Long-Term Transcriptomic Changes and Cardiomyocyte Hyperpolyploidy after Lactose Intolerance in Neonatal Rats
Abstract
1. Introduction
2. Results
2.1. Animals
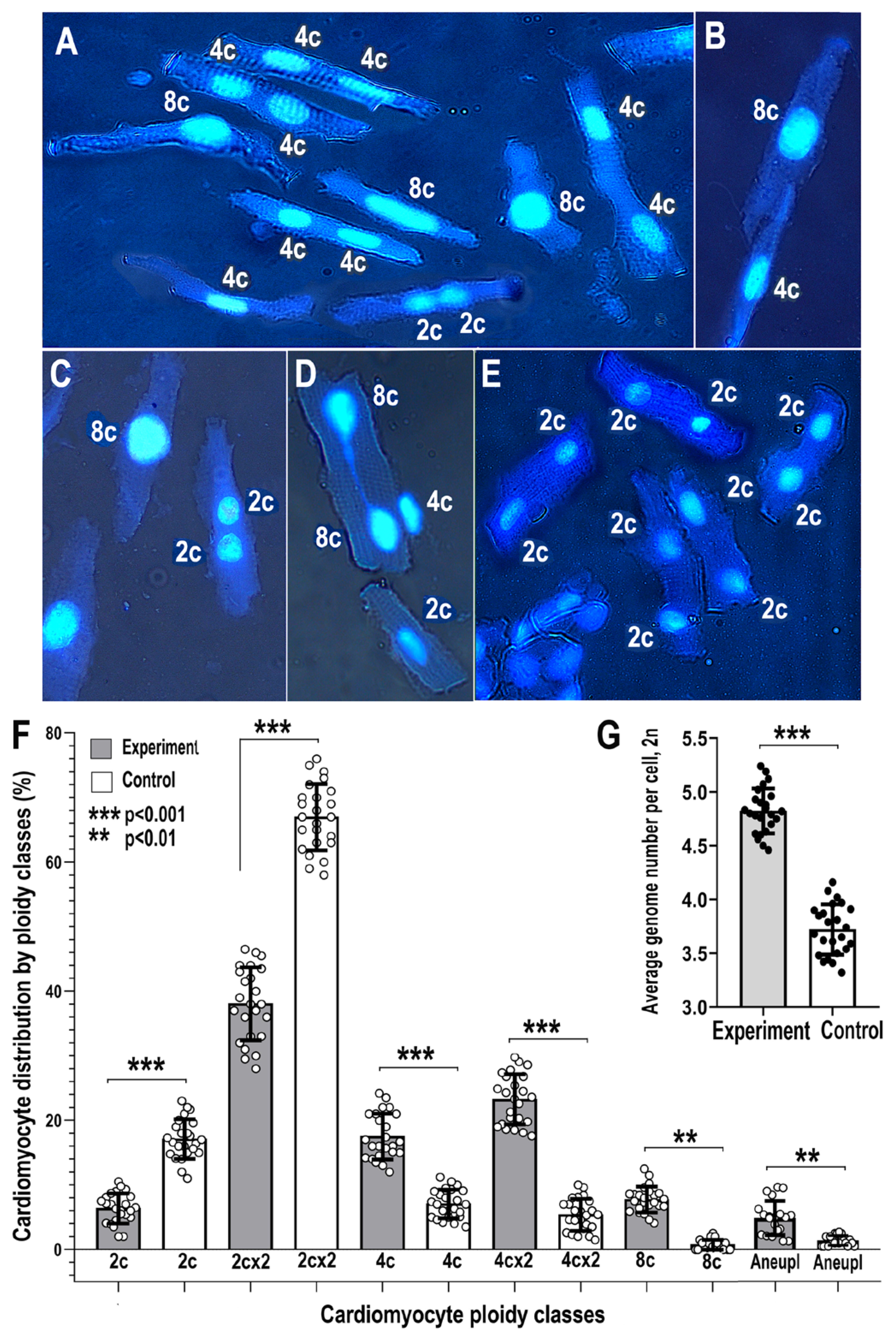
2.2. Cardiomyocyte DNA Instability and Polyploidy
2.3. Cardiomyocyte Protein Content Evaluation
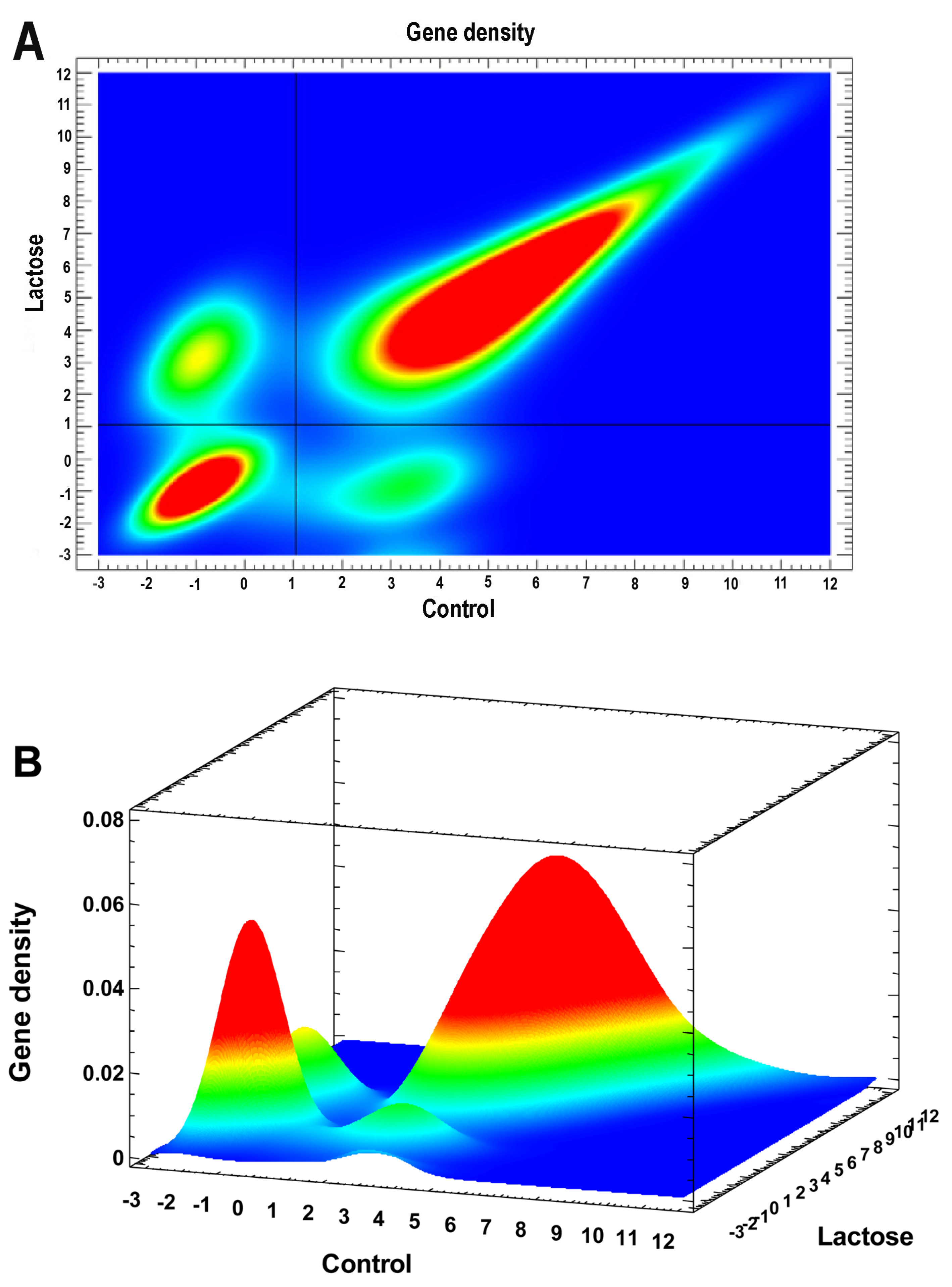
2.4. Cardiac Left Ventricle (LV) Transcriptome Changes in Adult Rats after NLI
2.4.1. General Picture
2.4.2. NLI Is Associated with Long-Term Persistence of Features of DNA Instability and DNA Damage Response
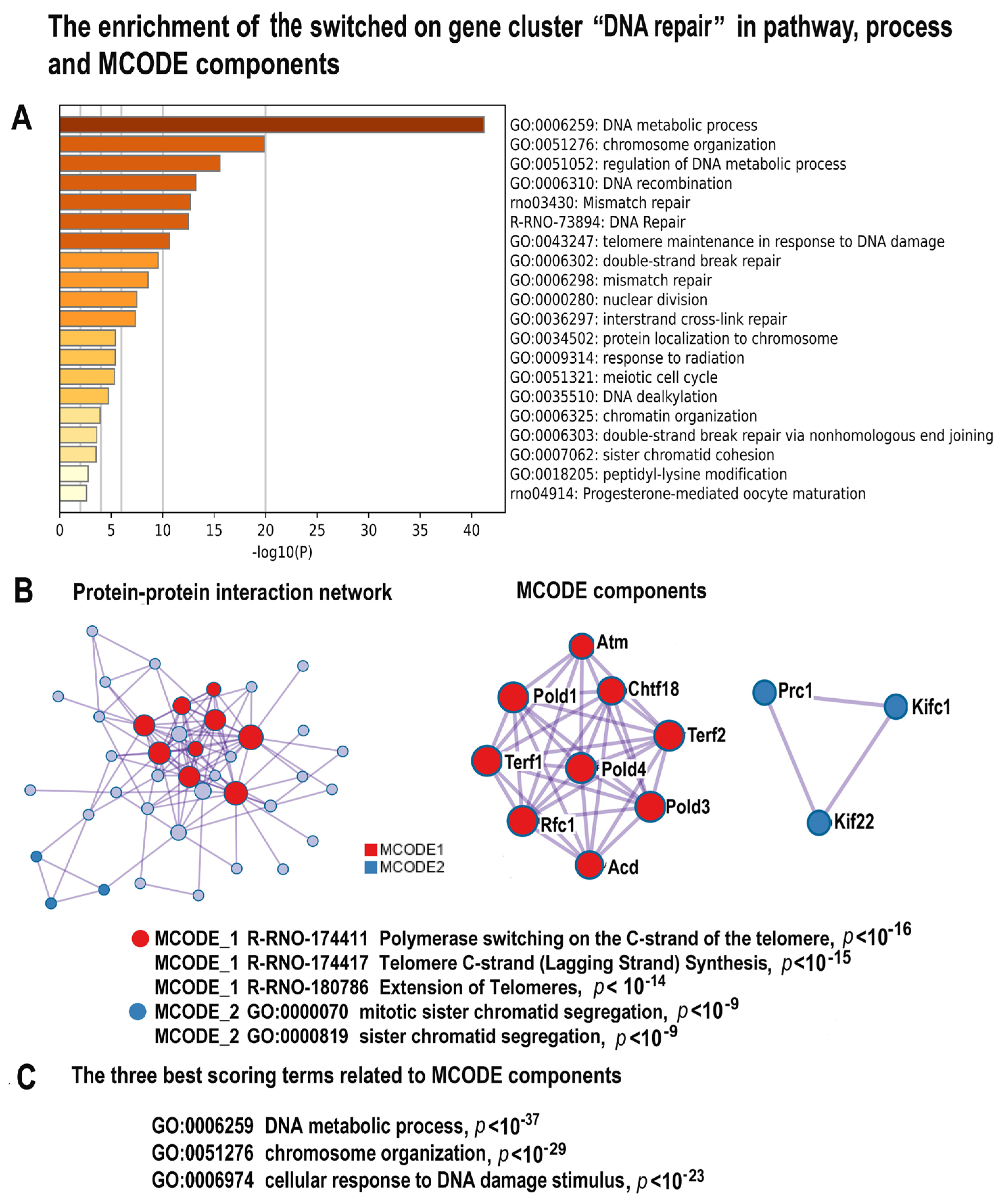
2.4.3. NLI Induces Gene Modules Related to Immunity and Inflammation
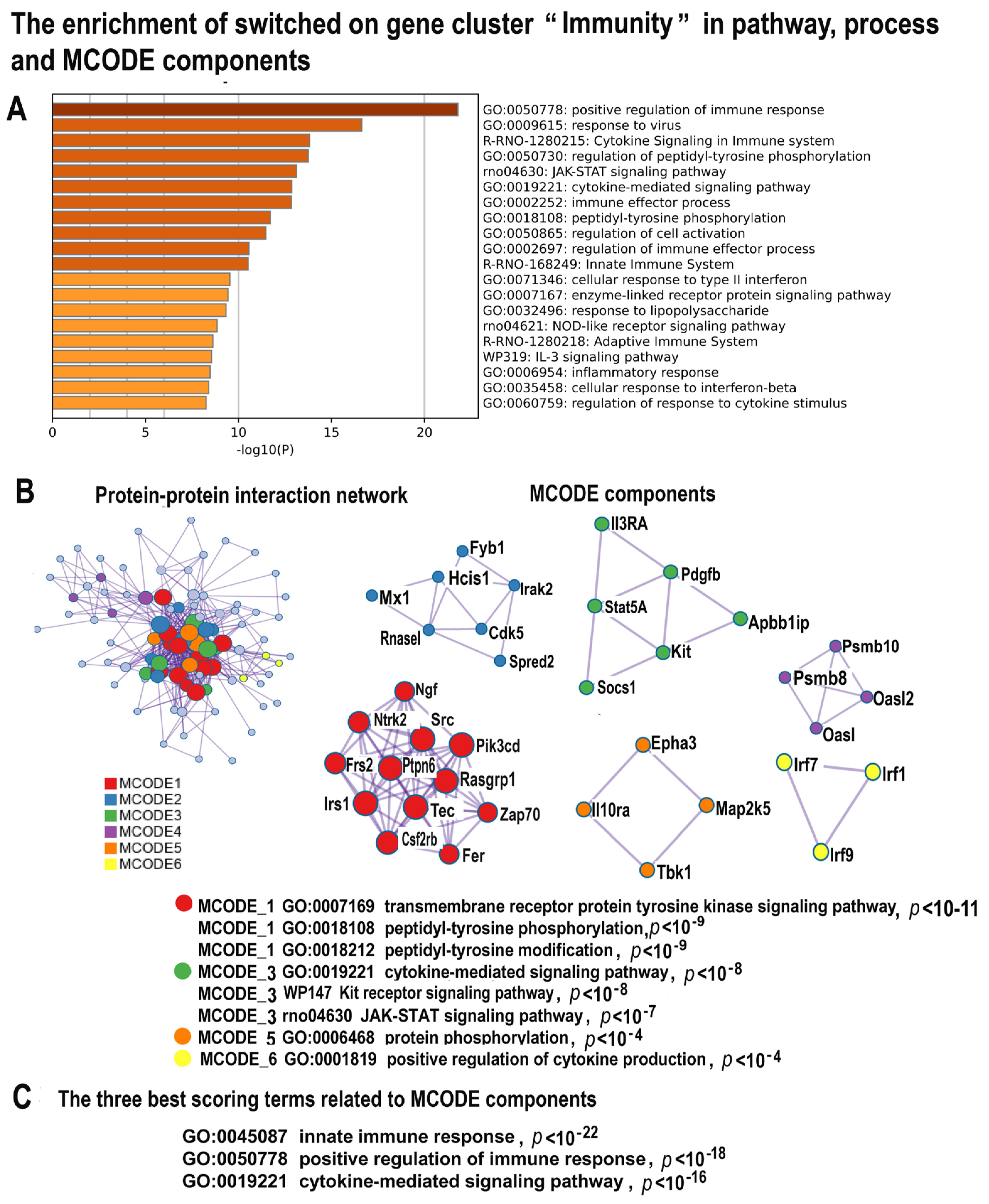
2.4.4. NLI Promotes Fibrosis
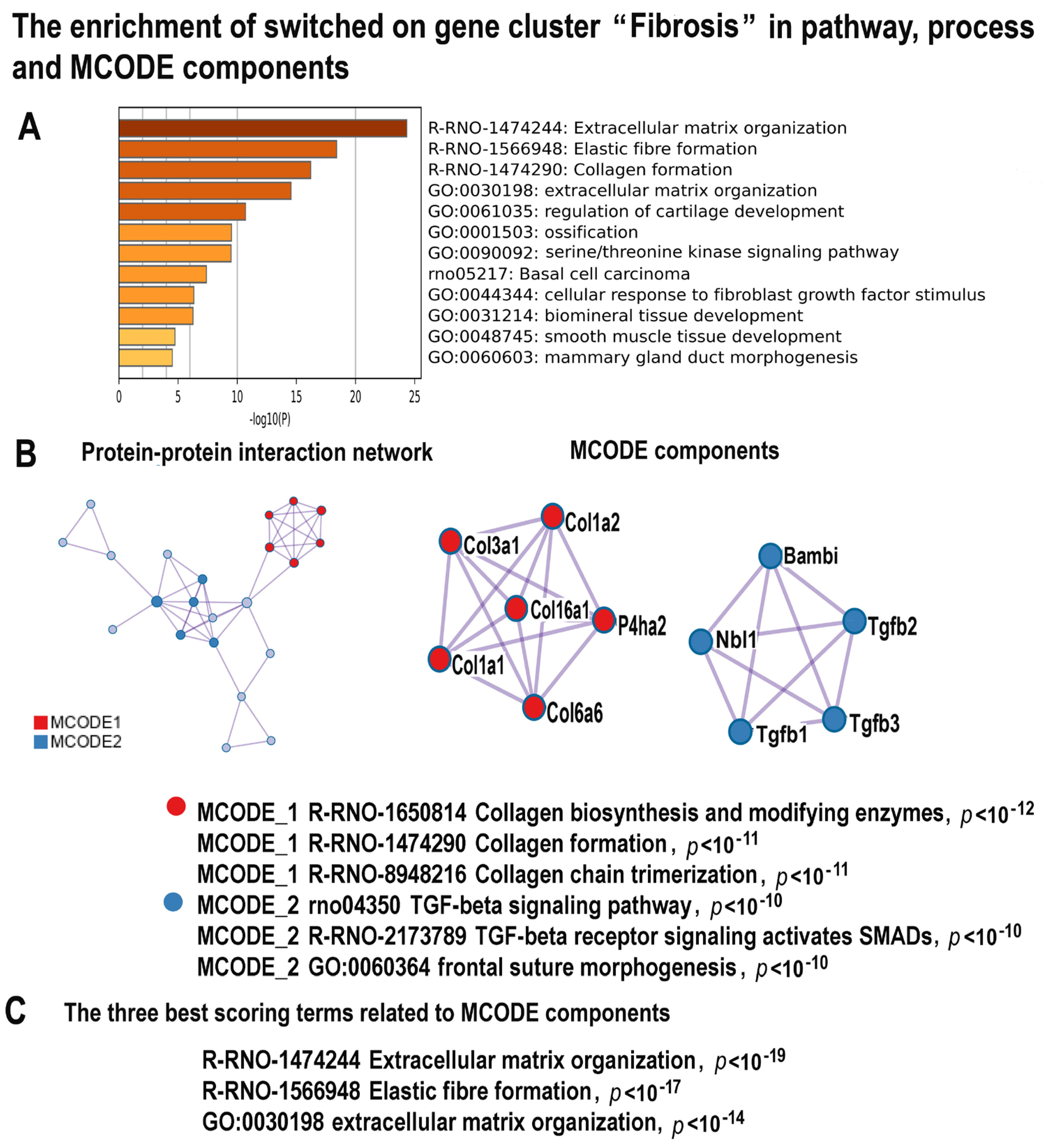
2.4.5. NLI Activates Gene Modules Related to Transcription and Ribosome Biogenesis
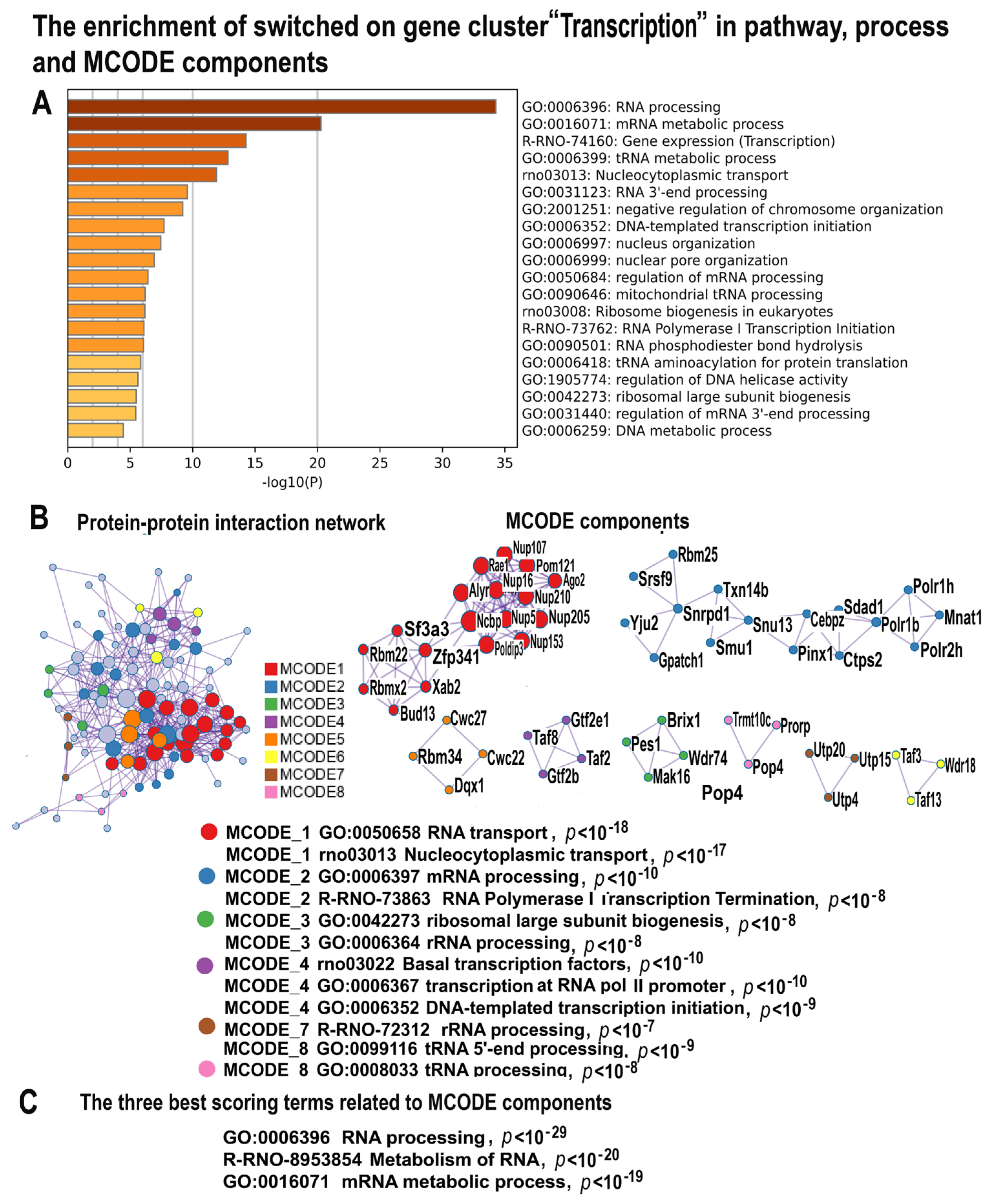
2.4.6. NLI Impairs Muscle Contraction via the Downregulation of Pathways Related to Calcium Signaling, Thyroid Hormone, and Circadian Clock
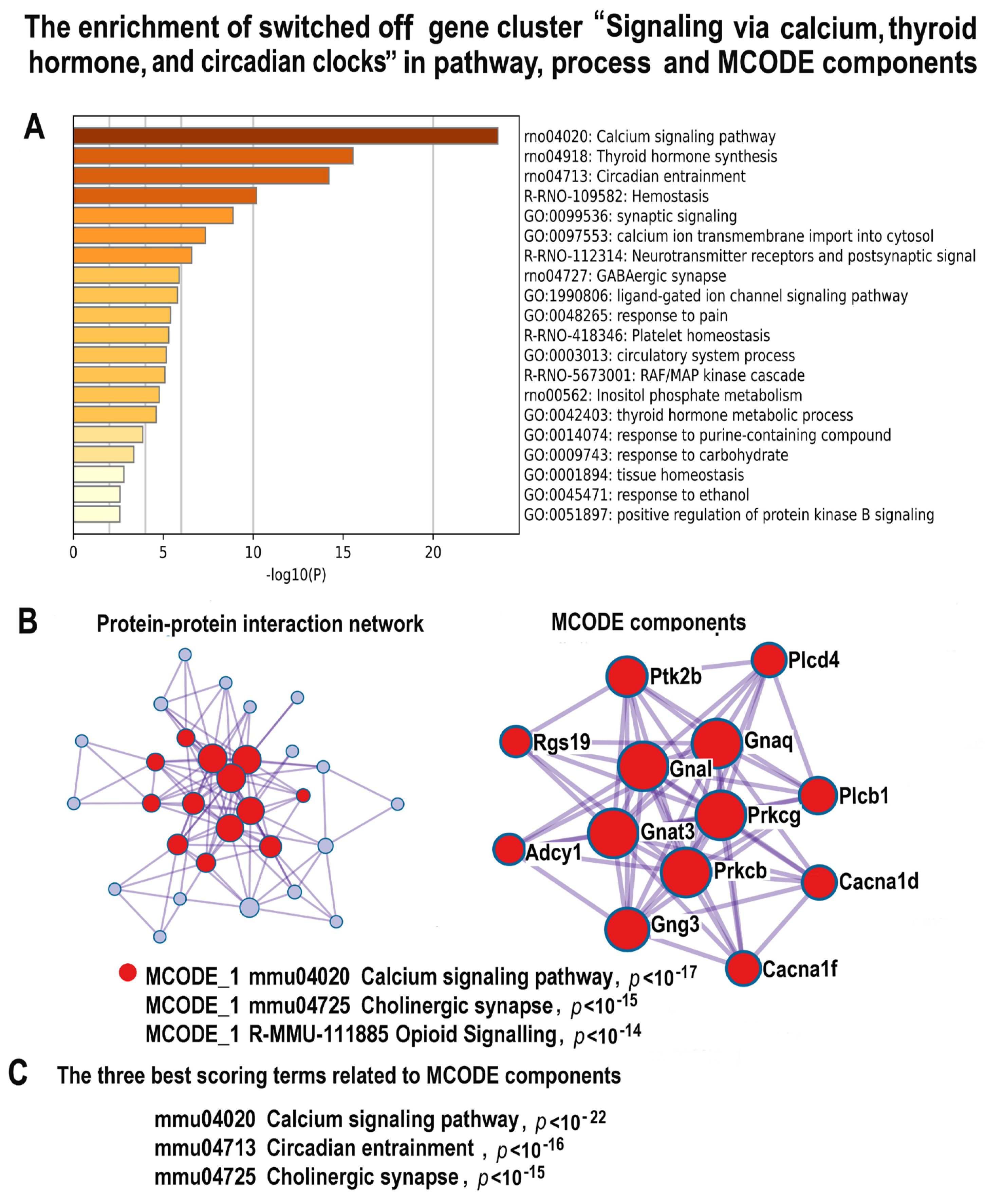
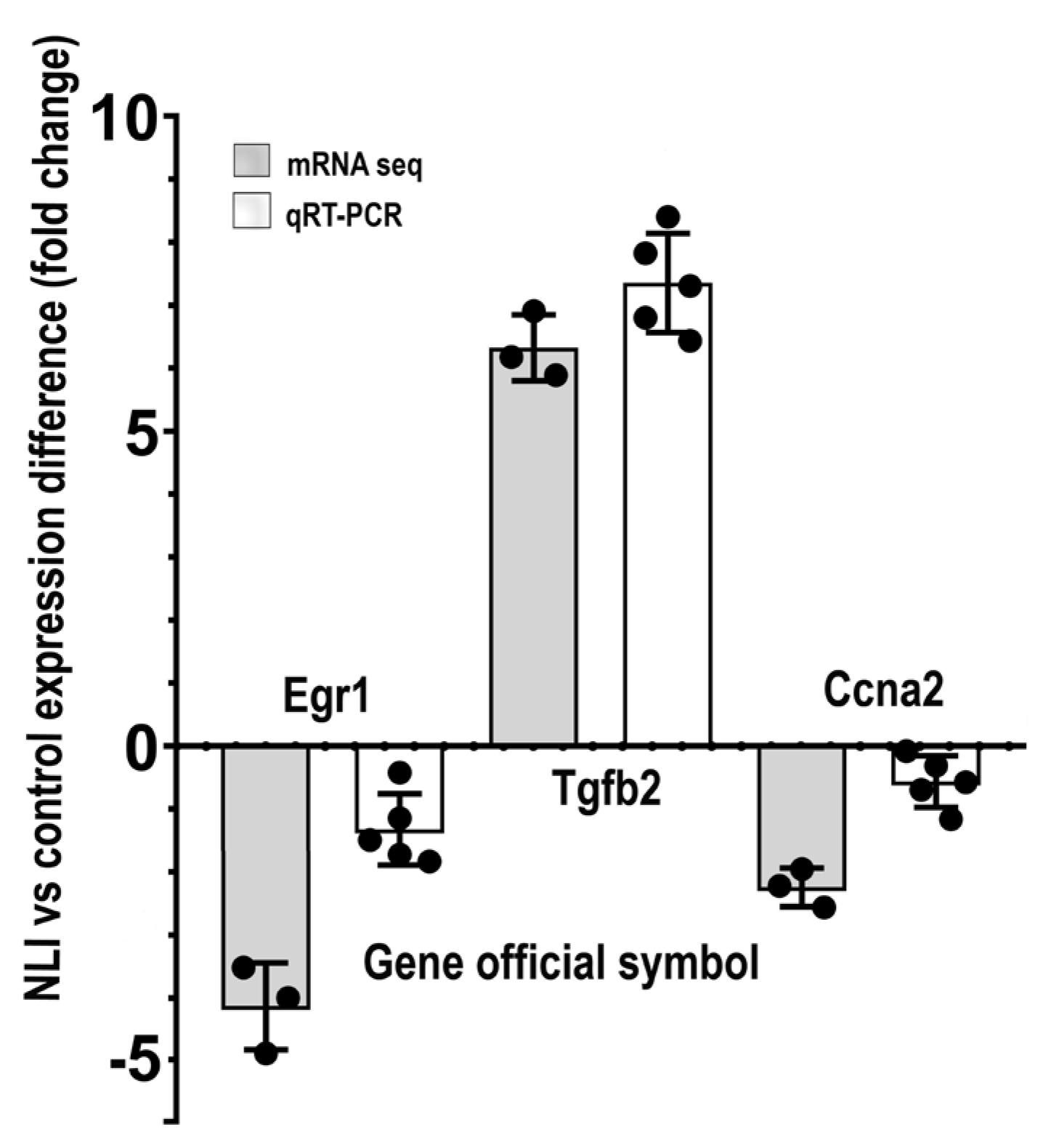
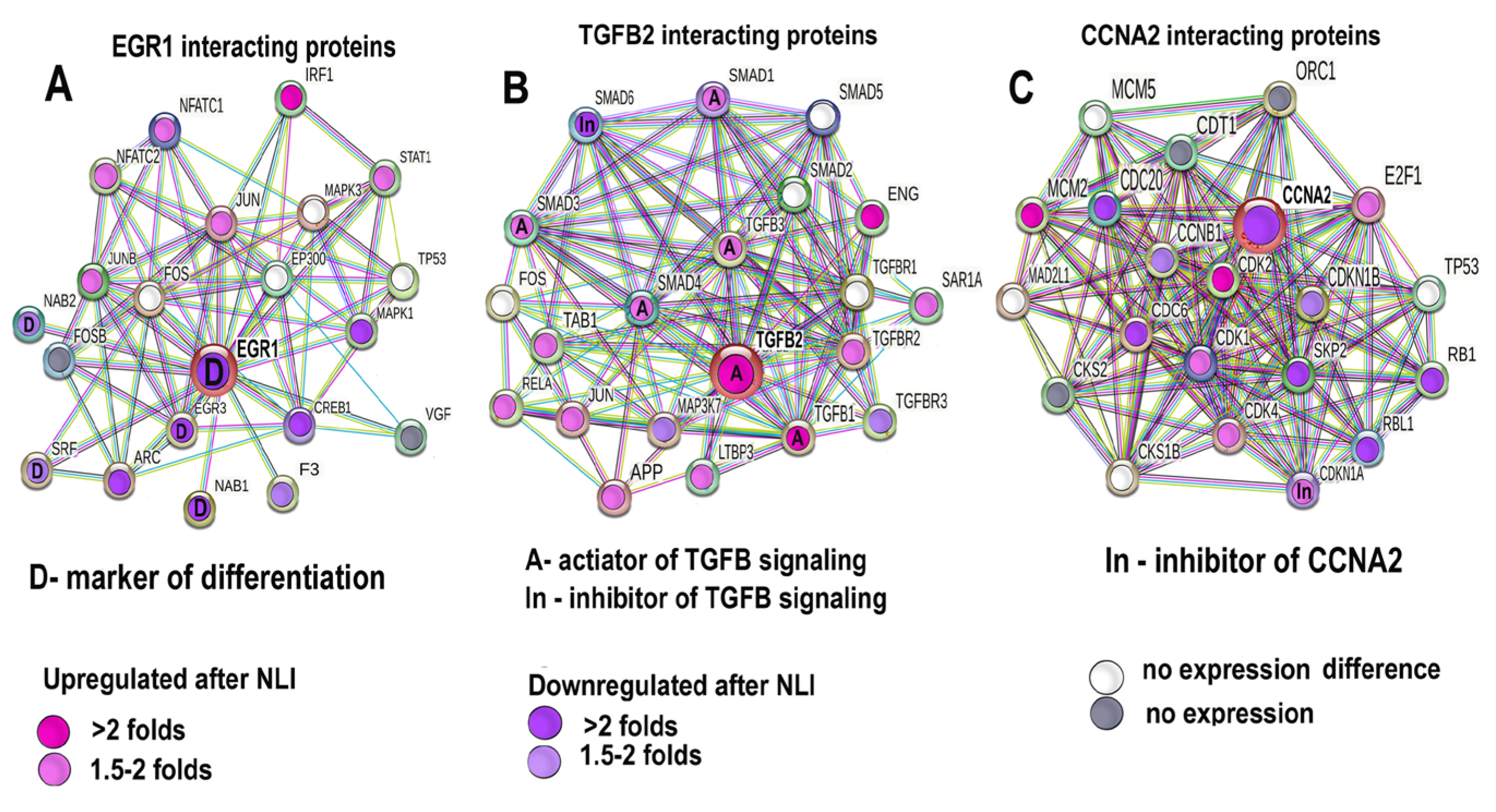
2.4.7. Validation of mRNA-Seq Data of the Experimental vs. Control Expression Difference for Egr1, Tgfb2, and Ccna2 by qRT-PCR and Protein Interaction Analysis
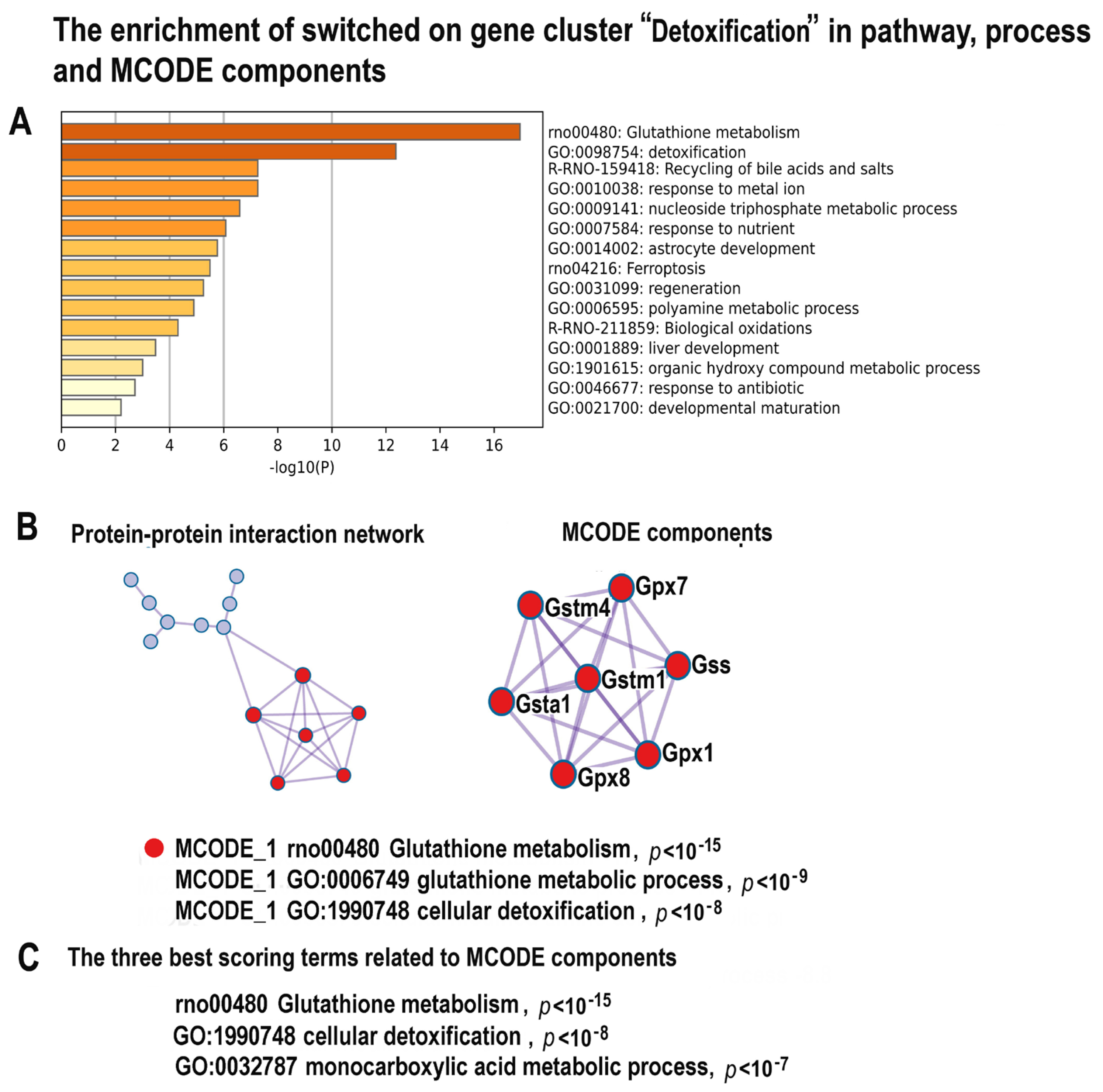
2.4.8. NLI Is Associated with Long-Term Deprivation of Glutathione Signaling, Detoxification, and Energy Metabolism
3. Discussion
3.1. Significance of NLI Studying, Main Findings, and Research Status
3.2. Strength, Possible Limitations, Future Directions, and Medical Applications of the Study
3.3. Conclusions
4. Materials and Methods
4.1. Animals
4.2. Statement of Ethics
4.3. Experimental Design and Lactose-Containing Diet
4.4. Cardiomyocyte Isolation
4.5. Cardiomyocyte Staining and Ploidy Evaluation
4.6. Cardiomyocyte Morphological Features of DNA Instability
4.7. Cardiomyocyte Protein Content Evaluation
4.8. mRNA Sequencing
4.9. Validation of mRNA-Seq by qRT-PCR
4.10. Data Purification from Noise and Gene Module Analysis
4.11. Tgfb2, Egr1, and Ccna2 Close-Interactants Network Construction
4.12. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harikrishnan, S.; Jeemon, P.; Mini, G.K.; Thankappan, K.R.; Sylaja, P. GBD 2017 Causes of Death Collaborators Global, Regional, and National Age-Sex-Specific Mortality for 282 Causes of Death in 195 Countries and Territories, 1980–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef]
- Mokdad, A.H.; Marks, J.S.; Stroup, D.F.; Gerberding, J.L. Actual Causes of Death in the United States, 2000. JAMA 2004, 291, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Segovia, S.A.; Vickers, M.H.; Harrison, C.J.; Patel, R.; Gray, C.; Reynolds, C.M. Maternal High-Fat and High-Salt Diets Have Differential Programming Effects on Metabolism in Adult Male Rat Offspring. Front. Nutr. 2018, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Lurbe, E.; Ingelfinger, J. Developmental and Early Life Origins of Cardiometabolic Risk Factors: Novel Findings and Implications. Hypertension 2021, 77, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Vrselja, A.; Pillow, J.J.; Black, M.J. Effect of Preterm Birth on Cardiac and Cardiomyocyte Growth and the Consequences of Antenatal and Postnatal Glucocorticoid Treatment. J. Clin. Med. 2021, 10, 3896. [Google Scholar] [CrossRef]
- Visker, J.R.; Ferguson, D.P. Postnatal undernutrition in mice causes cardiac arrhythmogenesis which is exacerbated when pharmacologically stressed. J. Dev. Orig. Heal. Dis. 2018, 9, 417–424. [Google Scholar] [CrossRef]
- Arima, Y.; Fukuoka, H. Developmental origins of health and disease theory in cardiology. J. Cardiol. 2020, 76, 14–17. [Google Scholar] [CrossRef]
- Hochberg, Z.; Belsky, J. Evo-devo of human adolescence: Beyond disease models of early puberty. BMC Med. 2013, 11, 113. [Google Scholar] [CrossRef]
- Koemel, N.A.; Skilton, M.R. Epigenetic Aging in Early Life: Role of Maternal and Early Childhood Nutrition. Curr. Nutr. Rep. 2022, 11, 318–328. [Google Scholar] [CrossRef]
- Hochberg, Z.; Feil, R.; Constancia, M.; Fraga, M.; Junien, C.; Carel, J.-C.; Boileau, P.; Le Bouc, Y.; Deal, C.L.; Lillycrop, K.; et al. Child Health, Developmental Plasticity, and Epigenetic Programming. Endocr. Rev. 2010, 32, 159–224. [Google Scholar] [CrossRef]
- Van Abeelen, A.F.; Elias, S.G.; Bossuyt, P.M.; Grobbee, D.E.; Van Der Schouw, Y.T.; Roseboom, T.J.; Uiterwaal, C.S. Cardiovascular consequences of famine in the young. Eur. Hear. J. 2011, 33, 538–545. [Google Scholar] [CrossRef]
- Ferguson, D.; Monroe, T.O.; Heredia, C.P.; Fleischman, R.E.; Rodney, G.; Taffet, G.E.; Fiorotto, M.L. Postnatal undernutrition alters adult female mouse cardiac structure and function leading to limited exercise capacity. J. Physiol. 2019, 597, 1855–1872. [Google Scholar] [CrossRef] [PubMed]
