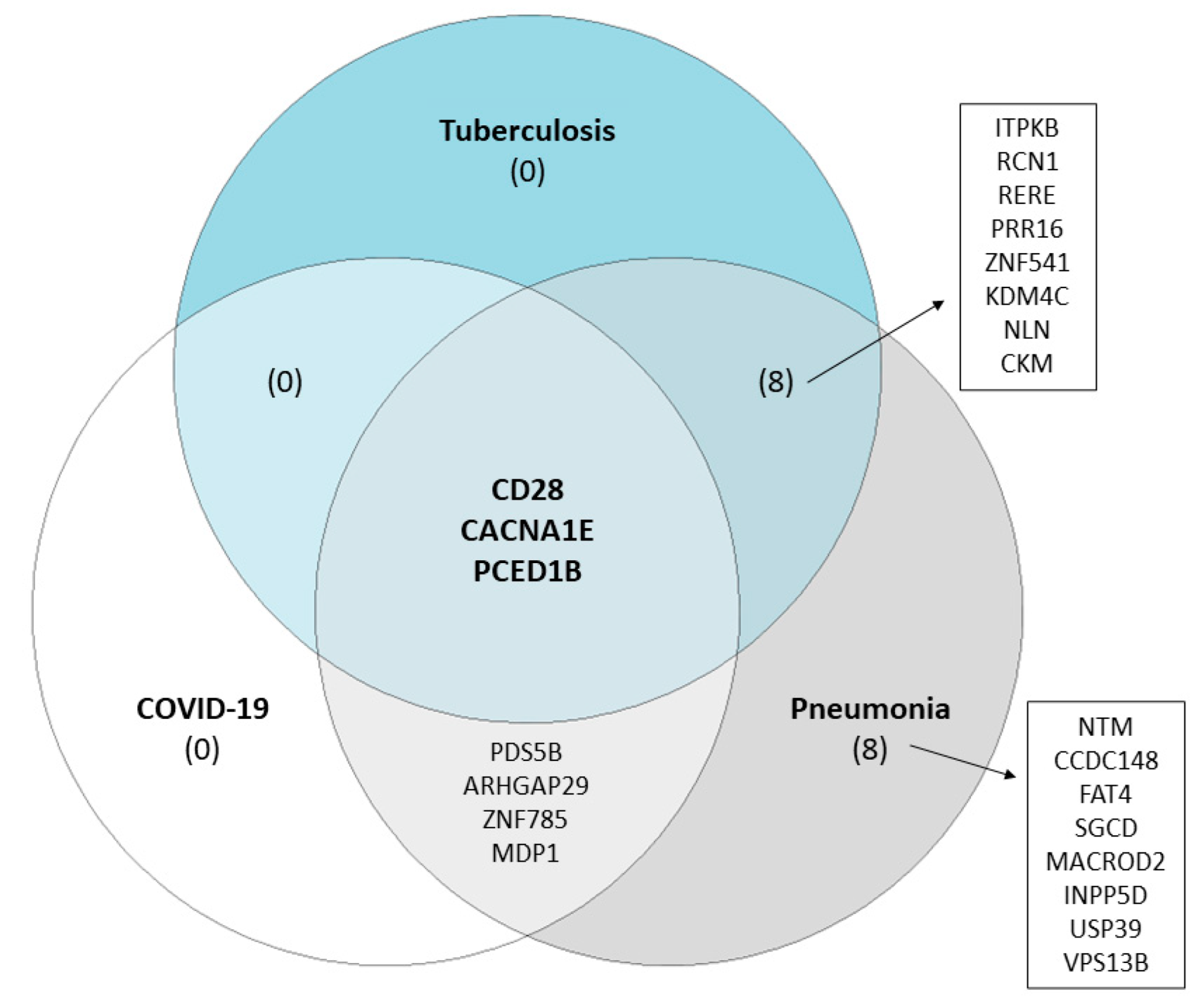
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Populations
4.2. Genotyping and SNP Imputation
4.3. Trait Definition
4.4. Statistical Analyses
4.4.1. Genome-Wide Association Analyses
4.4.2. Meta-Analyses
4.4.3. Proportion of Phenotypic Variance Explained by SNPs
4.4.4. SNP Function Annotation
4.4.5. Pathway Analysis
4.4.6. Validation with RNA-Seq Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fauci, A.S.; Morens, D.M. The Perpetual Challenge of Infectious Diseases. N. Engl. J. Med. 2012, 366, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M. Changing patterns of infectious disease. Nature 2000, 406, 762–767. [Google Scholar] [CrossRef]
- Karlsson, E.K.; Kwiatkowski, D.P.; Sabeti, P.C. Natural selection and infectious disease in human populations. Nat. Rev. Genet. 2014, 15, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Casanova, J.L.; Abel, L. The genetic theory of infectious diseases: A brief history and selected illustrations. Annu. Rev. Genom. Hum. Genet. 2013, 14, 215–243. [Google Scholar] [CrossRef]
- Burgner, D.; Jamieson, S.E.; Blackwell, J.M. Genetic susceptibility to infectious diseases: Big is beautiful, but will bigger be even better? Lancet Infect. Dis. 2006, 6, 653–663. [Google Scholar] [CrossRef]
- Patarcic, I.; Gelemanovic, A.; Kirin, M.; Kolcic, I.; Theodoratou, E.; Baillie, K.J.; de Jong, M.D.; Rudan, I.; Campbell, H.; Polasek, O. The role of host genetic factors in respiratory tract infectious diseases: Systematic review, meta-analyses and field synopsis. Sci. Rep. 2015, 5, 16119. [Google Scholar] [CrossRef] [PubMed]
- Chimusa, E.R.; Zaitlen, N.; Daya, M.; Moller, M.; van Helden, P.D.; Mulder, N.J.; Price, A.L.; Hoal, E.G. Genome-wide association study of ancestry-specific TB risk in the South African Coloured population. Hum. Mol. Genet. 2014, 23, 796–809. [Google Scholar] [CrossRef]
- Thye, T.; Owusu-Dabo, E.; Vannberg, F.O.; van Crevel, R.; Curtis, J.; Sahiratmadja, E.; Balabanova, Y.; Ehmen, C.; Muntau, B.; Ruge, G.; et al. Common variants at 11p13 are associated with susceptibility to tuberculosis. Nat. Genet. 2012, 44, 257–259. [Google Scholar] [CrossRef]
- Thye, T.; Vannberg, F.O.; Wong, S.H.; Owusu-Dabo, E.; Osei, I.; Gyapong, J.; Sirugo, G.; Sisay-Joof, F.; Enimil, A.; Chinbuah, M.A.; et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat. Genet. 2010, 42, 739–741. [Google Scholar] [CrossRef]
- Polasek, O. Future of biobanks—Bigger, longer, and more dimensional. Croat. Med. J. 2013, 54, 496–500. [Google Scholar] [CrossRef]
- Vitart, V.; Biloglav, Z.; Hayward, C.; Janicijevic, B.; Smolej-Narancic, N.; Barac, L.; Pericic, M.; Klaric, I.M.; Skaric-Juric, T.; Barbalic, M.; et al. 3000 years of solitude: Extreme differentiation in the island isolates of Dalmatia, Croatia. Eur. J. Hum. Genet. EJHG 2006, 14, 478–487. [Google Scholar] [CrossRef]
- Pattaro, C.; Kottgen, A.; Teumer, A.; Garnaas, M.; Boger, C.A.; Fuchsberger, C.; Olden, M.; Chen, M.H.; Tin, A.; Taliun, D.; et al. Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet. 2012, 8, e1002584. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Sim, X.; Go, M.J.; Wu, J.Y.; Gu, D.; Takeuchi, F.; Takahashi, A.; Maeda, S.; Tsunoda, T.; Chen, P.; et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat. Genet. 2012, 44, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Peloso, G.M.; Auer, P.L.; Bis, J.C.; Voorman, A.; Morrison, A.C.; Stitziel, N.O.; Brody, J.A.; Khetarpal, S.A.; Crosby, J.R.; Fornage, M.; et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet. 2014, 94, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Ruotsalainen, S.E.; Partanen, J.J.; Cichonska, A.; Lin, J.; Benner, C.; Surakka, I.; Reeve, M.P.; Palta, P.; Salmi, M.; Jalkanen, S.; et al. An expanded analysis framework for multivariate GWAS connects inflammatory biomarkers to functional variants and disease. Eur. J. Hum. Genet. EJHG 2021, 29, 309–324. [Google Scholar] [CrossRef]
