HIV-1/HBV Coinfection Accurate Multitarget Prediction Using a Graph Neural Network-Based Ensemble Predicting Model
Abstract
1. Introduction
2. Results
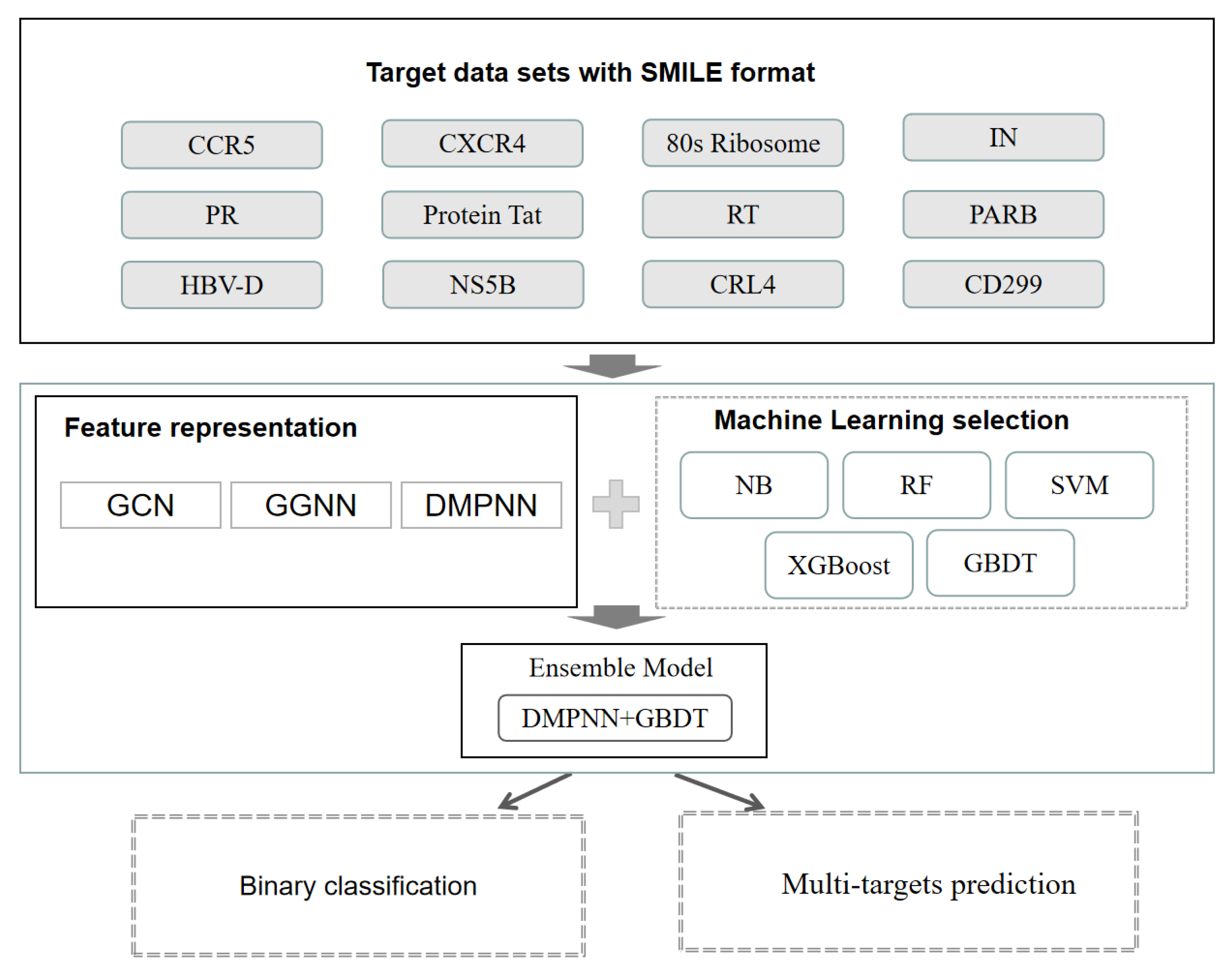
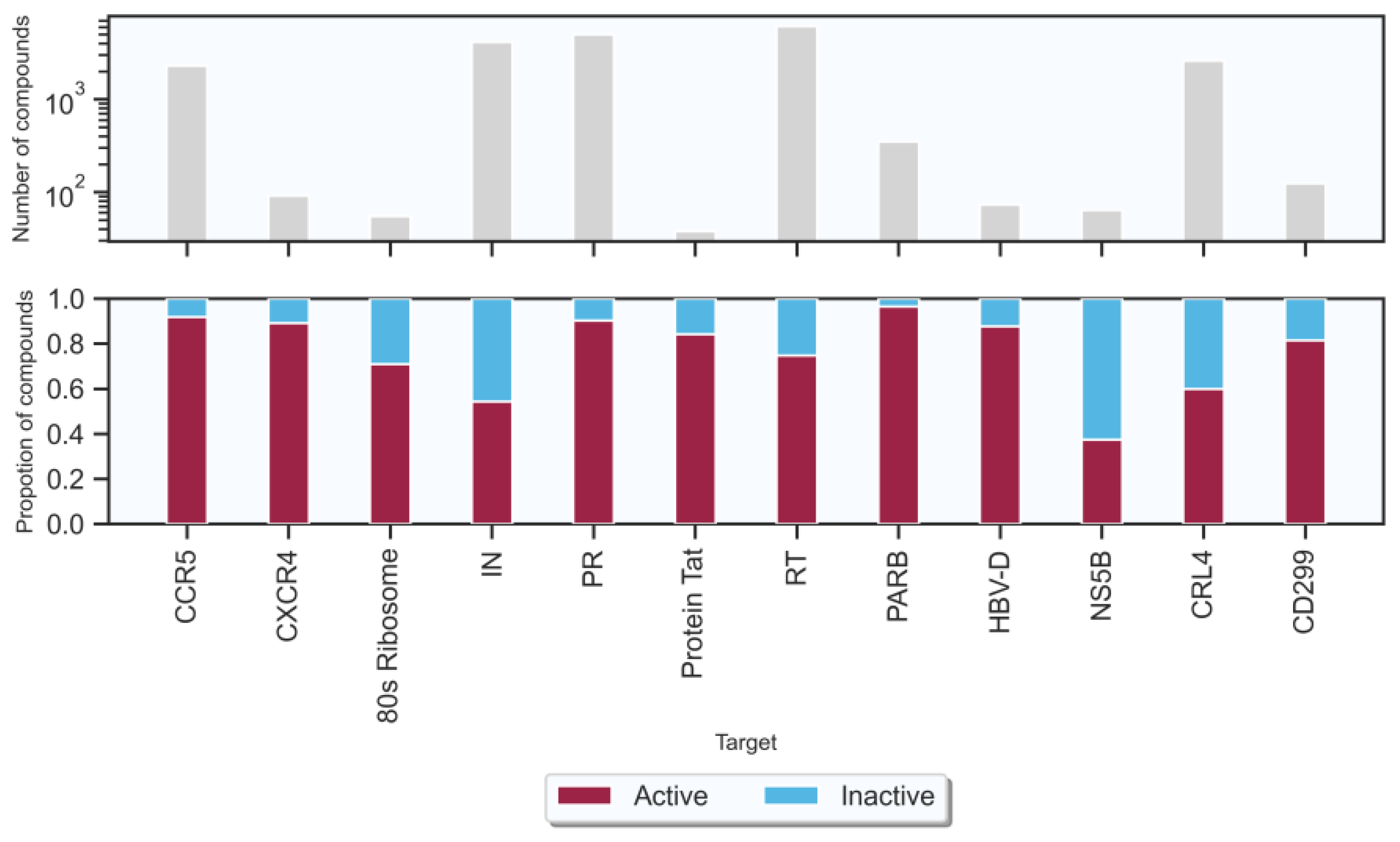
2.1. Experimental Data Sets
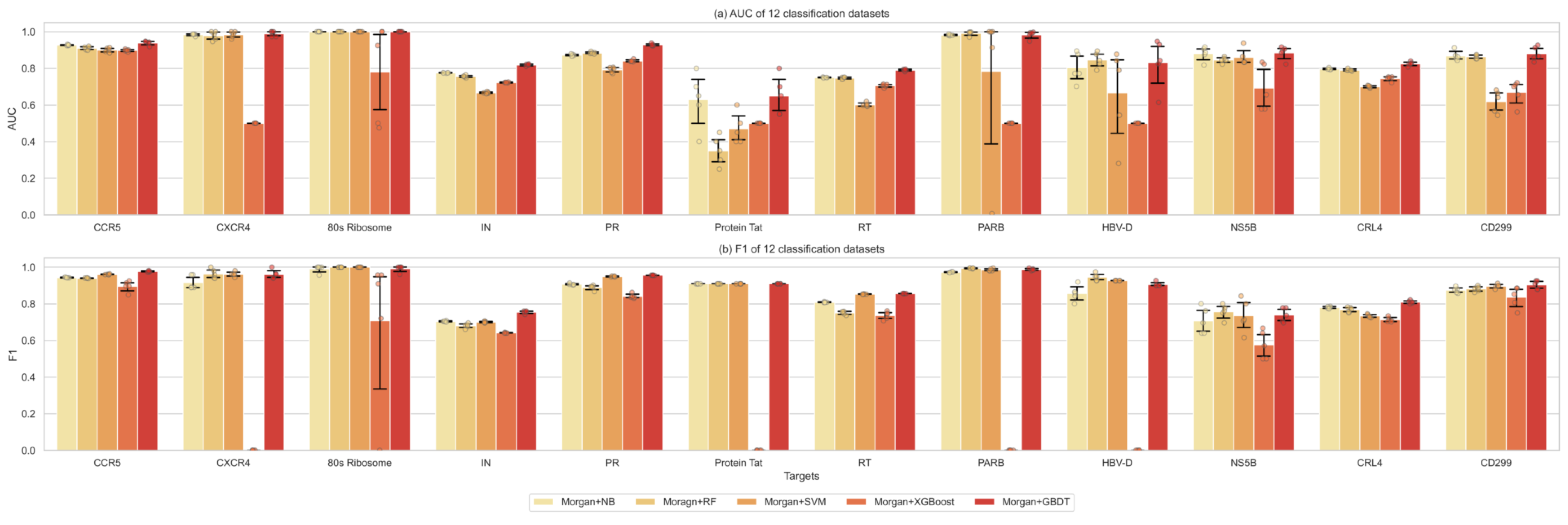
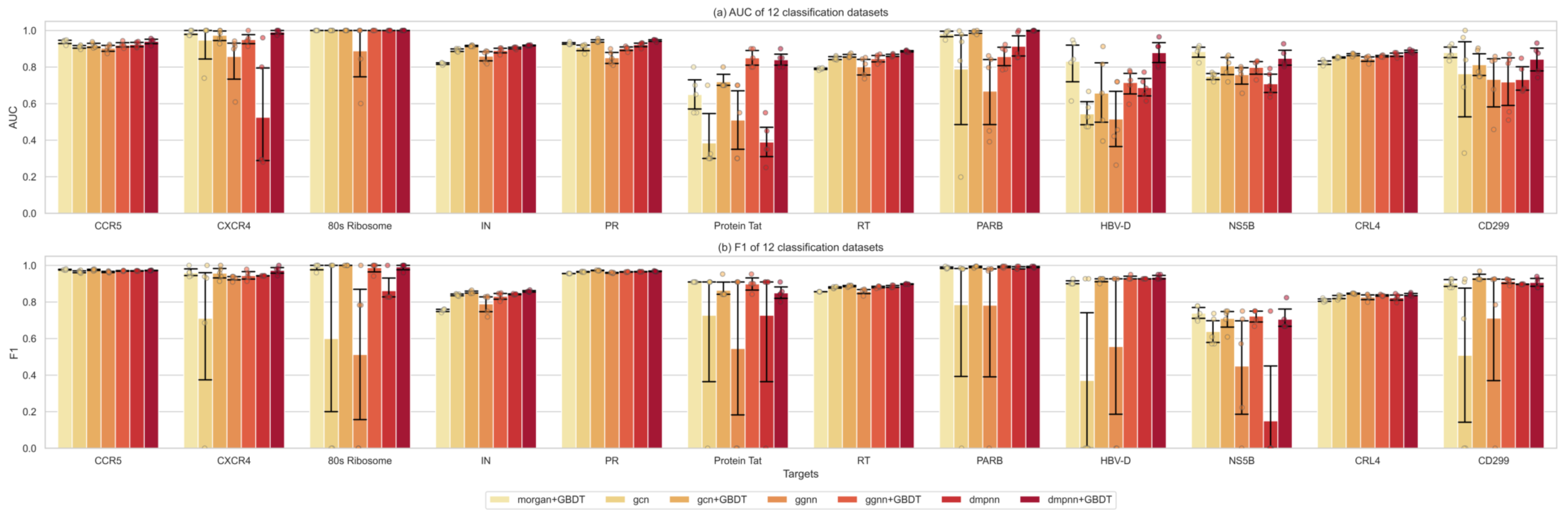
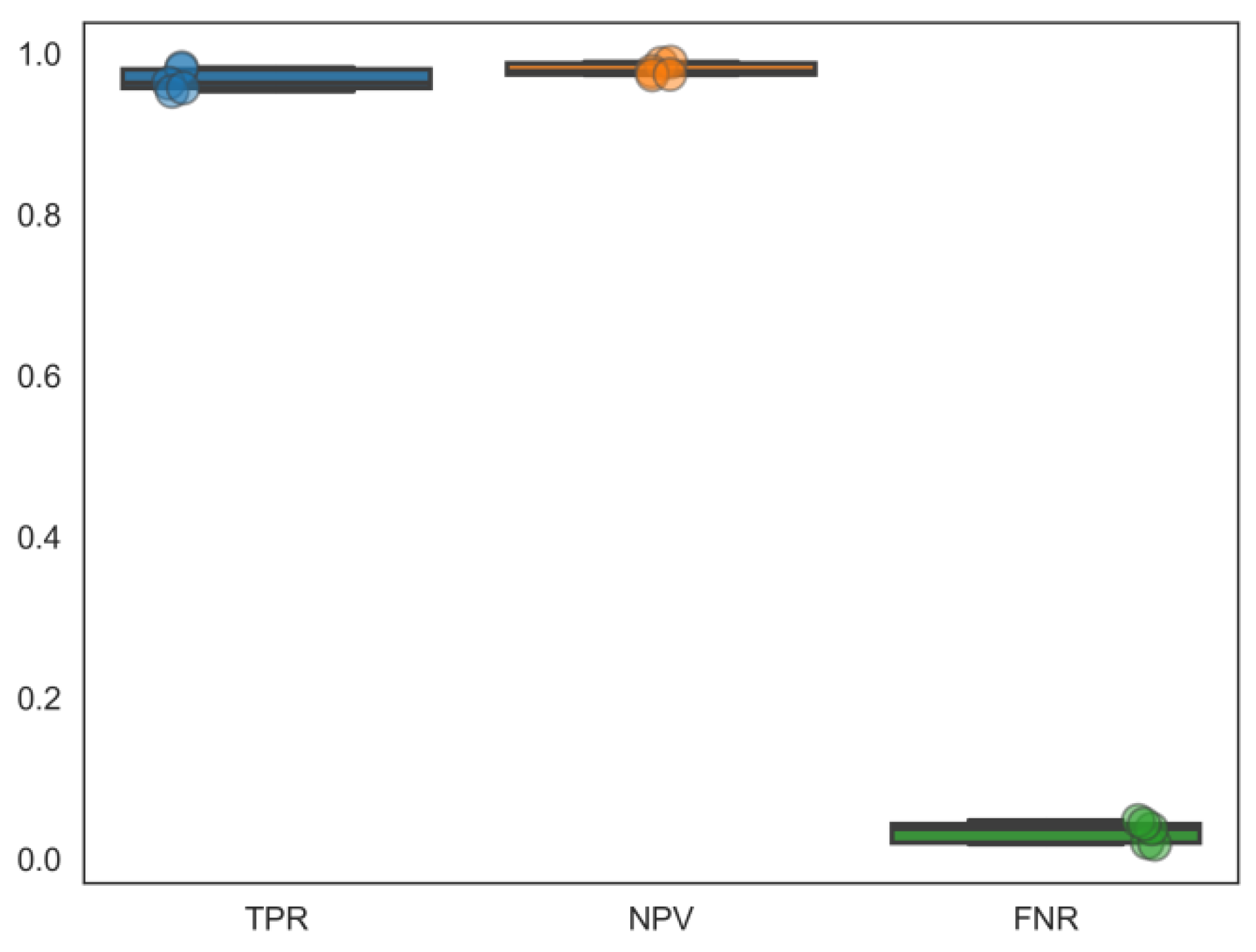
2.2. Comparison of the Classification Performance of Different Machine Learning Methods
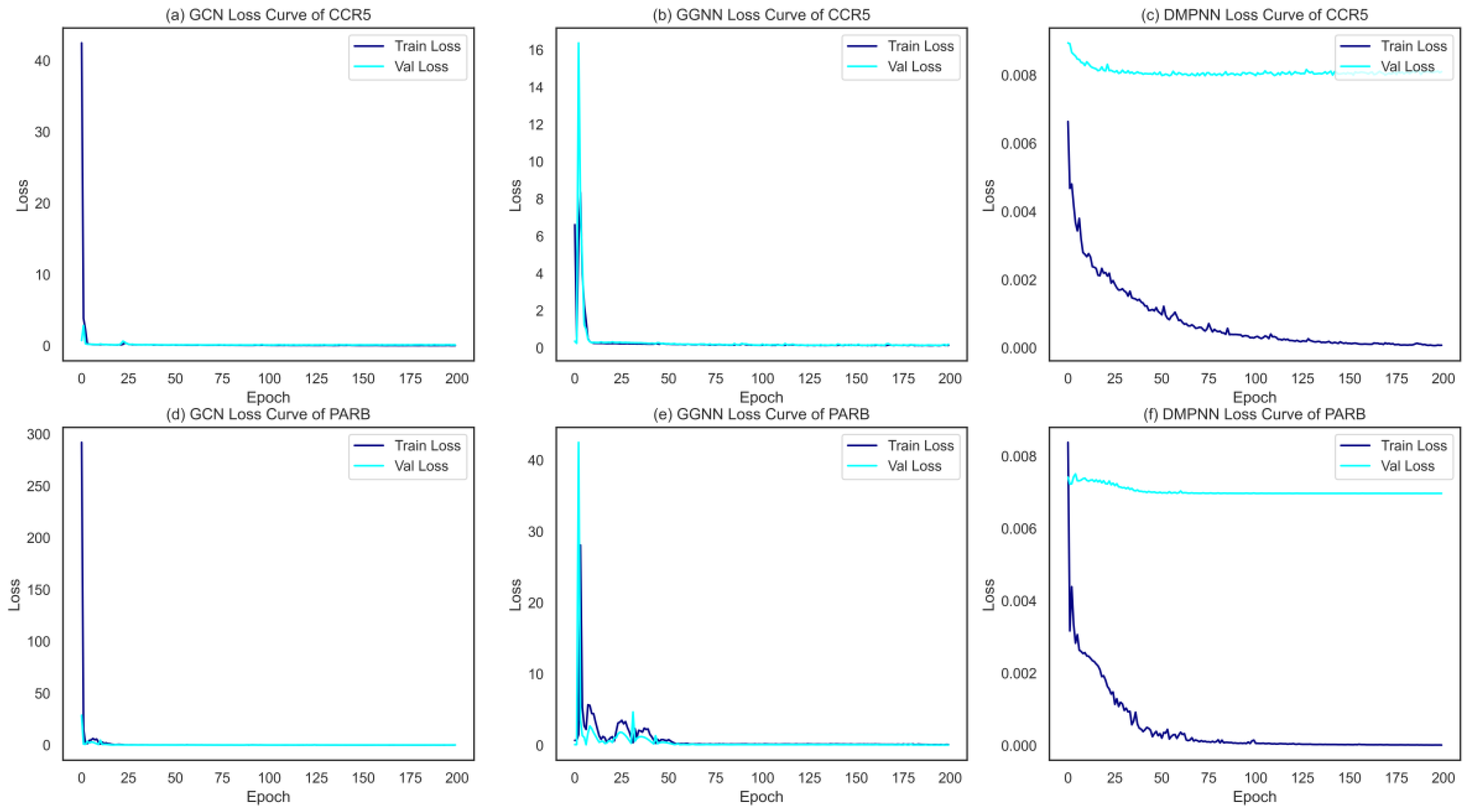
2.3. Performance Evaluation of Feature Extraction between Graph Representations and Supervised Learners
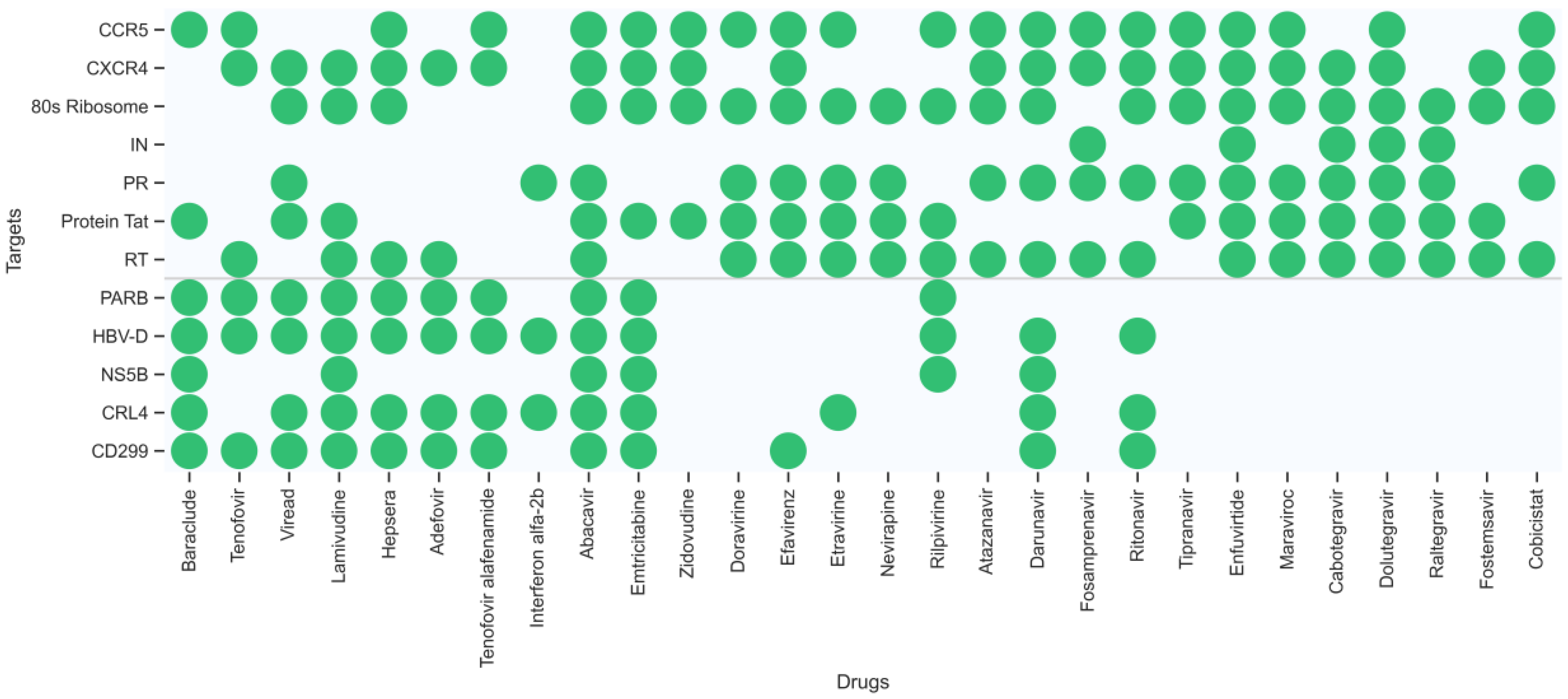
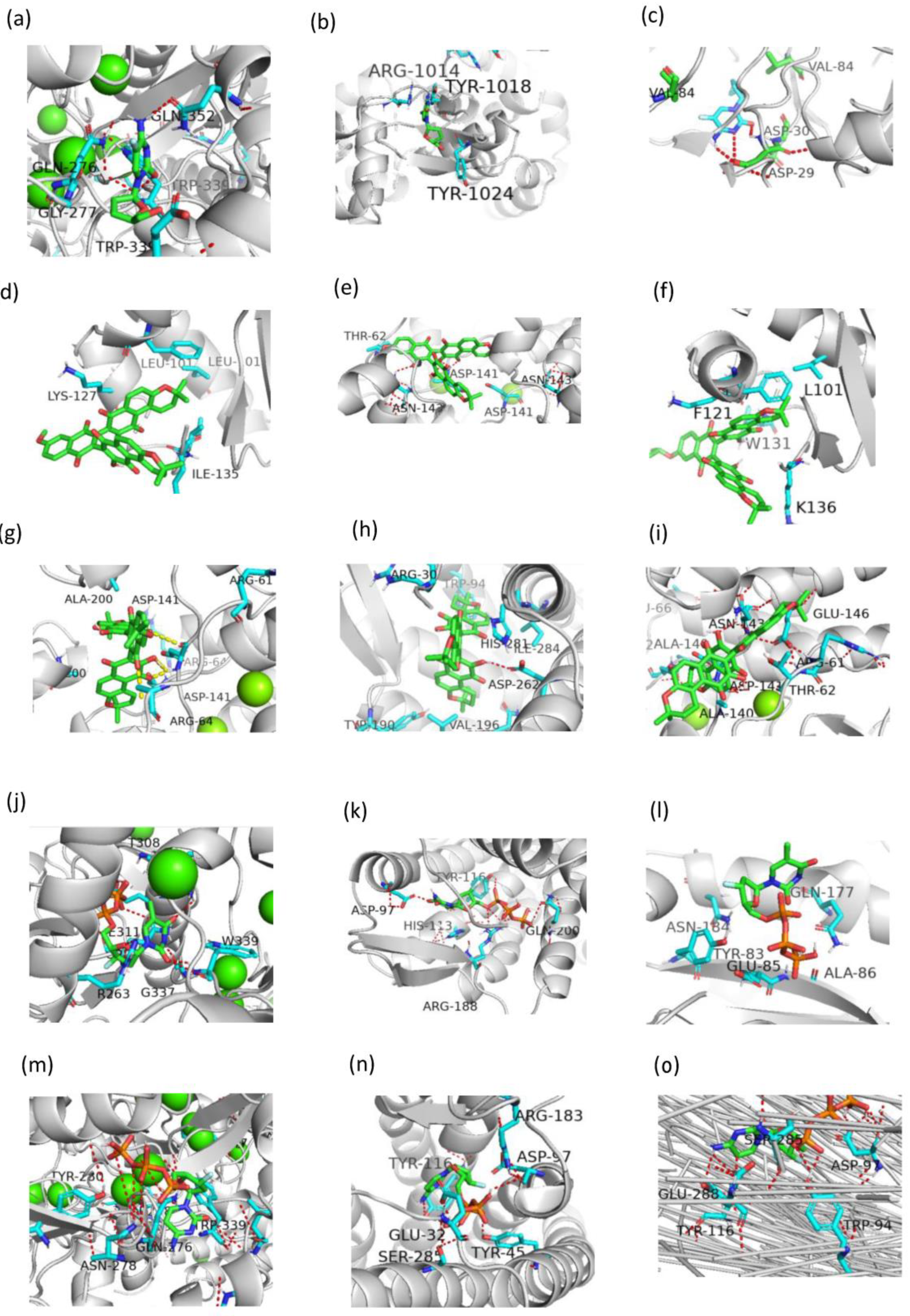
2.4. Multitarget Prediction
- Case 1: Retrospective polypharmacology prediction of known HIV-1/HBV drugs
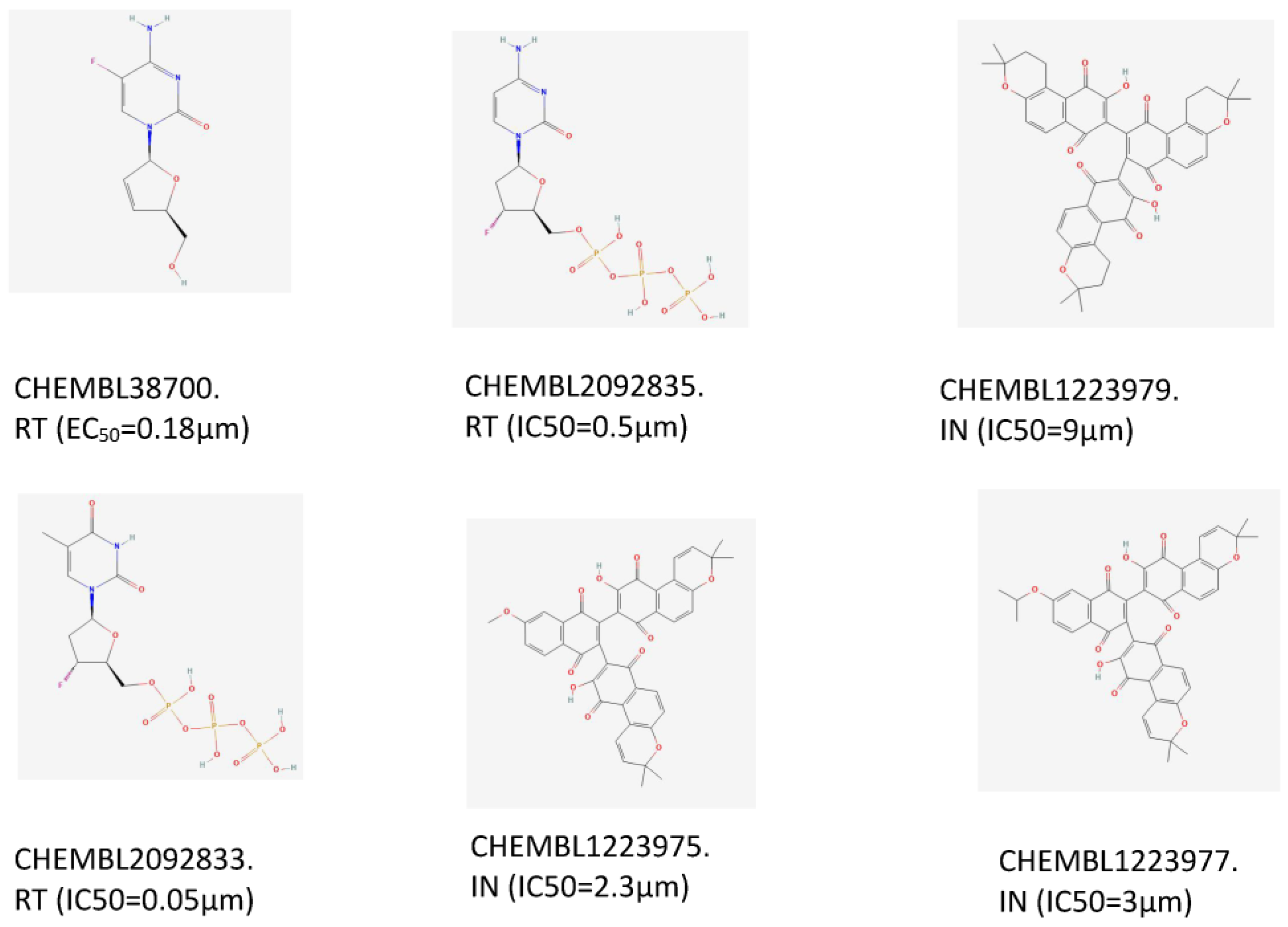
- Case 2: Multitarget prediction of new compounds
3. Discussion
4. Materials and Methods
4.1. Data Set Preparation
4.2. Molecular Feature Extraction by Graph Neural Network
4.3. Multitarget Prediction Model
4.4. Machine Learning Methods
4.5. Performance Metrics
4.6. Data Imbalance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- UNAIDS. AIDSinfo 2020. Available online: https://aidsinfo.unaids.org/ (accessed on 12 January 2022).
- Bendavid, E.; Young, S.D.; Katzenstein, D.A.; Bayoumi, A.M.; Sanders, G.D.; Owens, D.K. Cost-Effectiveness of HIV Monitoring Strategies in Resource-Limited Settings: A Southern African Analysis. Arch. Intern. Med. 2008, 168, 1910–1918. [Google Scholar] [CrossRef] [PubMed]
