Modulation of Disease-Associated Pathways in Hidradenitis Suppurativa by the Janus Kinase 1 Inhibitor Povorcitinib: Transcriptomic and Proteomic Analyses of Two Phase 2 Studies
Abstract
1. Introduction
2. Results
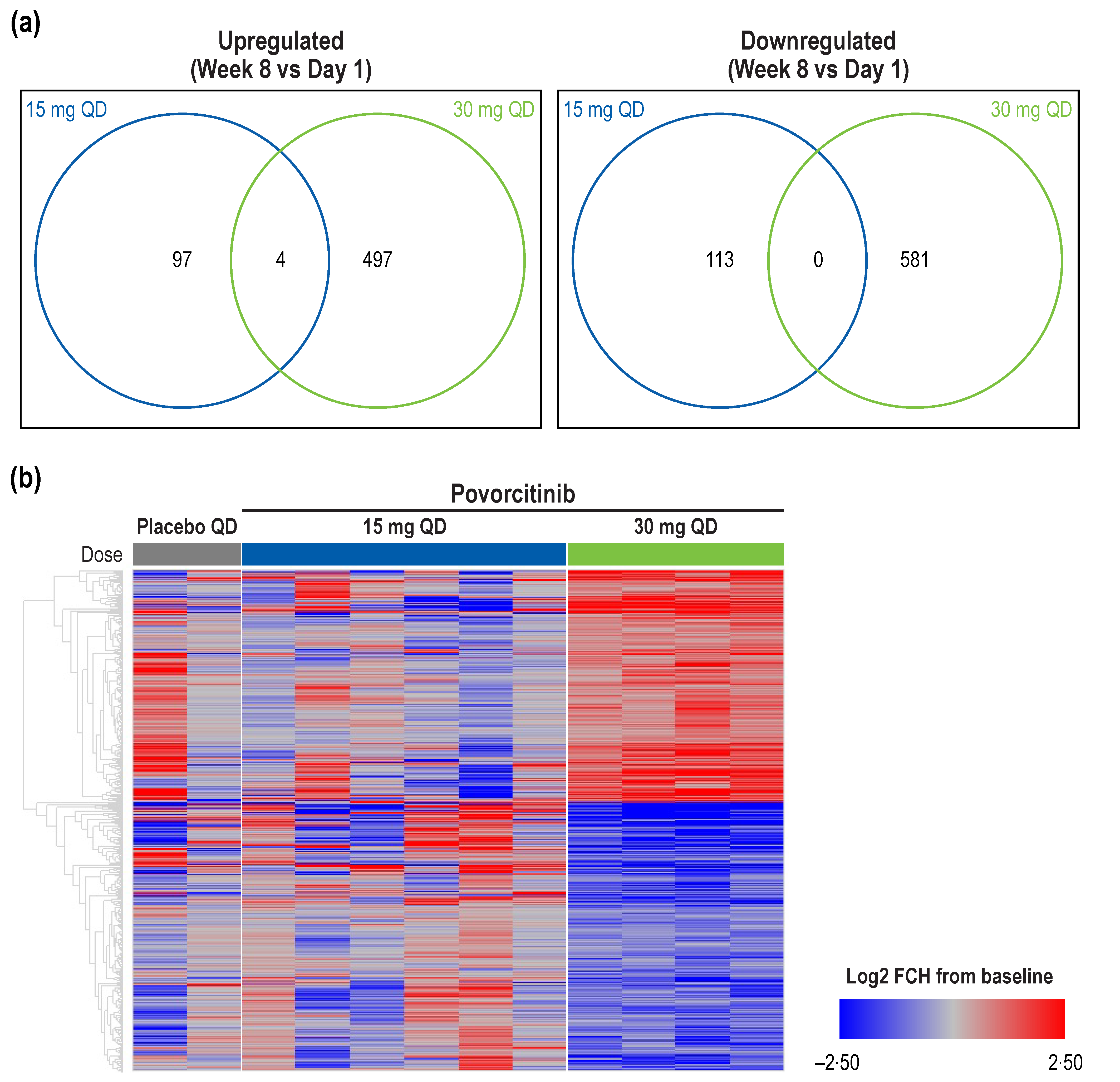
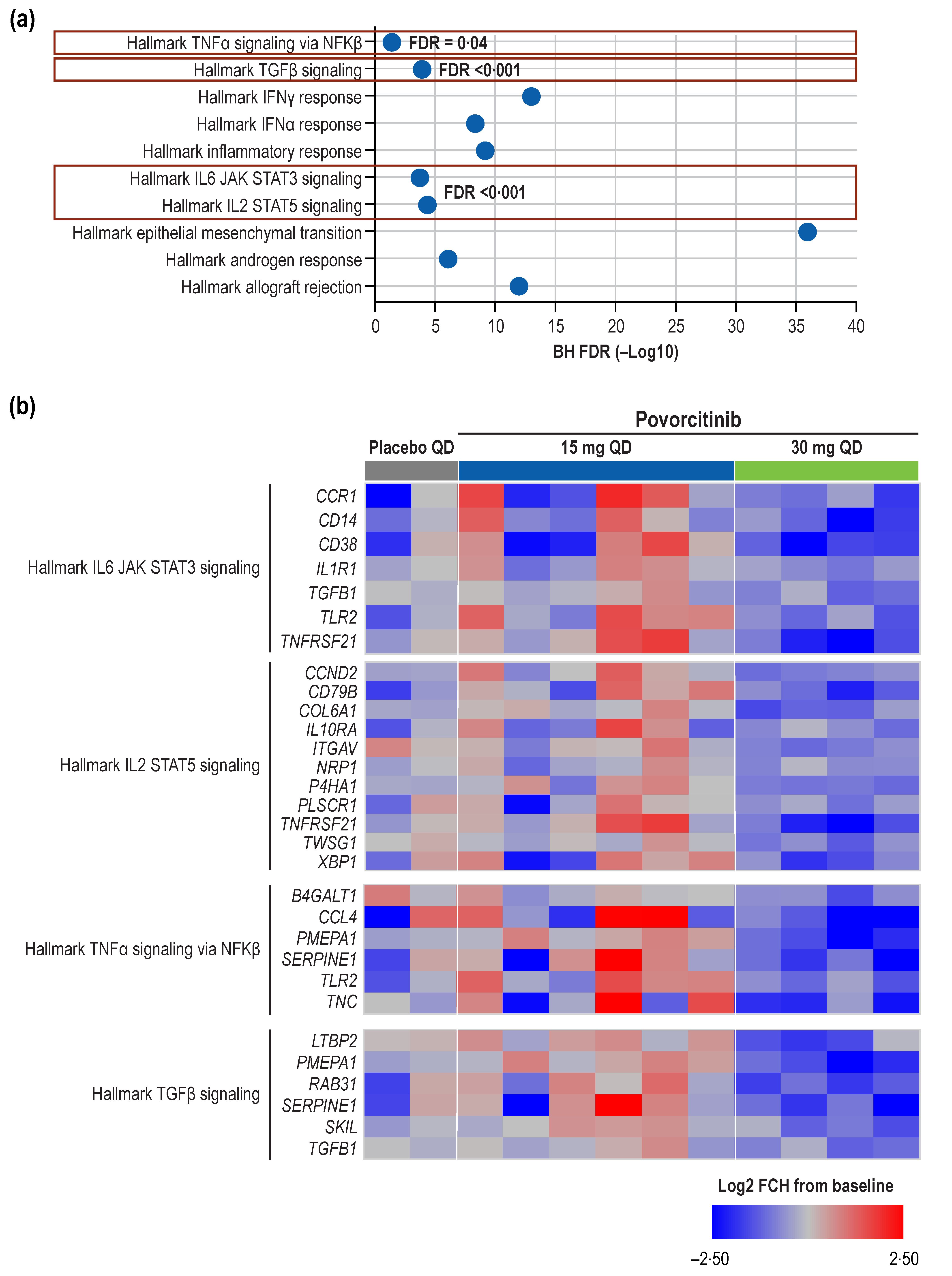
2.1. Transcriptomics
2.2. Proteomics
3. Discussion
4. Materials and Methods
4.1. Study Design and Patients
4.2. Transcriptomic Analyses
4.3. Proteomic Analyses
4.4. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sabat, R.; Jemec, G.B.E.; Matusiak, L.; Kimball, A.B.; Prens, E.; Wolk, K. Hidradenitis suppurativa. Nat. Rev. Dis. Prim. 2020, 6, 18. [Google Scholar] [CrossRef]
- Frew, J.W.; Lowes, M.A.; Goldfarb, N.; Butt, M.; Piguet, V.; O’Brien, E.; Ingram, J.; Jemec, G.B.E.; Tan, J.; Zouboulis, C.; et al. Global harmonization of morphological definitions in hidradenitis suppurativa for a proposed glossary. JAMA Dermatol. 2021, 157, 449–455. [Google Scholar] [CrossRef]
- Frew, J.W.; Hawkes, J.E.; Krueger, J.G. Topical, systemic and biologic therapies in hidradenitis suppurativa: Pathogenic insights by examining therapeutic mechanisms. Ther. Adv. Chronic Dis. 2019, 10, 2040622319830646. [Google Scholar] [CrossRef] [PubMed]
- Frew, J.W.; Marzano, A.V.; Wolk, K.; Join-Lambert, O.; Alavi, A.; Lowes, M.A.; Piguet, V. A systematic review of promising therapeutic targets in hidradenitis suppurativa: A critical evaluation of mechanistic and clinical relevance. J. Investig. Dermatol. 2021, 141, 316–324.e2. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Damiani, G.; Orenstein, L.A.V.; Hamzavi, I.; Jemec, G.B. Hidradenitis suppurativa: An update on epidemiology, phenotypes, diagnosis, pathogenesis, comorbidities and quality of life. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Zouboulis, C.C.; Benhadou, F.; Byrd, A.S.; Chandran, N.S.; Giamarellos-Bourboulis, E.J.; Fabbrocini, G.; Frew, J.W.; Fujita, H.; González-López, M.A.; Guillem, P.; et al. What causes hidradenitis suppurativa?-15 years after. Exp. Dermatol. 2020, 29, 1154–1170. [Google Scholar] [CrossRef]
- Navrazhina, K.; Renert-Yuval, Y.; Frew, J.W.; Grand, D.; Gonzalez, J.; Williams, S.C.; Garcet, S.; Krueger, J.G. Large-scale serum analysis identifies unique systemic biomarkers in psoriasis and hidradenitis suppurativa. Br. J. Dermatol. 2021, 186, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Navrazhina, K.; Garcet, S.; Zheng, X.; Hur, H.B.; Frew, J.W.; Krueger, J.G. High inflammation in hidradenitis suppurativa extends to perilesional skin and can be subdivided by lipocalin-2 expression. J. Allergy Clin. Immunol. 2022, 149, 135–144.e12. [Google Scholar] [CrossRef]