- Mallard, T.T.; Linner, R.K.; Grotzinger, A.D.; Sanchez-Roige, S.; Seidlitz, J.; Okbay, A.; de Vlaming, R.; Meddens, S.F.W.; Palmer, A.A.; Davis, L.K.; et al. Multivariate GWAS of psychiatric disorders and their cardinal symptoms reveal two dimensions of cross-cutting genetic liabilities. Cell Genom. 2022, 2, 100140. [Google Scholar] [CrossRef] [PubMed]
- Karlsson Linner, R.; Mallard, T.T.; Barr, P.B.; Sanchez-Roige, S.; Madole, J.W.; Driver, M.N.; Poore, H.E.; de Vlaming, R.; Grotzinger, A.D.; Tielbeek, J.J.; et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat. Neurosci. 2021, 24, 1367–1376. [Google Scholar] [CrossRef]
- Nicholls, H.L.; John, C.R.; Watson, D.S.; Munroe, P.B.; Barnes, M.R.; Cabrera, C.P. Reaching the End-Game for GWAS: Machine Learning Approaches for the Prioritization of Complex Disease Loci. Front. Genet. 2020, 11, 350. [Google Scholar] [CrossRef]
- Tian, C.; Hromatka, B.S.; Kiefer, A.K.; Eriksson, N.; Noble, S.M.; Tung, J.Y.; Hinds, D.A. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat. Commun. 2017, 8, 599. [Google Scholar] [CrossRef]
- Kleinstein, S.E.; Shea, P.R.; Allen, A.S.; Koelle, D.M.; Wald, A.; Goldstein, D.B. Genome-wide association study (GWAS) of human host factors influencing viral severity of herpes simplex virus type 2 (HSV-2). Genes Immun. 2019, 20, 112–120. [Google Scholar] [CrossRef]
- Moreau, K.; Clemenceau, A.; Le Moing, V.; Messika-Zeitoun, D.; Andersen, P.S.; Bruun, N.E.; Skov, R.L.; Couzon, F.; Bouchiat, C.; Erpelding, M.L.; et al. Human Genetic Susceptibility to Native Valve Staphylococcus aureus Endocarditis in Patients with S. aureus Bacteremia: Genome-Wide Association Study. Front. Microbiol. 2018, 9, 640. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, C.; Majumder, P.P.; Pandit, B. An exome wide association study of pulmonary tuberculosis patients and their asymptomatic household contacts. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019, 71, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Suliman, S.; Asgari, S.; Amariuta, T.; Baglaenko, Y.; Martinez-Bonet, M.; Ishigaki, K.; Gutierrez-Arcelus, M.; Calderon, R.; Lecca, L.; et al. Early progression to active tuberculosis is a highly heritable trait driven by 3q23 in Peruvians. Nat. Commun. 2019, 10, 3765. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Huang, S.; Chen, F.; Zhao, L.; Yuan, Y.; Francis, S.S.; Fang, L.; Li, Z.; Lin, L.; Liu, R.; et al. Genomic Analyses from Non-invasive Prenatal Testing Reveal Genetic Associations, Patterns of Viral Infections, and Chinese Population History. Cell 2018, 175, 347–359.e14. [Google Scholar] [CrossRef]
- Lees, J.A.; Ferwerda, B.; Kremer, P.H.C.; Wheeler, N.E.; Seron, M.V.; Croucher, N.J.; Gladstone, R.A.; Bootsma, H.J.; Rots, N.Y.; Wijmega-Monsuur, A.J.; et al. Joint sequencing of human and pathogen genomes reveals the genetics of pneumococcal meningitis. Nat. Commun. 2019, 10, 2176. [Google Scholar] [CrossRef]
- Omae, Y.; Toyo-Oka, L.; Yanai, H.; Nedsuwan, S.; Wattanapokayakit, S.; Satproedprai, N.; Smittipat, N.; Palittapongarnpim, P.; Sawanpanyalert, P.; Inunchot, W.; et al. Pathogen lineage-based genome-wide association study identified CD53 as susceptible locus in tuberculosis. J. Hum. Genet. 2017, 62, 1015–1022. [Google Scholar] [CrossRef]
- Zheng, R.; Li, Z.; He, F.; Liu, H.; Chen, J.; Xie, X.; Zhou, J.; Chen, H.; Wu, X.; Wu, J.; et al. Genome-wide association study identifies two risk loci for tuberculosis in Han Chinese. Nat. Commun. 2018, 9, 4072. [Google Scholar] [CrossRef]
- Rudan, I. Answering the initial 20 questions on COVID-19 (January–February 2020). J. Glob. Health 2020, 10, 010106. [Google Scholar] [CrossRef]
- Calisher, C.H. Good news or bad news? The coronavirus pandemic has sickened and killed only a relatively few people but has affected us all. Croat. Med. J. 2020, 61, 296–299. [Google Scholar] [CrossRef]