- Bensley, J.G.; Stacy, V.K.; De Matteo, R.; Harding, R.; Black, M.J. Cardiac remodelling as a result of pre-term birth: Implications for future cardiovascular disease. Eur. Heart J. 2010, 31, 2058–2066. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Sidorenko, N.V.; Beyer, T.V.; Vinogradov, A.E. Neonatal cardiomyocyte ploidy reveals critical windows of heart development. Int. J. Cardiol. 2010, 141, 81–91. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Irizarry, R.A. Evolution in Health and Medicine Sackler Colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. 1), 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Finch, C.E. Evolution in Health and Medicine Sackler Colloquium: Evolution of the human lifespan and diseases of aging: Roles of infection, inflammation, and nutrition. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. 1), 1718–1724. [Google Scholar] [CrossRef] [PubMed]
- Nesse, R.M.; Bergstrom, C.T.; Ellison, P.T.; Flier, J.S.; Gluckman, P.; Govindaraju, D.R.; Niethammer, D.; Omenn, G.S.; Perlman, R.L.; Schwartz, M.D.; et al. Evolution in Health and Medicine Sackler Colloquium: Making evolutionary biology a basic science for medicine. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. 1), 1800–1807. [Google Scholar] [CrossRef]
- Finch, C.E.; Crimmins, E.M. Inflammatory Exposure and Historical Changes in Human Life-Spans. Science 2004, 305, 1736–1739. [Google Scholar] [CrossRef]
- Crimmins, E.M.; Finch, C.E. Infection, inflammation, height, and longevity. Proc. Natl. Acad. Sci. USA 2005, 103, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Curione, M.; Aratari, A.; Amato, S.; Colotto, M.; Barbato, M.; Leone, S.; Tego, A.; Panetti, D.; Parlapiano, C. A study on QT interval in patients affected with inflammatory bowel disease without cardiac involvement. Intern. Emerg. Med. 2010, 5, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Gluckman, P.D.; Hanson, M.A.; Low, F.M. Evolutionary and developmental mismatches are consequences of adaptive developmental plasticity in humans and have implications for later disease risk. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180109. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E.; Kudryavtsev, B. Cardiomyocyte ploidy levels in birds with different growth rates. J. Exp. Zool. 2001, 289, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Anatskaya, O.V.; Vinogradov, A.E. Myocyte ploidy in heart chambers of birds with different locomotor activity. J. Exp. Zool. 2002, 293, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.E.; Anatskaya, O.V.; Kudryavtsev, B. Relationship of hepatocyte ploidy levels with body size and growth rate in mammals. Genome 2001, 44, 350–360. [Google Scholar] [CrossRef]
- Shiels, H.A. Avian cardiomyocyte architecture and what it reveals about the evolution of the vertebrate heart. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022, 377, 20210332. [Google Scholar] [CrossRef] [PubMed]
- Anatskaya, O.V.; Vinogradov, A.E. Heart and liver as developmental bottlenecks of mammal design: Evidence from cell polyploidization: Heart and liver as bottlenecks of mammal design. Biol. J. Linn. Soc. 2004, 83, 175–186. [Google Scholar] [CrossRef]
- Rodríguez, L.; Cervantes, E.; Ortiz, R. Malnutrition and Gastrointestinal and Respiratory Infections in Children: A Public Health Problem. Int. J. Environ. Res. Public Health 2011, 8, 1174–1205. [Google Scholar] [CrossRef]
- Alter, S.J.; Vidwan, N.K.; Sobande, P.O.; Omoloja, A.; Bennett, J.S. Common Childhood Bacterial Infections. Curr. Probl. Pediatr. Adolesc. Health Care 2011, 41, 256–283. [Google Scholar] [CrossRef]
- Guerrant, R.L.; Oriá, R.B.; Moore, S.R.; Oriá, M.O.; Lima, A.A. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr. Rev. 2008, 66, 487–505. [Google Scholar] [CrossRef]
- Khalil, I.A.; Troeger, C.; Rao, P.C.; Blacker, B.F.; Brown, A.; Brewer, T.G.; Colombara, D.V.; De Hostos, E.L.; Engmann, C.; Guerrant, R.L.; et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: A meta-analyses study. Lancet Glob. Health 2018, 6, e758–e768. [Google Scholar] [CrossRef]
- Matthews, S.B.; Waud, J.P.; Roberts, A.G.; Campbell, A.K. Systemic lactose intolerance: A new perspective on an old problem. Postgrad. Med. J. 2005, 81, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, X.; Zhou, X.; Cai, J. The Role and Mechanism of Intestinal Flora in Blood Pressure Regulation and Hypertension Development. Antioxid. Redox Signal. 2021, 34, 811–830. [Google Scholar] [CrossRef]
- Yano, Y.; Niiranen, T.J. Gut Microbiome over a Lifetime and the Association with Hypertension. Curr. Hypertens. Rep. 2021, 23, 15. [Google Scholar] [CrossRef]
- Crispel, Y.; Shaoul, R.; Khamaise, R.; Sabo, E.; Hochberg, Z. Effect of weaning age on the small intestine mucosa of rats. Appl. Physiol. Nutr. Metab. 2019, 44, 985–989. [Google Scholar] [CrossRef]
- Anatskaia, O.V.; Sidorenko, N.V.; Beĭer, T.V.; Vinogradov, A.E. Neonatal gastroenteritis triggers long-term cardiomyocyte atrophy, remodeling and irreversible hyperpolyploidization. Kardiologiia 2010, 50, 35–44. [Google Scholar] [PubMed]
- Anatskaia, O.V.; Matveev, I.V.; Sidorenko, N.V.; Kharchenko, M.V.; Kropotov, A.V.; Vinogradov, A.E. Changes in the heart of neonatal rats after cryptosporidial gastroenteritis of different degrees of severity. Zh. Evol. Biokhim. Fiziol. 2013, 49, 357–365. [Google Scholar] [CrossRef]