- Rumberger, B.E.; Boarder, E.L.; Owens, S.L.; Howell, M.D. Transcriptomic analysis of hidradenitis suppurativa skin suggests roles for multiple inflammatory pathways in disease pathogenesis. Inflamm. Res. 2020, 69, 967–973. [Google Scholar] [CrossRef]
- Coates, M.; Mariottoni, P.; Corcoran, D.L.; Kirshner, H.F.; Jaleel, T.; Brown, D.A.; Brooks, S.R.; Murray, J.; Morasso, M.I.; MacLeod, A.S. The skin transcriptome in hidradenitis suppurativa uncovers an antimicrobial and sweat gland gene signature which has distinct overlap with wounded skin. PLoS ONE 2019, 14, e0216249. [Google Scholar] [CrossRef]
- De Oliveira, A.; Bloise, G.; Moltrasio, C.; Coelho, A.; Agrelli, A.; Moura, R.; Tricarico, P.M.; Jamain, S.; Marzano, A.V.; Crovella, S.; et al. Transcriptome meta-analysis confirms the hidradenitis suppurativa pathogenic triad: Upregulated inflammation, altered epithelial organization, and dysregulated metabolic signaling. Biomolecules 2022, 12, 1371. [Google Scholar] [CrossRef] [PubMed]
- Jemec, G.B.E. Hidradenitis suppurativa and immune dysregulation. Br. J. Dermatol. 2012, 166, 237–238. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, A.S.; Mansbridge, J.N. The innate immune system in acute and chronic wounds. Adv. Wound Care 2015, 5, 65–78. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Frew, J.W.; Giamarellos-Bourboulis, E.J.; Jemec, G.B.E.; Del Marmol, V.; Marzano, A.V.; Nikolakis, G.; Sayed, C.J.; Tzellos, T.; Wolk, K.; et al. Target molecules for future hidradenitis suppurativa treatment. Exp. Dermatol. 2021, 30 (Suppl. S1), 8–17. [Google Scholar] [CrossRef]
- Howell, M.D.; Kuo, F.I.; Smith, P.A. Targeting the Janus kinase family in autoimmune skin diseases. Front. Immunol. 2019, 10, 2342. [Google Scholar] [CrossRef]
- Witte-Handel, E.; Wolk, K.; Tsaousi, A.; Irmer, M.L.; Mossner, R.; Shomroni, O.; Lingner, T.; Witte, K.; Kunkel, D.; Salinas, G.; et al. The IL-1 pathway is hyperactive in hidradenitis suppurativa and contributes to skin infiltration and destruction. J. Investig. Dermatol. 2019, 139, 1294–1305. [Google Scholar] [CrossRef] [PubMed]