- Ghosh, D.; Bernstein, J.A.; Mersha, T.B. COVID-19 pandemic: The African paradox. J. Glob. Health 2020, 10, e020347. [Google Scholar] [CrossRef]
- Liu, S.; Shen, J.; Fang, S.; Li, K.; Liu, J.; Yang, L.; Hu, C.D.; Wan, J. Genetic Spectrum and Distinct Evolution Patterns of SARS-CoV-2. Front. Microbiol. 2020, 11, 593548. [Google Scholar] [CrossRef]
- Anastassopoulou, C.; Gkizarioti, Z.; Patrinos, G.P.; Tsakris, A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum. Genom. 2020, 14, 40. [Google Scholar] [CrossRef]
- Choudhary, S.; Sreenivasulu, K.; Mitra, P.; Misra, S.; Sharma, P. Role of Genetic Variants and Gene Expression in the Susceptibility and Severity of COVID-19. Ann. Lab. Med. 2021, 41, 129–138. [Google Scholar] [CrossRef]
- Picard, C.; Casanova, J.L.; Abel, L. Mendelian traits that confer predisposition or resistance to specific infections in humans. Curr. Opin. Immunol. 2006, 18, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Honda, M.; Yamashita, T.; Arai, K.; Sakai, Y.; Sakai, A.; Nakamura, M.; Mizukoshi, E.; Kaneko, S. Peretinoin, an acyclic retinoid, improves the hepatic gene signature of chronic hepatitis C following curative therapy of hepatocellular carcinoma. BMC Cancer 2013, 13, 191. [Google Scholar] [CrossRef]
- Ramachandran, S.; Ilias Basha, H.; Sarma, N.J.; Lin, Y.; Crippin, J.S.; Chapman, W.C.; Mohanakumar, T. Hepatitis C virus induced miR200c down modulates FAP-1, a negative regulator of Src signaling and promotes hepatic fibrosis. PLoS ONE 2013, 8, e70744. [Google Scholar] [CrossRef] [PubMed]
- Dawood, R.M.; El-Meguid, M.A.; Ibrahim, M.K.; Bader El Din, N.G.; Barakat, A.; El-Wakeel, K.; Alla, M.; Wu, G.Y.; El Awady, M.K. Dysregulation of fibrosis related genes in HCV induced liver disease. Gene 2018, 664, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.Y.; Xing, Q.; Duan, M.; Wang, Z.C.; Yang, L.X.; Zhao, Y.J.; Wang, X.Y.; Liu, Y.; Deng, M.; Ding, Z.B.; et al. Inferring the progression of multifocal liver cancer from spatial and temporal genomic heterogeneity. Oncotarget 2016, 7, 2867–2877. [Google Scholar] [CrossRef]
- Huang, F.Y.; Wong, D.K.; Tsui, V.W.; Seto, W.K.; Mak, L.Y.; Cheung, T.T.; Lai, K.K.; Yuen, M.F. Targeted genomic profiling identifies frequent deleterious mutations in FAT4 and TP53 genes in HBV-associated hepatocellular carcinoma. BMC Cancer 2019, 19, 789. [Google Scholar] [CrossRef]
- Wei, X.L.; Qiu, M.Z.; Jin, Y.; Huang, Y.X.; Wang, R.Y.; Chen, W.W.; Wang, D.S.; Wang, F.; Luo, H.Y.; Zhang, D.S.; et al. Hepatitis B virus infection is associated with gastric cancer in China: An endemic area of both diseases. Brit. J. Cancer 2015, 112, 1283–1290. [Google Scholar] [CrossRef]
- Qiao, Y.T.; Chen, J.X.; Lim, Y.B.; Finch-Edmondson, M.L.; Seshachalam, V.P.; Qin, L.; Jiang, T.; Low, B.C.; Singh, H.; Lim, C.T.; et al. YAP Regulates Actin Dynamics through ARHGAP29 and Promotes Metastasis. Cell Rep. 2017, 19, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.; Churchill, N.D.; Mulrooney-Cousins, P.M.; Michalak, T.I. Initial sites of hepadnavirus integration into host genome in human hepatocytes and in the woodchuck model of hepatitis B-associated hepatocellular carcinoma. Oncogenesis 2017, 6, e317. [Google Scholar] [CrossRef] [PubMed]
- Starakis, I.; Panos, G.; Koutras, A.; Mazokopakis, E.E. Pathogens and chronic or long-term neurologic disorders. Cardiovasc. Hematol. Disord. Drug Targets 2011, 11, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Karim, S.; Mirza, Z.; Kamal, M.A.; Abuzenadah, A.M.; Azhar, E.I.; Al-Qahtani, M.H.; Damanhouri, G.A.; Ahmad, F.; Gan, S.H.; Sohrab, S.S. The role of viruses in neurodegenerative and neurobehavioral diseases. CNS Neurol. Disord. Drug Targets 2014, 13, 1213–1223. [Google Scholar] [CrossRef]