- Büller, H.; Rings, E.; Montgomery, R.K.; Grand, R.J. Clinical Aspects of Lactose Intolerance in Children and Adults. Scand. J. Gastroenterol. Suppl. 1991, 188, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Jasielska, M.; Grzybowska-Chlebowczyk, U. Lactose Malabsorption and Lactose Intolerance in Children with Inflammatory Bowel Diseases. Gastroenterol. Res. Pract. 2019, 2019, 2507242. [Google Scholar] [CrossRef]
- Al-Beltagi, M.; Saeed, N.K.; Bediwy, A.S.; Elbeltagi, R. Cow’s milk-induced gastrointestinal disorders: From infancy to adulthood. World J. Clin. Pediatr. 2022, 11, 437–454. [Google Scholar] [CrossRef]
- Grant, W.B. Commentary: Ecologic studies in identifying dietary risk factors for coronary heart disease and cancer. Int. J. Epidemiol. 2008, 37, 1209–1211. [Google Scholar] [CrossRef]
- Segall, J.J. Hypothesis: Is Lactose a Dietary Risk Factor for Ischaemic Heart Disease? Int. J. Epidemiol. 2008, 37, 1204–1208. [Google Scholar] [CrossRef] [PubMed]
- Hunter, R.J.; Patel, V.B.; Miell, J.P.; Wong, H.J.; Marway, J.S.; Richardson, P.J.; Preedy, V.R. Diarrhea Reduces the Rates of Cardiac Protein Synthesis in Myofibrillar Protein Fractions in Rats In Vivo. J. Nutr. 2001, 131, 1513–1519. [Google Scholar] [CrossRef] [PubMed]
- Campbell, A.K.; Wann, K.T.; Matthews, S.B. Lactose causes heart arrhythmia in the water flea Daphnia pulex. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2004, 139, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Bo-Htay, C.; Palee, S.; Apaijai, N.; Chattipakorn, S.C.; Chattipakorn, N. Effects of d-galactose-induced ageing on the heart and its potential interventions. J. Cell. Mol. Med. 2018, 22, 1392–1410. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-S.; Zhang, X.; Ke, Z.-Z.; Wen, X.-Y.; Li, W.-D.; Liu, W.-B.; Zhuang, X.-D.; Liao, L.-Z. D-galactose-induced cardiac ageing: A review of model establishment and potential interventions. J. Cell. Mol. Med. 2022, 26, 5335–5359. [Google Scholar] [CrossRef] [PubMed]
- Anatskaya, O.V.; Vinogradov, A.E.; Vainshelbaum, N.M.; Giuliani, A.; Erenpreisa, J. Phylostratic Shift of Whole-Genome Duplications in Normal Mammalian Tissues towards Unicellularity Is Driven by Developmental Bivalent Genes and Reveals a Link to Cancer. Int. J. Mol. Sci. 2020, 21, 8759. [Google Scholar] [CrossRef]
- Folguieri, M.S.; Franco, A.T.B.; Vieira, A.S.; Gontijo, J.A.R.; Boer, P.A. Transcriptome and morphological analysis on the heart in gestational protein-restricted aging male rat offspring. Front. Cell Dev. Biol. 2022, 10, 892322. [Google Scholar] [CrossRef]
- Mantzouratou, P.; Lavecchia, A.M.; Novelli, R.; Xinaris, C. Thyroid Hormone Signalling Alteration in Diabetic Nephropathy and Cardiomyopathy: A “Switch” to the Foetal Gene Programme. Curr. Diabetes Rep. 2020, 20, 58. [Google Scholar] [CrossRef]
- Spurrell, C.H.; Barozzi, I.; Kosicki, M.; Mannion, B.J.; Blow, M.J.; Fukuda-Yuzawa, Y.; Slaven, N.; Afzal, S.Y.; Akiyama, J.A.; Afzal, V.; et al. Genome-wide fetalization of enhancer architecture in heart disease. Cell Rep. 2022, 40, 111400. [Google Scholar] [CrossRef]
- D’Antonio, M.; Nguyen, J.P.; Arthur, T.D.; Matsui, H.; Donovan, M.K.R.; D’Antonio-Chronowska, A.; Frazer, K.A. In heart failure reactivation of RNA-binding proteins is associated with the expression of 1523 fetal-specific isoforms. PLoS Comput. Biol. 2022, 18, e1009918. [Google Scholar] [CrossRef]
- Bergmann, O. Cardiomyocytes in congenital heart disease: Overcoming cytokinesis failure in tetralogy of Fallot. J. Thorac. Cardiovasc. Surg. 2021, 161, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- Fanucchi, S.; Shibayama, Y.; Mhlanga, M.M. Are genes switched on when they kiss? Nucleus 2014, 5, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Trietsch, M.D.; Nooij, L.S.; Gaarenstroom, K.; van Poelgeest, M.I. Genetic and epigenetic changes in vulvar squamous cell carcinoma and its precursor lesions: A review of the current literature. Gynecol. Oncol. 2015, 136, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Biava, P.M. The Use of Stem Cell Differentiation Stage Factors (SCDSFs) Taken from Zebrafish Embryos during Organogenesis and Their Role in Regulating the Gene Expression of Normal and Pathological (Stem) Cells. Int. J. Mol. Sci. 2020, 21, 4914. [Google Scholar] [CrossRef] [PubMed]
- Petushkova, N.A.; Pyatnitskiy, M.; Rudenko, V.A.; Larina, O.V.; Trifonova, O.; Kisrieva, J.S.; Samenkova, N.F.; Kuznetsova, G.P.; Karuzina, I.I.; Lisitsa, A.V. Applying of Hierarchical Clustering to Analysis of Protein Patterns in the Human Cancer-Associated Liver. PLoS ONE 2014, 9, e103950. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.; Bui, T.T.; Helmy, M.; Selvarajoo, K. Identifying toggle genes from transcriptome-wide scatter: A new perspective for biological regulation. Genomics 2022, 114, 215–228. [Google Scholar] [CrossRef]
- Hong, Y.; Sonneville, R.; Wang, B.; Scheidt, V.; Meier, B.; Woglar, A.; Demetriou, S.; Labib, K.; Jantsch, V.; Gartner, A. LEM-3 is a midbody-tethered DNA nuclease that resolves chromatin bridges during late mitosis. Nat. Commun. 2018, 9, 728. [Google Scholar] [CrossRef]
- Kim, J.H.; Youn, Y.; Hwang, J.-H. NCAPH Stabilizes GEN1 in Chromatin to Resolve Ultra-Fine DNA Bridges and Maintain Chromosome Stability. Mol. Cells 2022, 45, 792–805. [Google Scholar] [CrossRef]