- Solimani, F.; Meier, K.; Ghoreschi, K. Emerging topical and systemic JAK inhibitors in dermatology. Front. Immunol. 2019, 10, 2847. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Gallo, D.; de la Varga-Martinez, R.; Ossorio-Garcia, L.; Albarran-Planelles, C.; Rodriguez, C.; Linares-Barrios, M. The clinical significance of increased serum proinflammatory cytokines, C-reactive protein, and erythrocyte sedimentation rate in patients with hidradenitis suppurativa. Mediators Inflamm. 2017, 2017, 2450401. [Google Scholar] [CrossRef]
- Alavi, A.; Hamzavi, I.; Brown, K.; Santos, L.L.; Zhu, Z.; Liu, H.; Howell, M.D.; Kirby, J. Janus kinase 1 inhibitor INCB054707 for patients with moderate-to-severe hidradenitis suppurativa: Results from two phase 2 studies. Br. J. Dermatol. 2022, 186, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Frew, J.W.; Navrazhina, K.; Byrd, A.S.; Garg, A.; Ingram, J.R.; Kirby, J.S.; Lowes, M.A.; Naik, H.; Piguet, V.; Prens, E.P. Defining lesional, perilesional and unaffected skin in hidradenitis suppurativa: Proposed recommendations for clinical trials and translational research studies. Br. J. Dermatol. 2019, 181, 1339–1341. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Fuchs, E. Sweat gland progenitors in development, homeostasis, and wound repair. Cold Spring Harb. Perspect. Med. 2014, 4, a015222. [Google Scholar] [CrossRef]
- Li, S.; Zheng, X.; Nie, Y.; Chen, W.; Liu, Z.; Tao, Y.; Hu, X.; Hu, Y.; Qiao, H.; Qi, Q.; et al. Defining key genes regulating morphogenesis of apocrine sweat gland in sheepskin. Front. Genet. 2018, 9, 739. [Google Scholar] [CrossRef]
- Moll, R.; Divo, M.; Langbein, L. The human keratins: Biology and pathology. Histochem. Cell Biol. 2008, 129, 705. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhang, X.; Chen, L.; Zhang, L.; Li, H. K31 as a novel marker for clear secretory cells in human eccrine sweat glands. J. Mol. Histol. 2020, 51, 47–53. [Google Scholar] [CrossRef]
- Cui, C.-Y.; Childress, V.; Piao, Y.; Michel, M.; Johnson, A.A.; Kunisada, M.; Ko, M.S.H.; Kaestner, K.H.; Marmorstein, A.D.; Schlessinger, D. Forkhead transcription factor FoxA1 regulates sweat secretion through Bestrophin 2 anion channel and Na-K-Cl cotransporter 1. Proc. Natl. Acad. Sci. USA 2012, 109, 1199. [Google Scholar] [CrossRef] [PubMed]
- Matusiak, Ł.; Bieniek, A.; Szepietowski, J.C. Soluble interleukin-2 receptor serum level is a useful marker of hidradenitis suppurativa clinical staging. Biomarkers 2009, 14, 432–437. [Google Scholar] [CrossRef]