- Charles, M.; Boyle, B.A. Excess and Access: The Continuing Controversy Regarding HIV and Health Care in Africa. AIDS Read. 2002, 12, 288–292. [Google Scholar]
- Xia, H.; Wang, Y.; Sun, H.L.; Gao, L.Y.; Cao, Y.; Zaongo, S.D.; Zeng, R.N.; Wu, H.; Zhang, M.J.; Ma, P. Safety and Effificacy of Allogeneic Natural Killer Cell Immunotherapy on Human Immunodefificiency Virus Type 1 Immunological Non-Responders: A Brief Report. Chin. Med. J. 2020, 133, 2803–2807. [Google Scholar] [CrossRef] [PubMed]
- Zaongo, S.D.; Xia, H.; Ma, P. HIV Gene Therapy Strategies and Safety: What Do We Know From the Recent Publications? AIDS Rev. 2020, 23, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Falkenhagen, A.; Joshi, S. Genetic Strategies for HIV Treatment and Prevention. Mol. Ther. Nucleic Acids 2018, 13, 514–533. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Travers, P.; Walport, M.; Shlomchik, M.J. Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Hoffmann, C.J.; Thio, C.L. Clinical Implications of HIV and Hepatitis B CoInfection in Asia and Africa. Lancet Infect. Dis. 2007, 7, 402–409. [Google Scholar] [CrossRef]
- Singh, K.P.; Crane, M.; Audsley, J.; Avihingsanon, A.; Sasadeusz, J.; Lewin, S.R. HIV-Hepatitis B Virus Coinfection: Epidemiology, Pathogenesis, and Treatment. AIDS 2017, 31, 2035–2052. [Google Scholar] [CrossRef]