- Das, L. Epigenetic alterations impede epithelial-mesenchymal transition by modulating centrosome amplification and Myc/RAS axis in triple negative breast cancer cells. Sci. Rep. 2023, 13, 2458. [Google Scholar] [CrossRef]
- Pertea, M. The Human Transcriptome: An Unfinished Story. Genes 2012, 3, 344–360. [Google Scholar] [CrossRef]
- Ham, L.; Jackson, M.; Stumpf, M.P. Pathway Dynamics Can Delineate the Sources of Transcriptional Noise in Gene Expression. eLife 2021, 10, e69324. [Google Scholar] [CrossRef]
- Johnstone, C.P.; Galloway, K.E. Engineering cellular symphonies out of transcriptional noise. Nat. Rev. Mol. Cell Biol. 2021, 22, 369–370. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.; Anatskaya, O. Evolutionary framework of the human interactome: Unicellular and multicellular giant clusters. Biosystems 2019, 181, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Kuchinsky, A.; Morris, J.H.; States, D.J.; Meng, F. GLay: Community structure analysis of biological networks. Bioinformatics 2010, 26, 3135–3137. [Google Scholar] [CrossRef]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. clusterMaker: A multi-algorithm clustering plugin for Cytoscape. BMC Bioinform. 2011, 12, 436. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Vinogradov, A.E. Accelerated pathway evolution in mouse-like rodents involves cell cycle control. Mamm. Genome 2015, 26, 609–618. [Google Scholar] [CrossRef]
- Vinogradov, A.E.; Anatskaya, O.V. Cell-cycle dependence of transcriptome gene modules: Comparison of regression lines. FEBS J. 2020, 287, 4427–4439. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.E. Consolidation of slow or fast but not moderately evolving genes at the level of pathways and processes. Gene 2015, 561, 30–34. [Google Scholar] [CrossRef]
- Vainshelbaum, N.M.; Salmina, K.; Gerashchenko, B.I.; Lazovska, M.; Zayakin, P.; Cragg, M.S.; Pjanova, D.; Erenpreisa, J. Role of the Circadian Clock “Death-Loop” in the DNA Damage Response Underpinning Cancer Treatment Resistance. Cells 2022, 11, 880. [Google Scholar] [CrossRef] [PubMed]
- Nomura, S.; Satoh, M.; Fujita, T.; Higo, T.; Sumida, T.; Ko, T.; Yamaguchi, T.; Tobita, T.; Naito, A.T.; Ito, M.; et al. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat. Commun. 2018, 9, 4435. [Google Scholar] [CrossRef] [PubMed]
- Caravia, X.M.; Ramirez-Martinez, A.; Gan, P.; Wang, F.; McAnally, J.R.; Xu, L.; Bassel-Duby, R.; Liu, N.; Olson, E.N. Loss of function of the nuclear envelope protein LEMD2 causes DNA damage–dependent cardiomyopathy. J. Clin. Investig. 2022, 132, e158897. [Google Scholar] [CrossRef]
- Sweet, M.E.; Cocciolo, A.; Slavov, D.; Jones, K.L.; Sweet, J.R.; Graw, S.L.; Reece, T.B.; Ambardekar, A.V.; Bristow, M.R.; Mestroni, L.; et al. Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure. BMC Genom. 2018, 19, 812. [Google Scholar] [CrossRef] [PubMed]
- Zhan, R.-Z.; Rao, L.; Chen, Z.; Strash, N.; Bursac, N. Loss of sarcomeric proteins via upregulation of JAK/STAT signaling underlies interferon-γ-induced contractile deficit in engineered human myocardium. Acta Biomater. 2021, 126, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Nakao, S.; Tsukamoto, T.; Ueyama, T.; Kawamura, T. STAT3 for Cardiac Regenerative Medicine: Involvement in Stem Cell Biology, Pathophysiology, and Bioengineering. Int. J. Mol. Sci. 2020, 21, 1937. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E. Whole-Genome Duplications in Evolution, Ontogeny, and Pathology: Complexity and Emergency Reserves. Mol. Biol. 2021, 55, 813–827. [Google Scholar] [CrossRef]
- Anatskaya, O. Polyploidy Activates Biological Pathways Related to Morphogenesis in Mammalian Tissues. MOJ 2018, 6, 90–93. [Google Scholar] [CrossRef]
- Sharma, S.; Ying, J.; Razeghi, P.; Stepkowski, S.; Taegtmeyer, H. Atrophic Remodeling of the Transplanted Rat Heart. Cardiology 2006, 105, 128–136. [Google Scholar] [CrossRef]
- Kishore, R.; Verma, S.K. Roles of STATs signaling in cardiovascular diseases. JAKSTAT 2012, 1, 118–124. [Google Scholar] [CrossRef]
- Broughton, K.M.; Sussman, M.A. Adult Cardiomyocyte Cell Cycle Detour: Off-ramp to Quiescent Destinations. Trends Endocrinol. Metab. 2019, 30, 557–567. [Google Scholar] [CrossRef]
- Segura, A.M.; Frazier, O.H.; Buja, L.M. Fibrosis and heart failure. Hear. Fail. Rev. 2012, 19, 173–185. [Google Scholar] [CrossRef]
- Ren, D.; Song, J.; Ni, M.; Kang, L.; Guo, W. Regulatory Mechanisms of Cell Polyploidy in Insects. Front. Cell Dev. Biol. 2020, 8, 361. [Google Scholar] [CrossRef]
- Ērenpreisa, J. Jānis Oļģerts Ērenpreiss and his School of Cancer Research: Commemorating the 90th Anniversary. Proc. Latv. Acad. Sci. B. Nat. Exact. Appl. Sci. 2019, 73, 533–537. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Ashman, T.-L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Salmina, K.; Anatskaya, O.; Cragg, M.S. Paradoxes of Cancer: Survival at the Brink. Semin. Cancer Biol. 2022, 81, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Anatskaya, O.V.; Vinogradov, A.E. Polyploidy as a Fundamental Phenomenon in Evolution, Development, Adaptation and Diseases. Int. J. Mol. Sci. 2022, 23, 3542. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The Evolutionary Significance of Polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E. Polyploidy and Myc Proto-Oncogenes Promote Stress Adaptation via Epigenetic Plasticity and Gene Regulatory Network Rewiring. Int. J. Mol. Sci. 2022, 23, 9691. [Google Scholar] [CrossRef] [PubMed]