- Alam, M.Z.; Alam, Q.; Kamal, M.A.; Jiman-Fatani, A.A.; Azhar, E.I.; Khan, M.A.; Haque, A. Infectious Agents and Neurodegenerative Diseases: Exploring the Links. Curr. Top. Med. Chem. 2017, 17, 1390–1399. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Seishima, M.; Saito, K.; Maeda, S.; Takemura, M.; Noma, A.; Kondo, A.; Manabe, M.; Urakami, K.; Nakashima, K. Apo A-I and apo E concentrations in cerebrospinal fluids of patients with acute meningitis. Ann. Clin. Biochem. 1998, 35 Pt 3, 408–414. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Wang, A.; Fu, P.; Yang, Y. The diagnostic value of apolipoprotein E in pediatric patients with invasive bacterial infections. Clin. Biochem. 2012, 45, 215–218. [Google Scholar] [CrossRef]
- Yao, X.; Gordon, E.M.; Figueroa, D.M.; Barochia, A.V.; Levine, S.J. Emerging Roles of Apolipoprotein E and Apolipoprotein A-I in the Pathogenesis and Treatment of Lung Disease. Am. J. Respir. Cell Mol. Biol. 2016, 55, 159–169. [Google Scholar] [CrossRef]
- Li, L.; Thompson, P.A.; Kitchens, R.L. Infection induces a positive acute phase apolipoprotein E response from a negative acute phase gene: Role of hepatic LDL receptors. J. Lipid Res. 2008, 49, 1782–1793. [Google Scholar] [CrossRef]
- Lin, Q.; Cao, Y.; Gao, J. Decreased expression of the APOA1-APOC3-APOA4 gene cluster is associated with risk of Alzheimer’s disease. Drug Des. Dev. Ther. 2015, 9, 5421–5431. [Google Scholar] [CrossRef]
- Heyes, S.; Pratt, W.S.; Rees, E.; Dahimene, S.; Ferron, L.; Owen, M.J.; Dolphin, A.C. Genetic disruption of voltage-gated calcium channels in psychiatric and neurological disorders. Prog. Neurobiol. 2015, 134, 36–54. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.T.; Wimmer, I.; Hoftberger, R.; Gerlach, S.; Haider, L.; Zrzavy, T.; Hametner, S.; Mahad, D.; Binder, C.J.; Krumbholz, M.; et al. Disease-specific molecular events in cortical multiple sclerosis lesions. Brain J. Neurol. 2013, 136 Pt 6, 1799–1815. [Google Scholar] [CrossRef] [PubMed]
- Perrin, P.J.; Lavi, E.; Rumbley, C.A.; Zekavat, S.A.; Phillips, S.M. Experimental autoimmune meningitis: A novel neurological disease in CD28-deficient mice. Clin. Immunol. 1999, 91, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Perrin, P.J.; June, C.H.; Maldonado, J.H.; Ratts, R.B.; Racke, M.K. Blockade of CD28 during in vitro activation of encephalitogenic T cells or after disease onset ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 1999, 163, 1704–1710. [Google Scholar] [CrossRef]
- Pouillon, V.; Marechal, Y.; Frippiat, C.; Erneux, C.; Schurmans, S. Inositol 1,4,5-trisphosphate 3-kinase B (Itpkb) controls survival, proliferation and cytokine production in mouse peripheral T cells. Adv. Biol. Regul. 2013, 53, 39–50. [Google Scholar] [CrossRef]
- Westernberg, L.; Conche, C.; Huang, Y.H.; Rigaud, S.; Deng, Y.; Siegemund, S.; Mukherjee, S.; Nosaka, L.; Das, J.; Sauer, K. Non-canonical antagonism of PI3K by the kinase Itpkb delays thymocyte beta-selection and renders it Notch-dependent. eLife 2016, 5, e10786. [Google Scholar] [CrossRef]
- Moris, A.; Murray, S.; Cardinaud, S. AID and APOBECs span the gap between innate and adaptive immunity. Front. Microbiol. 2014, 5, 534. [Google Scholar] [CrossRef]
- An, P.; Penugonda, S.; Thorball, C.W.; Bartha, I.; Goedert, J.J.; Donfield, S.; Buchbinder, S.; Binns-Roemer, E.; Kirk, G.D.; Zhang, W.; et al. Role of APOBEC3F Gene Variation in HIV-1 Disease Progression and Pneumocystis Pneumonia. PLoS Genet. 2016, 12, e1005921. [Google Scholar] [CrossRef]
- Yamamoto, N.; Kerfoot, S.M.; Hutchinson, A.T.; Dela Cruz, C.S.; Nakazawa, N.; Szczepanik, M.; Majewska-Szczepanik, M.; Nazimek, K.; Ohana, N.; Bryniarski, K.; et al. Expression of activation-induced cytidine deaminase enhances the clearance of pneumococcal pneumonia: Evidence of a subpopulation of protective anti-pneumococcal B1a cells. Immunology 2016, 147, 97–113. [Google Scholar] [CrossRef]
- Cai, L.; Wang, Y.; Wang, J.F.; Chou, K.C. Identification of proteins interacting with human SP110 during the process of viral infections. Med. Chem. 2011, 7, 121–126. [Google Scholar] [CrossRef]