- Cheng, Z.; Lin, P.; Cheng, N. HBV/HIV Coinfection: Impact on the Development and Clinical Treatment of Liver Diseases. Front. Med. 2021, 8, 713981. [Google Scholar] [CrossRef]
- Ferrante, N.D.; Lo Re, V. Epidemiology, Natural History, and Treatment of Hepatitis Delta Virus Infection in HIV/Hepatitis B Virus Coinfection. Curr. HIV/AIDS Rep. 2020, 17, 405–414. [Google Scholar] [CrossRef]
- Dong, M.-J.; Peng, B.; Liu, Z.-F.; Ye, Q.-N.; Liu, H.; Lu, X.-L.; Zhang, B.; Chen, J.-J. The Prevalence of HIV Among MSM in China: A Large-Scale Systematic Analysis. BMC Infect. Dis. 2019, 19, 1000. [Google Scholar] [CrossRef]
- McGrath, N.; Eaton, J.W.; Newell, M.L.; Hosegood, V. Migration, Sexual Behaviour, and HIV Risk: A General Population Cohort in Rural South Africa. Lancet HIV 2015, 2, e252–e259. [Google Scholar] [CrossRef] [PubMed]
- Karić, U.; Milošević, I.; Pešić-Pavlović, I.; Salemović, D.; Stojković, M.; Jevtović, D. The treatment outcome of chronic HBV infection among HBV/HIV co-infected and HBV mono-infected patients. Int. J. STD AIDS 2020, 31, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Archampong, T.N.; Boyce, C.L.; Lartey, M.; Sagoe, K.W.; Obo-Akwa, A.; Kenu, E.; Blackard, J.T.; Kwara, A. HBV genotypes and drug resistance mutations in antiretroviral treatment-naive and treatment-experienced HBV-HIV-coinfected patients. Antivir. Ther. 2017, 22, 13–20. [Google Scholar] [CrossRef]