- Banerjee, A.; McNish, S.; Shanmugam, V.K. Interferon-gamma (IFN-γ) is elevated in wound exudate from hidradenitis suppurativa. Immunol. Investig. 2017, 46, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Prenzler, S.; Rudrawar, S.; Waespy, M.; Kelm, S.; Anoopkumar-Dukie, S.; Haselhorst, T. The role of sialic acid-binding immunoglobulin-like-lectin-1 (siglec-1) in immunology and infectious disease. Int. Rev. Immunol. 2021, 42, 113–138. [Google Scholar] [CrossRef]
- Zheng, Q.; Hou, J.; Zhou, Y.; Yang, Y.; Xie, B.; Cao, X. Siglec1 suppresses antiviral innate immune response by inducing TBK1 degradation via the ubiquitin ligase TRIM27. Cell Res. 2015, 25, 1121–1136. [Google Scholar] [CrossRef]
- Page, T.H.; Charles, P.J.; Piccinini, A.M.; Nicolaidou, V.; Taylor, P.C.; Midwood, K.S. Raised circulating tenascin-C in rheumatoid arthritis. Arthritis Res. Ther. 2012, 14, R260. [Google Scholar] [CrossRef] [PubMed]
- Závada, J.; Uher, M.; Svobodová, R.; Olejárová, M.; Hušáková, M.; Ciferská, H.; Hulejová, H.; Tomčík, M.; Šenolt, L.; Vencovský, J. Serum tenascin-C discriminates patients with active SLE from inactive patients and healthy controls and predicts the need to escalate immunosuppressive therapy: A cohort study. Arthritis Res. Ther. 2015, 17, 341. [Google Scholar] [CrossRef]
- Yuan, W.; Zhang, W.; Yang, X.; Zhou, L.; Hanghua, Z.; Xu, K. Clinical significance and prognosis of serum tenascin-C in patients with sepsis. BMC Anesthesiol. 2018, 18, 170. [Google Scholar] [CrossRef]
- Kalia, N.; Singh, J.; Kaur, M. The role of dectin-1 in health and disease. Immunobiology 2021, 226, 152071. [Google Scholar] [CrossRef]
- Der Sarkissian, S.; Hessam, S.; Kirby, J.S.; Lowes, M.A.; Mintoff, D.; Naik, H.B.; Ring, H.C.; Chandran, N.S.; Frew, J.W. Identification of biomarkers and critical evaluation of biomarker validation in hidradenitis suppurativa: A systematic review. JAMA Dermatol. 2022, 158, 300–313. [Google Scholar] [CrossRef]
- Lowe, M.M.; Naik, H.B.; Clancy, S.; Pauli, M.; Smith, K.M.; Bi, Y.; Dunstan, R.; Gudjonsson, J.E.; Paul, M.; Harris, H.; et al. Immunopathogenesis of hidradenitis suppurativa and response to anti-TNF-α therapy. J. Clin. Investig. 2020, 5, e139932. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Hong, F.; Conlon, D.M.; Sidur, L.; Smith, K.M.; Fang, Y.; Cuff, C.A.; Kaymakcalan, Z.; Ruzek, M.C. Potential predictive biomarkers of adalimumab response in patients with hidradenitis suppurativa. Br. J. Dermatol. 2021, 185, 804–814. [Google Scholar] [CrossRef]