- Shamay, M.; Barak, O.; Doitsh, G.; Ben-Dor, I.; Shaul, Y. Hepatitis B virus pX interacts with HBXAP, a PHD finger protein to coactivate transcription. J. Biol. Chem. 2002, 277, 9982–9988. [Google Scholar] [CrossRef] [PubMed]
- Khong, J.J.; Burdon, K.P.; Lu, Y.; Leonardos, L.; Laurie, K.J.; Walsh, J.P.; Gajdatsy, A.D.; Ebeling, P.R.; McNab, A.A.; Hardy, T.G.; et al. Association of Polymorphisms in MACRO Domain Containing 2 with Thyroid-Associated Orbitopathy. Investig. Ophthalmol. Vis. Sci. 2016, 57, 3129–3137. [Google Scholar] [CrossRef]
- Chamberlain, T.C.; Cheung, S.T.; Yoon, J.S.J.; Ming-Lum, A.; Gardill, B.R.; Shakibakho, S.; Dzananovic, E.; Ban, F.; Samiea, A.; Jawanda, K.; et al. Interleukin-10 and Small Molecule SHIP1 Allosteric Regulators Trigger Anti-inflammatory Effects through SHIP1/STAT3 Complexes. iScience 2020, 23, 101433. [Google Scholar] [CrossRef] [PubMed]
- Samiea, A.; Yoon, J.S.J.; Cheung, S.T.; Chamberlain, T.C.; Mui, A.L. Interleukin-10 contributes to PGE2 signalling through upregulation of EP4 via SHIP1 and STAT3. PLoS ONE 2020, 15, e0230427. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, H.; Xiao, J.; Yuan, T.; Shu, Z.; Min, Y.; Xu, W.; Yin, Y.; Zhang, X. Streptococcus pneumoniae Endopeptidase O Promotes the Clearance of Staphylococcus aureus and Streptococcus pneumoniae via SH2 Domain-Containing Inositol Phosphatase 1-Mediated Complement Receptor 3 Upregulation. Front. Cell. Infect. Microbiol. 2020, 10, 358. [Google Scholar] [CrossRef]
- Qin, S.; Li, J.; Zhou, C.; Privratsky, B.; Schettler, J.; Deng, X.; Xia, Z.; Zeng, Y.; Wu, H.; Wu, M. SHIP-1 Regulates Phagocytosis and M2 Polarization Through the PI3K/Akt-STAT5-Trib1 Circuit in Pseudomonas aeruginosa Infection. Front. Immunol. 2020, 11, 307. [Google Scholar] [CrossRef]
- Chen, Y.; Fachko, D.; Ivanov, N.S.; Skinner, C.M.; Skalsky, R.L. Epstein-Barr virus microRNAs regulate B cell receptor signal transduction and lytic reactivation. PLoS Pathog. 2019, 15, e1007535. [Google Scholar] [CrossRef] [PubMed]
- Cojohari, O.; Mahmud, J.; Altman, A.M.; Peppenelli, M.A.; Miller, M.J.; Chan, G.C. Human Cytomegalovirus Mediates Unique Monocyte-to-Macrophage Differentiation through the PI3K/SHIP1/Akt Signaling Network. Viruses 2020, 12, 652. [Google Scholar] [CrossRef]
- Mahmud, J.; Miller, M.J.; Altman, A.M.; Chan, G.C. Human Cytomegalovirus Glycoprotein-Initiated Signaling Mediates the Aberrant Activation of Akt. J. Virol. 2020, 94, e00167-20. [Google Scholar] [CrossRef]
- Cojohari, O.; Peppenelli, M.A.; Chan, G.C. Human Cytomegalovirus Induces an Atypical Activation of Akt To Stimulate the Survival of Short-Lived Monocytes. J. Virol. 2016, 90, 6443–6452. [Google Scholar] [CrossRef]
- Gold, M.J.; Antignano, F.; Hughes, M.R.; Zaph, C.; McNagny, K.M. Dendritic-cell expression of Ship1 regulates Th2 immunity to helminth infection in mice. Eur. J. Immunol. 2016, 46, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Soini, Y.; Kosma, V.M.; Pirinen, R. KDM4A, KDM4B and KDM4C in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 12922–12928. [Google Scholar] [PubMed]
- Zhu, J.; Deng, L.; Chen, B.; Huang, W.; Lin, X.; Chen, G.; Tzeng, C.M.; Ying, M.; Lu, Z. Magnesium-dependent Phosphatase (MDP) 1 is a Potential Suppressor of Gastric Cancer. Curr. Cancer Drug Targets 2019, 19, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cui, J.; Niu, W.; Huang, J.; Feng, T.; Sun, B.; Yao, H. Long non-coding PCED1B-AS1 regulates macrophage apoptosis and autophagy by sponging miR-155 in active tuberculosis. Biochem. Biophys. Res. Commun. 2019, 509, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.V.S. Evolution, revolution and heresy in the genetics of infectious disease susceptibility. Philos. Trans. R. Soc. B 2012, 367, 840–849. [Google Scholar] [CrossRef]
- Chapman, S.J.; Hill, A.V. Human genetic susceptibility to infectious disease. Nat. Rev. Genet. 2012, 13, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Alcais, A.; Quintana-Murci, L.; Thaler, D.S.; Schurr, E.; Abel, L.; Casanova, J.L. Life-threatening infectious diseases of childhood: Single-gene inborn errors of immunity? Ann. N. Y. Acad. Sci. 2010, 1214, 18–33. [Google Scholar] [CrossRef]