- Xue, J.; Guijas, C.; Benton, H.P.; Warth, B.; Siuzdak, G. METLIN MS(2) molecular standards database: A broad chemical and biological resource. Nat. Methods 2020, 17, 953–954. [Google Scholar] [CrossRef] [PubMed]
- Capecchi, A.; Awale, M.; Probst, D.; Reymond, J.L. PubChem and ChEMBL beyond Lipinski. Mol. Inf. 2019, 38, 1900016. [Google Scholar] [CrossRef]
- Tropsha, A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inf. 2010, 29, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.-C.; Rensi, S.E.; Torng, W.; Altman, R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 2018, 23, 1538–1546. [Google Scholar] [CrossRef] [PubMed]
- Miljkovic, F.; Rodríguez-Pérez, R.; Bajorath, J. Machine learning models for accurate prediction of kinase inhibitors with different binding modes. J. Med. Chem. 2020, 63, 8738–8748. [Google Scholar] [CrossRef]
- Li, H.; Sze, K.; Lu, G.; Ballester, P.J. Machine-learning scoring functions for structure-based drug lead optimization. WIREs Comput. Mol. Sci. 2020, 10, e1465. [Google Scholar] [CrossRef]
- Yao, X.J.; Panaye, A.; Doucet, J.P.; Zhang, R.S.; Chen, H.F.; Liu, M.C.; Hu, Z.D.; Fan, B.T. Comparative study of QSAR/QSPR correlations using support vector machines, radial basis function neural networks, and multiple linear regression. J. Chem. Inf. Comput. Sci. 2004, 44, 1257–1266. [Google Scholar] [CrossRef]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random forest: A classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Porwal, A.; Carranza, E.J.M.; Hale, M. Bayesian network classififiers for mineral potential mapping. Comput. Geosci. 