- Bulut-Karslioglu, A.; Macrae, T.A.; Oses-Prieto, J.A.; Covarrubias, S.; Percharde, M.; Ku, G.; Diaz, A.; McManus, M.T.; Burlingame, A.L.; Ramalho-Santos, M. The Transcriptionally Permissive Chromatin State of Embryonic Stem Cells Is Acutely Tuned to Translational Output. Cell Stem Cell 2018, 22, 369–383.e8. [Google Scholar] [CrossRef] [PubMed]
- Maslov, D.L.; Zemskaya, N.V.; Trifonova, O.P.; Lichtenberg, S.; Balashova, E.E.; Lisitsa, A.V.; Moskalev, A.A.; Lokhov, P.G. Comparative Metabolomic Study of Drosophila Species with Different Lifespans. Int. J. Mol. Sci. 2021, 22, 12873. [Google Scholar] [CrossRef] [PubMed]
- Montalvo, D.; Pérez-Treviño, P.; Madrazo-Aguirre, K.; González-Mondellini, F.A.; Miranda-Roblero, H.O.; Ramonfaur-Gracia, D.; Jacobo-Antonio, M.; Mayorga-Luna, M.; Gómez-Víquez, N.L.; García, N.; et al. Underlying mechanism of the contractile dysfunction in atrophied ventricular myocytes from a murine model of hypothyroidism. Cell Calcium 2018, 72, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wei, X.; Zhang, W.; Wang, F.; Li, Q. MiR-326 targets MDK to regulate the progression of cardiac hypertrophy through blocking JAK/STAT and MAPK signaling pathways. Eur. J. Pharmacol. 2020, 872, 172941. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Nomura, S.; Harada, M.; Yamaguchi, T.; Ko, T.; Sumida, T.; Toko, H.; Naito, A.T.; Takeda, N.; Tobita, T.; et al. High-throughput single-molecule RNA imaging analysis reveals heterogeneous responses of cardiomyocytes to hemodynamic overload. J. Mol. Cell. Cardiol. 2019, 128, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Kinugawa, K.; Minobe, W.A.; Wood, W.M.; Ridgway, E.C.; Baxter, J.D.; Ribeiro, R.C.; Tawadrous, M.F.; Lowes, B.A.; Long, C.S.; Bristow, M.R. Signaling Pathways Responsible for Fetal Gene Induction in the Failing Human Heart: Evidence for Altered Thyroid Hormone Receptor Gene Expression. Circulation 2001, 103, 1089–1094. [Google Scholar] [CrossRef]
- Cui, M.; Atmanli, A.; Morales, M.G.; Tan, W.; Chen, K.; Xiao, X.; Xu, L.; Liu, N.; Bassel-Duby, R.; Olson, E.N. Nrf1 promotes heart regeneration and repair by regulating proteostasis and redox balance. Nat. Commun. 2021, 12, 5270. [Google Scholar] [CrossRef]
- Durgan, D.J.; Young, M.E. The Cardiomyocyte Circadian Clock: Emerging Roles in Health and Disease. Circ. Res. 2010, 106, 647–658. [Google Scholar] [CrossRef]
- Gilbert, G.; Demydenko, K.; Dries, E.; Puertas, R.D.; Jin, X.; Sipido, K.; Roderick, H.L. Calcium Signaling in Cardiomyocyte Function. Cold Spring Harb. Perspect. Biol. 2020, 12, a035428. [Google Scholar] [CrossRef]
- Coenye, T. Do results obtained with RNA-sequencing require independent verification? Biofilm 2021, 3, 100043. [Google Scholar] [CrossRef] [PubMed]
- Azhar, M.; Runyan, R.B.; Gard, C.; Sanford, L.P.; Miller, M.L.; Andringa, A.; Pawlowski, S.; Rajan, S.; Doetschman, T. Ligand-specific function of transforming growth factor beta in epithelial-mesenchymal transition in heart development. Dev. Dyn. 2009, 238, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.; Alcoreza, N.; Delgado, M.; Joddar, B.; Chattopadhyay, M. Cardioprotective Effect of Glycyrrhizin on Myocardial Remodeling in Diabetic Rats. Biomolecules 2021, 11, 569. [Google Scholar] [CrossRef]
- Parker, T.G.; Packer, S.E.; Schneider, M.D. Peptide growth factors can provoke “fetal” contractile protein gene expression in rat cardiac myocytes. J. Clin. Investig. 1990, 85, 507–514. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. Transforming growth factor-β in myocardial disease. Nat. Rev. Cardiol. 2022, 19, 435–455. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.P.; Gupta, M.; Zak, R.; Sukhatme, V. Egr-1, a serum-inducible zinc finger protein, regulates transcription of the rat cardiac alpha-myosin heavy chain gene. J. Biol. Chem. 1991, 266, 12813–12816. [Google Scholar] [CrossRef] [PubMed]
- Koneva, L.; Vyas, A.; McEachin, R.; Puttabyatappa, M.; Wang, H.-S.; Sartor, M.; Padmanabhan, V. Developmental programming: Interaction between prenatal BPA and postnatal overfeeding on cardiac tissue gene expression in female sheep. Environ. Mol. Mutagen. 2017, 58, 4–18. [Google Scholar] [CrossRef]
- Laggner, M.; Oberndorfer, F.; Golabi, B.; Bauer, J.; Zuckermann, A.; Hacker, P.; Lang, I.; Skoro-Sajer, N.; Gerges, C.; Taghavi, S.; et al. EGR1 Is Implicated in Right Ventricular Cardiac Remodeling Associated with Pulmonary Hypertension. Biology 2022, 11, 677. [Google Scholar] [CrossRef]
- Lyn, D.; Liu, X.; Bennett, N.A.; Emmett, N.L. Gene expression profile in mouse myocardium after ischemia. Physiol. Genom. 2000, 2, 93–100. [Google Scholar] [CrossRef]
- Pacini, L.; Suffredini, S.; Ponti, D.; Coppini, R.; Frati, G.; Ragona, G.; Cerbai, E.; Calogero, A. Altered calcium regulation in isolated cardiomyocytes from Egr-1 knock-out mice. Can. J. Physiol. Pharmacol. 2013, 91, 1135–1142. [Google Scholar] [CrossRef]