- Rudan, I.; Marusic, A.; Jankovic, S.; Rotim, K.; Boban, M.; Lauc, G.; Grkovic, I.; Dogas, Z.; Zemunik, T.; Vatavuk, Z.; et al. “10001 Dalmatians”: Croatia launches its national biobank. Croat. Med. J. 2009, 50, 4–6. [Google Scholar] [CrossRef]
- Joshi, P.K.; Esko, T.; Mattsson, H.; Eklund, N.; Gandin, I.; Nutile, T.; Jackson, A.U.; Schurmann, C.; Smith, A.V.; Zhang, W.; et al. Directional dominance on stature and cognition in diverse human populations. Nature 2015, 523, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Delaneau, O.; Zagury, J.F.; Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 2013, 10, 5–6. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, J.; Gurdasani, D.; Delaneau, O.; Pirastu, N.; Ulivi, S.; Cocca, M.; Traglia, M.; Huang, J.; Huffman, J.E.; Rudan, I.; et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014, 10, e1004234. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.; Das, S.; Kretzschmar, W.; Delaneau, O.; Wood, A.R.; Teumer, A.; Kang, H.M.; Fuchsberger, C.; Danecek, P.; Sharp, K.; et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar] [PubMed]
- Cheng, C.Y.; Schache, M.; Ikram, M.K.; Young, T.L.; Guggenheim, J.A.; Vitart, V.; MacGregor, S.; Verhoeven, V.J.; Barathi, V.A.; Liao, J.; et al. Nine loci for ocular axial length identified through genome-wide association studies, including shared loci with refractive error. Am. J. Hum. Genet. 2013, 93, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Nettleton, J.A.; McKeown, N.M.; Kanoni, S.; Lemaitre, R.N.; Hivert, M.F.; Ngwa, J.; van Rooij, F.J.; Sonestedt, E.; Wojczynski, M.K.; Ye, Z.; et al. Interactions of dietary whole-grain intake with fasting glucose- and insulin-related genetic loci in individuals of European descent: A meta-analysis of 14 cohort studies. Diabetes Care 2010, 33, 2684–2691. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Marroni, F.; Hayward, C.; Franklin, C.S.; Kirichenko, A.V.; Jonasson, I.; Hicks, A.A.; Vitart, V.; Isaacs, A.; Axenovich, T.; et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum. Mol. Genet. 2009, 18, 373–380. [Google Scholar] [CrossRef]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef]
- Sikorska, K.; Lesaffre, E.; Groenen, P.F.; Eilers, P.H. GWAS on your notebook: Fast semi-parallel linear and logistic regression for genome-wide association studies. BMC Bioinform. 2013, 14, 166. [Google Scholar] [CrossRef] [PubMed]
- Haller, T.; Kals, M.; Esko, T.; Magi, R.; Fischer, K. RegScan: A GWAS tool for quick estimation of allele effects on continuous traits and their combinations. Brief. Bioinform. 2015, 16, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Svishcheva, G.R.; Axenovich, T.I.; Belonogova, N.M.; van Duijn, C.M.; Aulchenko, Y.S. Rapid variance components-based method for whole-genome association analysis. Nat. Genet. 2012, 44, 1166–1170. [Google Scholar] [CrossRef] [PubMed]
- Winkler, T.W.; Day, F.R.; Croteau-Chonka, D.C.; Wood, A.R.; Locke, A.E.; Magi, R.; Ferreira, T.; Fall, T.; Graff, M.; Justice, A.E.; et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014, 9, 1192–1212. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef] [PubMed]
- Tin, A.; Marten, J.; Halperin Kuhns, V.L.; Li, Y.; Wuttke, M.; Kirsten, H.; Sieber, K.B.; Qiu, C.; Gorski, M.; Yu, Z.; et al. Target genes, variants, tissues and transcriptional pathways influencing human serum urate levels. Nat. Genet. 2019, 51, 1459–1474. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.R.; Blackshaw, J.; Kamat, M.A.; Ellis, S.; Surendran, P.; Sun, B.B.; Paul, D.S.; Freitag, D.; Burgess, S.; Danesh, J.; et al. PhenoScanner: A database of human genotype-phenotype associations. Bioinformatics 2016, 32, 3207–3209. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
| Vis | Korčula | Split | TOTAL | p * | |