2006, 32, 1–16. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Elman, J.L. Finding Structure in Time. Cogn. Sci. 1990, 14, 179–211. [Google Scholar] [CrossRef]
- Lin, X.; Quan, Z.; Wang, Z.-J.; Ma, T.; Zeng, X. Kgnn: Knowledge graph neural network for drug-drug interaction prediction. In Proceedings of the Twenty-Ninth International Joint Conference on Artifificial Intelligence, Yokohama, Japan, 11–17 July 2020; pp. 2739–2745. [Google Scholar]
- Li, B.; Wang, W.; Sun, Y.; Zhang, L.; Ali, M.A.; Wang, Y. GraphER: Token-Centric Entity Resolution with Graph Convolutional Neural Networks. Proc. Conf. AAAI Artif. Intell. 2020, 34, 8172–8179. [Google Scholar] [CrossRef]
- Ruiz, L.; Gama, F.; Ribeiro, A. Gated graph recurrent neural networks. arXiv 2020, arXiv:2002.01038. [Google Scholar] [CrossRef]
- Gilmer, J.; Schoenholz, S.S.; Riley, P.F.; Vinyals, O.; Dahl, G.E. Neural message passing for quantum chemistry. arXiv 2017, arXiv:1704.01212. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Additive logistic regression: A statistical view of boosting (with discussion and a rejoinder by the authors). Ann. Stat. 2000, 28, 337–407. [Google Scholar] [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y. Xgboost: Extreme Gradient Boosting; R Package Version 0.4-2; CRAN: Vienna, Austria, 2015; pp. 1–4. [Google Scholar]