- Vermeulen, S.; Roumans, N.; Honig, F.; Carlier, A.; Hebels, D.G.; Eren, A.D.; Dijke, P.T.; Vasilevich, A.; de Boer, J. Mechanotransduction is a context-dependent activator of TGF-β signaling in mesenchymal stem cells. Biomaterials 2020, 259, 120331. [Google Scholar] [CrossRef] [PubMed]
- Dobaczewski, M.; Chen, W.; Frangogiannis, N.G. Transforming growth factor (TGF)-β signaling in cardiac remodeling. J. Mol. Cell. Cardiol. 2011, 51, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, S.; Ahmad, H. Molecular switch model for cardiomyocyte proliferation. Cell Regen. 2019, 8, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Bajic, V.P.; Van Neste, C.; Obradovic, M.; Zafirovic, S.; Radak, D.; Bajic, V.B.; Essack, M.; Isenovic, E.R. Glutathione “Redox Homeostasis” and Its Relation to Cardiovascular Disease. Oxid. Med. Cell. Longev. 2019, 2019, 5028181. [Google Scholar] [CrossRef] [PubMed]
- Skvortsova, E.V.; Nazarov, I.B.; Tomilin, A.N.; Sinenko, S.A. Dual Mode of Mitochondrial ROS Action during Reprogramming to Pluripotency. Int. J. Mol. Sci. 2022, 23, 10924. [Google Scholar] [CrossRef]
- Selenina, A.V.; Tsimokha, A.S.; Tomilin, A.N. Proteasomes in Protein Homeostasis of Pluripotent Stem Cells. Acta Nat. 2017, 9, 39–47. [Google Scholar] [CrossRef]
- Sinenko, S.A.; Starkova, T.Y.; Kuzmin, A.A.; Tomilin, A.N. Physiological Signaling Functions of Reactive Oxygen Species in Stem Cells: From Flies to Man. Front. Cell Dev. Biol. 2021, 9, 714370. [Google Scholar] [CrossRef]
- Damy, T.; Kirsch, M.; Khouzami, L.; Caramelle, P.; Le Corvoisier, P.; Roudot-Thoraval, F.; Dubois-Randé, J.-L.; Hittinger, L.; Pavoine, C.; Pecker, F. Glutathione Deficiency in Cardiac Patients Is Related to the Functional Status and Structural Cardiac Abnormalities. PLoS ONE 2009, 4, e4871. [Google Scholar] [CrossRef]
- Matuz-Mares, D.; Riveros-Rosas, H.; Vilchis-Landeros, M.; Vázquez-Meza, H. Glutathione Participation in the Prevention of Cardiovascular Diseases. Antioxidants 2021, 10, 1220. [Google Scholar] [CrossRef]
- Rodondi, N.; Newman, A.B.; Vittinghoff, E.; De Rekeneire, N.; Satterfield, S.; Harris, T.B.; Bauer, D.C. Subclinical Hypothyroidism and the Risk of Heart Failure, Other Cardiovascular Events, and Death. Arch. Intern. Med. 2005, 165, 2460–2466. [Google Scholar] [CrossRef]
- Biondi, B.; Cappola, A.R.; Cooper, D.S. Subclinical Hypothyroidism: A Review. JAMA 2019, 322, 153–160. [Google Scholar] [CrossRef]
- Bachhawat, A.K.; Yadav, S.; Jainarayanan, A.K.; Dubey, P. Heart failure and the glutathione cycle: An integrated view. Biochem. J. 2020, 477, 3123–3130. [Google Scholar] [CrossRef]
- Morciano, G.; Rimessi, A.; Patergnani, S.; Vitto, V.A.; Danese, A.; Kahsay, A.; Palumbo, L.; Bonora, M.; Wieckowski, M.R.; Giorgi, C.; et al. Calcium dysregulation in heart diseases: Targeting calcium channels to achieve a correct calcium homeostasis. Pharmacol. Res. 2022, 177, 106119. [Google Scholar] [CrossRef] [PubMed]
- Lazzeri, E.; Angelotti, M.L.; Conte, C.; Anders, H.-J.; Romagnani, P. Surviving Acute Organ Failure: Cell Polyploidization and Progenitor Proliferation. Trends Mol. Med. 2019, 25, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Cappola, A.R.; Desai, A.S.; Medici, M.; Cooper, L.S.; Egan, D.; Sopko, G.; Fishman, G.I.; Goldman, S.; Cooper, D.S.; Mora, S.; et al. Thyroid and Cardiovascular Disease Research Agenda for Enhancing Knowledge, Prevention, and Treatment. Circulation 2019, 139, 2892–2909. [Google Scholar] [CrossRef] [PubMed]
- Mercier, G.; Turque, N.; Schumacher, M. Rapid effects of triiodothyronine on immediate-early gene expression in Schwann cells. Glia 2001, 35, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Kanaan, G.N.; Harper, M.-E. Cellular redox dysfunction in the development of cardiovascular diseases. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 2822–2829. [Google Scholar] [CrossRef]
- Liang, Q.; Xu, H.; Liu, M.; Qian, L.; Yan, J.; Yang, G.; Chen, L. Postnatal Deletion of Bmal1 in Cardiomyocyte Promotes Pressure Overload Induced Cardiac Remodeling in Mice. J. Am. Hear. Assoc. 2022, 11, e025021. [Google Scholar] [CrossRef]
- Bensley, J.G.; Moore, L.; De Matteo, R.; Harding, R.; Black, M.J. Impact of preterm birth on the developing myocardium of the neonate. Pediatr. Res. 2018, 83, 880–888. [Google Scholar] [CrossRef]
- Bensley, J.G.; De Matteo, R.; Harding, R.; Black, M.J. Three-dimensional direct measurement of cardiomyocyte volume, nuclearity, and ploidy in thick histological sections. Sci. Rep. 2016, 6, 23756. [Google Scholar] [CrossRef]
- Sukhacheva, T.V.; Serov, R.A.; Nizyaeva, N.V.; Burov, A.A.; Pavlovich, S.V.; Podurovskaya, Y.L.; Samsonova, M.V.; Chernyaev, A.L.; Shchegolev, A.I.; Kim, A.I.; et al. Accelerated Growth, Differentiation, and Ploidy with Reduced Proliferation of Right Ventricular Cardiomyocytes in Children with Congenital Heart Defect Tetralogy of Fallot. Cells 2022, 11, 175. [Google Scholar] [CrossRef]
- Davoli, T.; de Lange, T. Telomere-Driven Tetraploidization Occurs in Human Cells Undergoing Crisis and Promotes Transformation of Mouse Cells. Cancer Cell 2012, 21, 765–776. [Google Scholar] [CrossRef]