|---|---|---|---|---|---|
| N | 960 | 2698 | 966 | 4624 | |
| Gender, women; n (%) | 558 (58.12) | 1712 (63.45) | 587 (60.77) | 2857 (61.79) | 0.010 KV |
| Age in years, median (IQR) | 56 (24.00) | 55 (23.00) | 52 (21.00) | 55 (22.25) | <0.001 KV, KS, VS |
| Years of schooling; n (%) | |||||
| Elementary school [0–8] | 390 (40.63) | 711 (26.35) | 62 (6.42) | 1163 (25.15) | <0.001 KV, KS, VS |
| High school [9–12] | 413 (43.02) | 1461 (54.15) | 473 (48.96) | 2347 (50.76) | |
| University [≥13] | 157 (16.35) | 526 (19.50) | 431 (44.62) | 1114 (24.09) | |
| Socioeconomic status; n (%) | |||||
| 1st quartile [0–8] | 350 (36.46) | 662 (24.54) | 134 (13.87) | 1146 (24.78) | <0.001 KV, KS, VS |
| 2nd quartile [9–10] | 254 (26.46) | 769 (28.50) | 200 (20.70) | 1223 (26.45) | |
| 3rd quartile [11–12] | 224 (23.33) | 737 (27.32) | 301 (31.16) | 1262 (27.29) | |
| 4th quartile [≥13] | 132 (13.75) | 530 (19.64) | 331 (34.27) | 993 (21.48) | |
| Infectious disease count; n (%) | |||||
| None | 576 (60.00) | 1677 (62.16) | 622 (64.39) | 2875 (62.18) | 0.114 |
| One | 293 (30.52) | 752 (27.87) | 277 (28.68) | 1322 (28.59) | |
| Two | 77 (8.02) | 231 (8.56) | 61 (6.31) | 369 (7.98) | |
| Three | 11 (1.15) | 33 (1.22) | 6 (0.62) | 50 (1.08) | |
| Four | 3 (0.31) | 5 (0.19) | NA | 8 (0.17) | |
| Recurrent pneumonia cases; n (%) | 9 (0.94) | 16 (0.59) | 1 (0.10) | 26 (0.56) | 0.047 VS |
| Infectious traits; n (%) | |||||
| Tuberculosis | 12 (1.25) | 16 (0.59) | 6 (0.62) | 34 (0.74) | 0.110 |
| Pneumonia | 105 (10.94) | 235 (8.71) | 27 (2.80) | 367 (7.94) | <0.001 KS, VS |
| Hepatitis | 26 (2.71) | 30 (1.11) | 20 (2.07) | 76 (1.64) | 0.002 KV |
| Meningitis | 7 (0.73) | 16 (0.59) | 7 (0.73) | 30 (0.65) | 0.854 |
| Respiratory infections | 161 (16.77) | 535 (19.83) | 111 (11.49) | 807 (17.45) | <0.001 KS, VS |
| Gastrointestinal infections | 39 (4.06) | 55 (2.04) | 33 (3.42) | 127 (2.75) | 0.002 KV |
| Systemic infections | 26 (2.71) | 36 (1.33) | 25 (2.59) | 87 (1.88) | 0.005 KV, KS |
| Bacterial infections | 186 (19.38) | 571 (21.16) | 134 (13.87) | 891 (19.27) | <0.001 KS, VS |
| Viral infections | 74 (7.71) | 66 (2.45) | 42 (4.35) | 182 (3.94) | <0.001 KV, KS, VS |
| Appendectomy | 69 (7.19) | 169 (6.26) | 74 (7.66) | 312 (6.75) | 0.273 |
| Tonsillectomy | 168 (17.50) | 491 (18.20) | 160 (16.56) | 819 (17.71) | 0.517 |
| Infectious burden | |||||
| 0 | 576 (60.00) | 1677 (62.16) | 622 (64.39) | 2875 (62.18) | 0.001 KS, VS |
| 0.5 | 151 (15.73) | 385 (14.27) | 168 (17.39) | 704 (15.22) | |
| 1 | 149 (15.52) | 388 (14.38) | 116 (12.01) | 653 (14.12) | |
| 1.5 | 41 (4.27) | 164 (6.08) | 38 (3.93) | 243 (5.25) | |
| 2 | 26 (2.71) | 48 (1.78) | 19 (1.97) | 93 (2.01) | |
| 2.5 | 8 (0.83) | 22 (0.81) | 2 (0.21) | 32 (0.69) | |
| 3 | 8 (0.83) | 7 (0.26) | 1 (0.10) | 16 (0.35) | |
| 3.5 | 1 (0.11) | 3 (0.11) | NA | 4 (0.09) | |
| 4 | NA | 4 (0.15) | NA | 4 (0.09) | |
| Annual cold frequency, survey response | 244 | 771 | NA | 1015 | |
| Several times per year | 67 (27.46) | 160 (20.75) | NA | 227 (22.36) | 0.274 |
| Once a year | 83 (34.01) | 305 (39.56) | NA | 388 (38.23) | |
| Less than once a year | 67 (27.46) | 229 (29.70) | NA | 296 (29.16) | |
| Never or almost never | 27 (11.07) | 77 (9.99) | NA | 104 (10.25) | |
| Influenza frequency in the last 10 years, survey response | 197 | 655 | NA | 852 | |
| Every year | 9 (4.57) | 16 (2.44) | NA | 25 (2.93) | 0.857 |
| Several times | 48 (24.37) | 178 (27.18) | NA | 226 (26.53) | |
| Once | 57 (28.93) | 180 (27.48) | NA | 237 (27.82) | |
| Never | 83 (42.13) | 281 (42.90) | NA | 364 (42.72) |
| Trait | SNP | Location | Alleles * | EAF | OR (95% CI) | p | Gene | Variant Type |
|---|---|---|---|---|---|---|---|---|
| Hepatitis | rs188290902 | 11:131868501 | G/A | 0.006 | 1.13 (1.09–1.16) | 9.05 × 10−12 | NTM | Intron |
| rs72936092 | 2:159097061 | A/G | 0.004 | 1.16 (1.11–1.20) | 8.68 × 10−11 | CCDC148 | Intron | |