- Rogers, D.; Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010, 50, 742. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards directdeposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, J.; Suzek, T.O.; Zhang, J.; Wang, J.; Zhou, Z.; Han, L.; Karapetyan, K.; Dracheva, S.; Shoemaker, B.A.; et al. PubChem’s BioAssay Database. Nucleic Acids Res. 2012, 40, D400–D412. [Google Scholar] [CrossRef] [PubMed]
- DORA: A Doravirine-Based First-Line Antiretroviral Therapy for Women of Reproductive Potential Living with HIV (DORA). Available online: https://clinicaltrials.gov/show/NCT04433780 (accessed on 16 June 2020).
- A Multicenter, Open-Label, Randomized Clinical Study to Assess Efficacy and Safety of 3 Doses of Myrcludex B for 24 Weeks in Combination with Tenofovir Compared to Tenofovir Alone to Suppress HBV Replication in Patients with Chronic Hepatitis D. Available online: https://clinicaltrials.gov/show/NCT03546621 (accessed on 30 December 2019).
- Safety and Efficacy of Celecoxib Plus Nucleos(t)Ide Analogues on the Hepatitis B Surface Antigen of Virally Suppressed Subjects with Chronic Hepatitis B. Available online: https://clinicaltrials.gov/show/NCT05256823 (accessed on 25 February 2022).
- Study of INO-1800 with or without INO-9112 + EP in Chronic Hepatitis B Subjects. Available online: https://clinicaltrials.gov/show/NCT02431312 (accessed on 1 May 2015).