- Zheng, L.; Dai, H.; Zhou, M.; Li, X.; Liu, C.; Guo, Z.; Wu, X.; Wu, J.; Wang, C.; Zhong, J.; et al. Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumour progression. Nat. Commun. 2012, 3, 815. [Google Scholar] [CrossRef]
- Vainshelbaum, N.M.; Giuliani, A.; Salmina, K.; Pjanova, D.; Erenpreisa, J. The Transcriptome and Proteome Networks of Malignant Tumours Reveal Atavistic Attractors of Polyploidy-Related Asexual Reproduction. Int. J. Mol. Sci. 2022, 23, 14930. [Google Scholar] [CrossRef]
- Vinogradov, A.E.; Anatskaya, O.V. Cellular Biogenetic Law and Its Distortion by Protein Interactions: A Possible Unified Framework for Cancer Biology and Regenerative Medicine. Int. J. Mol. Sci. 2022, 23, 11486. [Google Scholar] [CrossRef]
- Kasperski, A. Life Entrapped in a Network of Atavistic Attractors: How to Find a Rescue. Int. J. Mol. Sci. 2022, 23, 4017. [Google Scholar] [CrossRef]
- Niculescu, V.F. aCLS cancers: Genomic and epigenetic changes transform the cell of origin of cancer into a tumorigenic pathogen of unicellular organization and lifestyle. Gene 2020, 726, 144174. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E. Genome multiplication as adaptation to tissue survival: Evidence from gene expression in mammalian heart and liver. Genomics 2007, 89, 70–80. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E. Somatic polyploidy promotes cell function under stress and energy depletion: Evidence from tissue-specific mammal transcriptome. Funct. Integr. Genom. 2010, 10, 433–446. [Google Scholar] [CrossRef]
- Vazquez-Martin, A.; Anatskaya, O.V.; Giuliani, A.; Erenpreisa, J.; Huang, S.; Salmina, K.; Inashkina, I.; Huna, A.; Nikolsky, N.N.; Vinogradov, A.E. Somatic polyploidy is associated with the upregulation of c-MYC interacting genes and EMT-like signature. Oncotarget 2016, 7, 75235–75260. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Cragg, M.S. Three steps to the immortality of cancer cells: Senescence, polyploidy and self-renewal. Cancer Cell Int. 2013, 13, 92. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. The “life code”: A theory that unifies the human life cycle and the origin of human tumors. Semin. Cancer Biol. 2020, 60, 380–397. [Google Scholar] [CrossRef]
- Liu, J.; Erenpreisa, J.; Sikora, E. Polyploid giant cancer cells: An emerging new field of cancer biology. Semin. Cancer Biol. 2021, 81, 1–4. [Google Scholar] [CrossRef]
- Niculescu, V.F. Cancer genes and cancer stem cells in tumorigenesis: Evolutionary deep homology and controversies. Genes Dis. 2022, 9, 1234–1247. [Google Scholar] [CrossRef]
- Zybina, T.G. Genome Modifications Involved in Developmental Programs of the Placental Trophoblast. In Cytogenetics-Classical and Molecular Strategies for Analysing Heredity Material; Larramendy, M.L., Soloneski, S., Eds.; IntechOpen: London, UK, 2021; ISBN 978-1-83968-941-3. [Google Scholar]
- Ghiraldini, F.G.; Silva, I.S.; Mello, M.L.S. Polyploidy and chromatin remodeling in hepatocytes from insulin-dependent diabetic and normoglycemic aged mice. Cytom. Part A 2012, 81A, 755–764. [Google Scholar] [CrossRef]
- Silva, I.S.; Ghiraldini, F.G.; Veronezi, G.M.; Mello, M.L.S. Polyploidy and nuclear phenotype characteristics of cardiomyocytes from diabetic adult and normoglycemic aged mice. Acta Histochem. 2018, 120, 84–94. [Google Scholar] [CrossRef]
- Pienta, K.J.; Hammarlund, E.U.; Brown, J.S.; Amend, S.R.; Axelrod, R.M. Cancer recurrence and lethality are enabled by enhanced survival and reversible cell cycle arrest of polyaneuploid cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2020838118. [Google Scholar] [CrossRef]
- Pienta, K.; Hammarlund, E.; Austin, R.; Axelrod, R.; Brown, J.; Amend, S. Cancer cells employ an evolutionarily conserved polyploidization program to resist therapy. Semin. Cancer Biol. 2022, 81, 145–159. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Sidorenko, N.V.; Vinogradov, A.E.; Beyer, T.V. Impact of neonatal cryptosporidial gastroenteritis on epigenetic programming of rat hepatocytes. Cell Biol. Int. 2007, 31, 420–427. [Google Scholar] [CrossRef]
- Hirose, K.; Payumo, A.Y.; Cutie, S.; Hoang, A.; Zhang, H.; Guyot, R.; Lunn, D.; Bigley, R.B.; Yu, H.; Wang, J.; et al. Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science 2019, 364, 184–188. [Google Scholar] [CrossRef]
- Golubnitschaja, O.; Costigliola, V. Predictive, Preventive and Personalised Medicine as the Medicine of the Future: Anticipatory Scientific Innovation and Advanced Medical Services. In Anticipation and Medicine; Nadin, M., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 69–85. ISBN 978-3-319-45140-4. [Google Scholar]
- Koklesova, L.; Liskova, A.; Samec, M.; Zhai, K.; Al-Ishaq, R.K.; Bugos, O.; Šudomová, M.; Biringer, K.; Pec, M.; Adamkov, M.; et al. Protective Effects of Flavonoids Against Mitochondriopathies and Associated Pathologies: Focus on the Predictive Approach and Personalized Prevention. Int. J. Mol. Sci. 2021, 22, 8649. [Google Scholar] [CrossRef] [PubMed]
- Liskova, A.; Samec, M.; Koklesova, L.; Kudela, E.; Kubatka, P.; Golubnitschaja, O. Mitochondriopathies as a Clue to Systemic Disorders—Analytical Tools and Mitigating Measures in Context of Predictive, Preventive, and Personalized (3P) Medicine. Int. J. Mol. Sci. 2021, 22, 2007. [Google Scholar] [CrossRef] [PubMed]