- Montaudié, H.; Seitz-Polski, B.; Cornille, A.; Benzaken, S.; Lacour, J.P.; Passeron, T. Interleukin 6 and high-sensitivity C-reactive protein are potential predictive markers of response to infliximab in hidradenitis suppurativa. J. Am. Acad. Dermatol. 2017, 76, 156–158. [Google Scholar] [CrossRef] [PubMed]
- Blok, J.L.; Li, K.; Brodmerkel, C.; Horvátovich, P.; Jonkman, M.F.; Horváth, B. Ustekinumab in hidradenitis suppurativa: Clinical results and a search for potential biomarkers in serum. Br. J. Dermatol. 2016, 174, 839–846. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Iglesias-Bartolome, R.; Uchiyama, A.; Molinolo, A.A.; Abusleme, L.; Brooks, S.R.; Callejas-Valera, J.L.; Edwards, D.; Doci, C.; Asselin-Labat, M.L.; Onaitis, M.W.; et al. Transcriptional signature primes human oral mucosa for rapid wound healing. Sci. Transl. Med. 2018, 10, eaap8798. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Santos, L.L.; Smith, S.H. Modulation of Disease-Associated Pathways in Hidradenitis Suppurativa by the Janus Kinase 1 Inhibitor Povorcitinib: Transcriptomic and Proteomic Analyses of Two Phase 2 Studies. Int. J. Mol. Sci. 2023, 24, 7185. https://doi.org/10.3390/ijms24087185
Liu H, Santos LL, Smith SH. Modulation of Disease-Associated Pathways in Hidradenitis Suppurativa by the Janus Kinase 1 Inhibitor Povorcitinib: Transcriptomic and Proteomic Analyses of Two Phase 2 Studies. International Journal of Molecular Sciences. 2023; 24(8):7185. https://doi.org/10.3390/ijms24087185
Chicago/Turabian StyleLiu, Huiqing, Leandro L. Santos, and Susan H. Smith. 2023. "Modulation of Disease-Associated Pathways in Hidradenitis Suppurativa by the Janus Kinase 1 Inhibitor Povorcitinib: Transcriptomic and Proteomic Analyses of Two Phase 2 Studies" International Journal of Molecular Sciences 24, no. 8: 7185. https://doi.org/10.3390/ijms24087185
APA StyleLiu, H., Santos, L. L., & Smith, S. H. (2023). Modulation of Disease-Associated Pathways in Hidradenitis Suppurativa by the Janus Kinase 1 Inhibitor Povorcitinib: Transcriptomic and Proteomic Analyses of Two Phase 2 Studies. International Journal of Molecular Sciences, 24(8), 7185. https://doi.org/10.3390/ijms24087185