- Safety and Effectiveness of Giving Adefovir Dipivoxil Plus Abacavir Plus Efavirenz Plus Amprenavir to HIV-Infected Patients Who Have Failed to Respond to Previous Protease Inhibitor Treatment. Available online: https://clinicaltrials.gov/show/NCT00002419 (accessed on 31 August 2001).
- The Safety and Effectiveness of Zidovudine Plus Adefovir in HIV-Infected Patients. Available online: https://clinicaltrials.gov/show/NCT00002326 (accessed on 31 August 2001).
- Relative Bioavailability Study to Investigate a Potential Interaction between DTG DT and F/TAF TOS. Available online: https://clinicaltrials.gov/show/NCT05489406 (accessed on 5 August 2022).
- A Single and Multiple Ascending Dose Study of JNJ-64457744. Available online: https://clinicaltrials.gov/ct2/show/NCT05423106 (accessed on 21 June 2022).
- Structured Treatment Interruptions with or without Pegylated Interferon Alpha for HIV-Infected Patients after Prolonged Viral Suppression. Available online: https://clinicaltrials.gov/show/NCT00125814 (accessed on 2 August 2005).
- Combination Therapy with Interferon Plus Interleukin 2 and Hepatitis B Vaccine in Chronic Hepatitis B Patients. Available online: https://clinicaltrials.gov/show/NCT02360592 (accessed on 10 February 2015).
- ROCKET I—Randomized Open Label Switch for Cholesterol Elevation on Kivexa Evaluation Trial (ROCKET I). Available online: https://clinicaltrials.gov/show/NCT00615810 (accessed on 14 February 2008).
- BFTAF Elderly Switch Study (BFTAF). Available online: https://clinicaltrials.gov/show/NCT05243602 (accessed on 17 February 2022).