| rs17077736 | 13:33251708 | A/G | 0.017 | 1.06 (1.04–1.08) | 3.19 × 10−10 | PDS5B | Intron | |
| rs34447953 | 1:94730687 | A/C | 0.006 | 1.12 (1.08–1.15) | 7.99 × 10−10 | ARHGAP29 | Intron | |
| rs78111295 | 4:126203327 | T/C | 0.031 | 1.04 (1.03–1.06) | 1.21 × 10−9 | FAT4 | Upstream | |
| rs145607180 | 18:3452913 | T/C | 0.004 | 1.15 (1.10–1.19) | 2.97 × 10−9 | TGIF1 | Intron | |
| Meningitis | rs13358188 | 5:155325029 | G/A | 0.005 | 1.10 (1.08–1.13) | 8.46 × 10−13 | SGCD | Intron |
| rs17587821 | 1:226820605 | C/T | 0.013 | 1.06 (1.04–1.08) | 4.14 × 10−12 | ITPKB | 3′-UTR | |
| rs189257688 | 2:204544219 | T/C | 0.013 | 1.06 (1.04–1.07) | 1.31 × 10−11 | CD28 | Upstream | |
| rs188530871 | 15:76781806 | T/C | 0.006 | 1.08 (1.06–1.10) | 7.57 × 10−11 | SCAPER | Intron | |
| rs61878814 | 11:32119462 | C/T | 0.026 | 1.05 (1.03–1.06) | 1.41 × 10−10 | RCN1 | Intron | |
| rs116886525 | 11:116671391 | T/C | 0.006 | 1.09 (1.06–1.12) | 2.29 × 10−10 | APOA5 | Upstream | |
| rs35608792 | 1:8450425 | G/A | 0.023 | 1.04 (1.03–1.05) | 4.87 × 10−10 | RERE | Intron | |
| rs116306652 | 1:181673900 | T/G | 0.014 | 1.05 (1.04–1.07) | 1.46 × 10−9 | CACNA1E | Intron | |
| Pneumonia | rs187624194 | 12:7775204 | C/T | 0.009 | 1.15 (1.10–1.20) | 2.15 × 10−8 | APOBEC1 | Downstream |
| Systemic infections | rs146072725 | 11:77519509 | T/C | 0.008 | 1.13 (1.09–1.16) | 7.50 × 10−13 | RSF1 | Intron |
| rs142441889 | 20:16052044 | T/G | 0.006 | 1.13 (1.09–1.17) | 1.63 × 10−10 | MACROD2 | Downstream | |
| rs76931343 | 5:119876729 | G/A | 0.005 | 1.14 (1.10–1.18) | 3.70 × 10−10 | PRR16 | Intron | |
| rs58219087 | 19:48061409 | C/T | 0.004 | 1.20 (1.14–1.26) | 7.57 × 10−10 | ZNF541 | Upstream | |
| rs138336976 | 2:234090697 | A/G | 0.010 | 1.10 (1.07–1.12) | 1.26 × 10−9 | INPP5D | Intron | |
| rs192437130 | 2:85840200 | T/C | 0.004 | 1.21 (1.15–1.27) | 1.34 × 10−9 | USP39 | Intron | |
| rs6565193 | 16:30600260 | C/T | 0.028 | 1.05 (1.04–1.07) | 2.38 × 10−9 | ZNF785 | Upstream | |
| Tuberculosis | rs554596237 | 9:6953063 | G/T | 0.006 | 1.09 (1.07–1.12) | 2.06 × 10−11 | KDM4C | Intron |
| rs145254894 | 14:24683304 | A/C | 0.011 | 1.07 (1.05–1.09) | 1.17 × 10−10 | MDP1 | Missense | |
| rs570545343 | 5:65171990 | A/G | 0.015 | 1.06 (1.04–1.07) | 1.24 × 10−10 | NLN | Downstream | |
| rs117768315 | 12:47562000 | A/G | 0.010 | 1.06 (1.04–1.07) | 1.48 × 10−10 | PCED1B | Intron | |
| rs182320411 | 19:45821257 | A/G | 0.011 | 1.08 (1.05–1.10) | 9.69 × 10−10 | CKM | Intron | |
| rs140511699 | 8:102628803 | C/A | 0.008 | 1.07 (1.05–1.10) | 2.37 × 10−9 | GRHL2 | Intron | |
| rs140782448 | 8:100743166 | C/T | 0.004 | 1.10 (1.07–1.13) | 2.99 × 10−9 | VPS13B | Intron |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gelemanović, A.; Ćatipović Ardalić, T.; Pribisalić, A.; Hayward, C.; Kolčić, I.; Polašek, O. Genome-Wide Meta-Analysis Identifies Multiple Novel Rare Variants to Predict Common Human Infectious Diseases Risk. Int. J. Mol. Sci. 2023, 24, 7006. https://doi.org/10.3390/ijms24087006
Gelemanović A, Ćatipović Ardalić T, Pribisalić A, Hayward C, Kolčić I, Polašek O. Genome-Wide Meta-Analysis Identifies Multiple Novel Rare Variants to Predict Common Human Infectious Diseases Risk. International Journal of Molecular Sciences. 2023; 24(8):7006. https://doi.org/10.3390/ijms24087006
Chicago/Turabian StyleGelemanović, Andrea, Tatjana Ćatipović Ardalić, Ajka Pribisalić, Caroline Hayward, Ivana Kolčić, and Ozren Polašek. 2023. "Genome-Wide Meta-Analysis Identifies Multiple Novel Rare Variants to Predict Common Human Infectious Diseases Risk" International Journal of Molecular Sciences 24, no. 8: 7006. https://doi.org/10.3390/ijms24087006
APA StyleGelemanović, A., Ćatipović Ardalić, T., Pribisalić, A., Hayward, C., Kolčić, I., & Polašek, O. (2023). Genome-Wide Meta-Analysis Identifies Multiple Novel Rare Variants to Predict Common Human Infectious Diseases Risk. International Journal of Molecular Sciences, 24(8), 7006. https://doi.org/10.3390/ijms24087006