- Zedan, M.M.; Mansour, A.K.; Bakr, A.A.; Sobh, M.A.; Khodadadi, H.; Salles, E.L.; Alhashim, A.; Baban, B.; Golubnitschaja, O.; Elmarakby, A.A. Effect of Everolimus versus Bone Marrow-Derived Stem Cells on Glomerular Injury in a Rat Model of Glomerulonephritis: A Preventive, Predictive and Personalized Implication. Int. J. Mol. Sci. 2021, 23, 344. [Google Scholar] [CrossRef] [PubMed]
- Gan, P.; Patterson, M.; Velasquez, A.; Wang, K.; Tian, D.; Windle, J.J.; Tao, G.; Judge, D.P.; Makita, T.; Park, T.J.; et al. Tnni3k alleles influence ventricular mononuclear diploid cardiomyocyte frequency. PLoS Genet. 2019, 15, e1008354. [Google Scholar] [CrossRef]
- Gan, P.; Patterson, M.; Sucov, H.M. Cardiomyocyte Polyploidy and Implications for Heart Regeneration. Annu. Rev. Physiol. 2020, 82, 45–61. [Google Scholar] [CrossRef]
- Auchampach, J.; Han, L.; Huang, G.N.; Kühn, B.; Lough, J.W.; O’Meara, C.C.; Payumo, A.Y.; Rosenthal, N.A.; Sucov, H.M.; Yutzey, K.E.; et al. Measuring cardiomyocyte cell-cycle activity and proliferation in the age of heart regeneration. Am. J. Physiol. Circ. Physiol. Heart 2022, 322, H579–H596. [Google Scholar] [CrossRef]
- Sedmera, D. Function and form in the developing cardiovascular system. Cardiovasc. Res. 2011, 91, 252–259. [Google Scholar] [CrossRef]
- Chaithra, S.; Agarwala, S.; Ramachandra, N. High-risk genes involved in common septal defects of congenital heart disease. Gene 2022, 840, 146745. [Google Scholar] [CrossRef]
- O’Connor, T.P.; Diamond, J. Ontogeny of intestinal safety factors: Lactase capacities and lactose loads. Am. J. Physiol. 1999, 276, R753–R765. [Google Scholar] [CrossRef]
- National Research Council (US). Institute for Laboratory Animal Research Guide for the Care and Use of Laboratory Animals; National Academies Press (US): Washington DC, USA, 1996; ISBN 978-0-309-05377-8. [Google Scholar]
- Siedlecka, U.; Arora, M.; Kolettis, T.; Soppa, G.K.R.; Lee, J.; Stagg, M.A.; Harding, S.E.; Yacoub, M.H.; Terracciano, C.M.N. Effects of clenbuterol on contractility and Ca2+ homeostasis of isolated rat ventricular myocytes. Am. J. Physiol. Circ. Physiol. 2008, 295, H1917–H1926. [Google Scholar] [CrossRef]
- Auguste, G.; Gurha, P.; Lombardi, R.; Coarfa, C.; Willerson, J.T.; Marian, A.J. Suppression of Activated FOXO Transcription Factors in the Heart Prolongs Survival in a Mouse Model of Laminopathies. Circ. Res. 2018, 122, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Rumyantsev, P.P. Interrelations of the proliferation and differentiation processes during cardiact myogenesis and regeneration. Int. Rev. Cytol. 1977, 51, 186–273. [Google Scholar] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021, 50, D20–D26. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Lindner, R.; Friedel, C.C. A Comprehensive Evaluation of Alignment Algorithms in the Context of RNA-Seq. PLoS ONE 2012, 7, e52403. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Zhou, X.; Lindsay, H.; Robinson, M.D. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. 2014, 42, e91. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T.; Russell, D.W.; Green, M.R. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA; Cold Spring Harbor: New York, NY, USA, 1989; ISBN 978-0-87969-309-1. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]

| Gene | Sequence |
|---|---|
| Glyceraldehyde triphosphate Dehydrogenase, (Gapdh); Gene ID 24383 | Forward: 5′-GGGGGCTCTCTGCTCCTCCC-3′ Reverse: 5′-CAGGCGTCCGATACGGCCAA-3′ |
| Early growth response, (Egr1); Gene ID 24383 | Forward: 5′-CACCAGACCATGCTTCAGTGAGA-3′ Reverse: 5′-GTTGCATGGCTGTTCACAGGA-3′ |
| Transforming growth factor β2, (Tgfβ2), Gene ID 81809 | Forward: 5′-CTCCACATATGCCAGTGGTG-3′ Reverse: 5′-CTAAAGCAATAGGCGGCATC-3′ |
| Cyclin A2, (Ccna2); Gene ID: 114494 | Forward: 5′-ATGTCACCGTTCCTCCTTG-3′ Reverse: 5′-GGGCATCTTCACGCTCTATT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anatskaya, O.V.; Runov, A.L.; Ponomartsev, S.V.; Vonsky, M.S.; Elmuratov, A.U.; Vinogradov, A.E. Long-Term Transcriptomic Changes and Cardiomyocyte Hyperpolyploidy after Lactose Intolerance in Neonatal Rats. Int. J. Mol. Sci. 2023, 24, 7063. https://doi.org/10.3390/ijms24087063
Anatskaya OV, Runov AL, Ponomartsev SV, Vonsky MS, Elmuratov AU, Vinogradov AE. Long-Term Transcriptomic Changes and Cardiomyocyte Hyperpolyploidy after Lactose Intolerance in Neonatal Rats. International Journal of Molecular Sciences. 2023; 24(8):7063. https://doi.org/10.3390/ijms24087063
Chicago/Turabian StyleAnatskaya, Olga V., Andrey L. Runov, Sergey V. Ponomartsev, Maxim S. Vonsky, Artem U. Elmuratov, and Alexander E. Vinogradov. 2023. "Long-Term Transcriptomic Changes and Cardiomyocyte Hyperpolyploidy after Lactose Intolerance in Neonatal Rats" International Journal of Molecular Sciences 24, no. 8: 7063. https://doi.org/10.3390/ijms24087063
APA StyleAnatskaya, O. V., Runov, A. L., Ponomartsev, S. V., Vonsky, M. S., Elmuratov, A. U., & Vinogradov, A. E. (2023). Long-Term Transcriptomic Changes and Cardiomyocyte Hyperpolyploidy after Lactose Intolerance in Neonatal Rats. International Journal of Molecular Sciences, 24(8), 7063. https://doi.org/10.3390/ijms24087063