- Rollover Protocol Continued Access to Emtricitabine/Tenofovir Disoproxil Fumarate for Adults in United States. Available online: https://clinicaltrials.gov/show/NCT00936715 (accessed on 10 July 2009).
- DAAs Treatment for Chronic HCV/HBV Co-Infection Patients(DASCO) (DASCO). Available online: https://clinicaltrials.gov/show/NCT02555943 (accessed on 22 September 2015).
- Darunavir and Rilpivirine Interactions with Levonorgestrel Implant (DRIVE-2). Available online: https://clinicaltrials.gov/show/NCT03589027 (accessed on 17 July 2018).
- Li, X.; Carmichael, E.; Feng, M.; King, I.; Doyle, T.W.; Chen, S.H. Bis-S-acyl-2-thioethyl (SATE)-bearing monophosphate prodrug of beta-L-FD4C as potent anti-HBV agent. Bioorg. Med. Chem. Lett. 1998, 8, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Lin, S.; King, I.; Spinka, T.; Dutschman, G.E.; Gullen, E.A.; Cheng, Y.C.; Doyle, T.W. Synthesis and comparative evaluation of two antiviral agents: Beta-L-Fd4C and beta-D-Fd4C. Bioorg. Med. Chem. Lett. 1998, 8, 3245–3250. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. PubChem Bioassay Record for AID 87578, Source: ChEMBL. 2023. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/87578 (accessed on 27 March 2023).
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Bento, A.P.; Hersey, A.; Félix, E.; Landrum, G.; Gaulton, A.; Atkinson, F.; Bellis, L.J.; De Veij, M.; Leach, A.R. An open source chemical structure curation pipeline using RDKit. J. Cheminform. 2020, 12, 51. [Google Scholar] [CrossRef]
- Cho, K.; Van Merrienboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using rnn encoder-decoder for statistical machine translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Jo, J.; Kwak, B.; Choi, H.S.; Yoon, S. The message passing neural networks for chemical property prediction on SMILES. Methods 2020, 179, 65–72. [Google Scholar] [CrossRef]


| Object | Target | Total | Training Set | Test Set | ||||
|---|---|---|---|---|---|---|---|---|
| Active | Inactive | Total | Active | Inactive | Total | |||
| HIV-1 | CCR5: C-C chemokine receptor type 5 | 2296 | 1478 | 129 | 1607 | 633 | 56 | 689 |
| CXCR4: C-X-C chemokine receptor type 4 | 92 | 57 | 7 | 64 | 25 | 3 | 28 | |
| 80s Ribosome | 55 | 27 | 11 | 38 | 12 | 5 | 17 | |
| IN: Integrase | 4161 | 1584 | 1328 | 2912 | 679 | 570 | 1249 | |
| PR: Protease | 5005 | 3162 | 341 | 3503 | 1356 | 146 | 1502 | |
| Protein Tat: Human immunodeficiency virus | 38 | 22 | 4 | 26 | 10 | 2 | 12 | |
| RT: Reverse transcriptase | 6122 | 3207 | 1078 | 4285 | 1375 | 462 | 1837 | |
| HBV | PARB | 350 | 237 | 8 | 245 | 101 | 4 | 105 |
| HBV-D: HBV genotype D | 73 | 45 | 6 | 51 | 19 | 3 | 22 | |
| NS5B: RNA-dependent RNA polymerase | 64 | 16 | 28 | 44 | 8 | 12 | 20 | |
| CRL4: E3 ubiquitin ligase | 2614 | 1096 | 733 | 1829 | 470 | 315 | 785 | |
| CD299: Core antigen of hepatitis | 124 | 70 | 16 | 86 | 31 | 7 | 38 | |
| Drugs | Experimental Factor Ontology (EFO) Terms | Max Phase for Indication * | References |
|---|---|---|---|
| tenofovir | HIV-1 infection | 3 | [37] |
| tenofovir | Hepatitis B virus infection | 3 | [38], FDA |
| hepsera | Chronic hepatitis B virus infection | 4 | [39], |
| hepsera | Hepatitis B virus infection | 4 | [40], FDA |
| hepsera | HIV infection | 3 | [41] |
| adefovir | HIV infection | 1 | [42] |
| adefovir | Hepatitis B virus infection | 3 | [40], FDA |
| tenofovir alafenamide | HIV-1 infection | 4 | [43] |
| tenofovir alafenamide | Hepatitis B virus infection | 4 | [44], FDA |
| interferon alfa-2b | HIV-1 infection | 3 | [45] |
| interferon alfa-2b | Hepatitis B virus infection | 4 | [46], FDA |
| abacavir | HIV-1 infection | 4 | [47], FDA |
| emtricitabine | HIV-1 infection | 4 | [48], FDA |
| emtricitabine | Hepatitis B virus infection | 3 | [49] |
| ritonavir | Hepatitis B virus infection | 2 | [50] |
| ritonavir | HIV-1 infection | 4 | [51], FDA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, Y.; Chen, X.; Zhao, L. HIV-1/HBV Coinfection Accurate Multitarget Prediction Using a Graph Neural Network-Based Ensemble Predicting Model. Int. J. Mol. Sci. 2023, 24, 7139. https://doi.org/10.3390/ijms24087139
Wang Y, Li Y, Chen X, Zhao L. HIV-1/HBV Coinfection Accurate Multitarget Prediction Using a Graph Neural Network-Based Ensemble Predicting Model. International Journal of Molecular Sciences. 2023; 24(8):7139. https://doi.org/10.3390/ijms24087139
Chicago/Turabian StyleWang, Yishu, Yue Li, Xiaomin Chen, and Lutao Zhao. 2023. "HIV-1/HBV Coinfection Accurate Multitarget Prediction Using a Graph Neural Network-Based Ensemble Predicting Model" International Journal of Molecular Sciences 24, no. 8: 7139. https://doi.org/10.3390/ijms24087139
APA StyleWang, Y., Li, Y., Chen, X., & Zhao, L. (2023). HIV-1/HBV Coinfection Accurate Multitarget Prediction Using a Graph Neural Network-Based Ensemble Predicting Model. International Journal of Molecular Sciences, 24(8), 7139. https://doi.org/10.3390/ijms24087139




