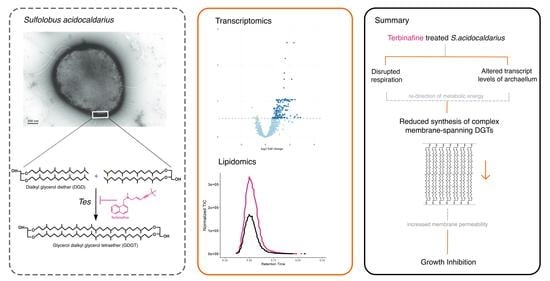
Membrane Adaptations and Cellular Responses of Sulfolobus acidocaldarius to the Allylamine Terbinafine
Abstract
:1. Introduction
2. Results
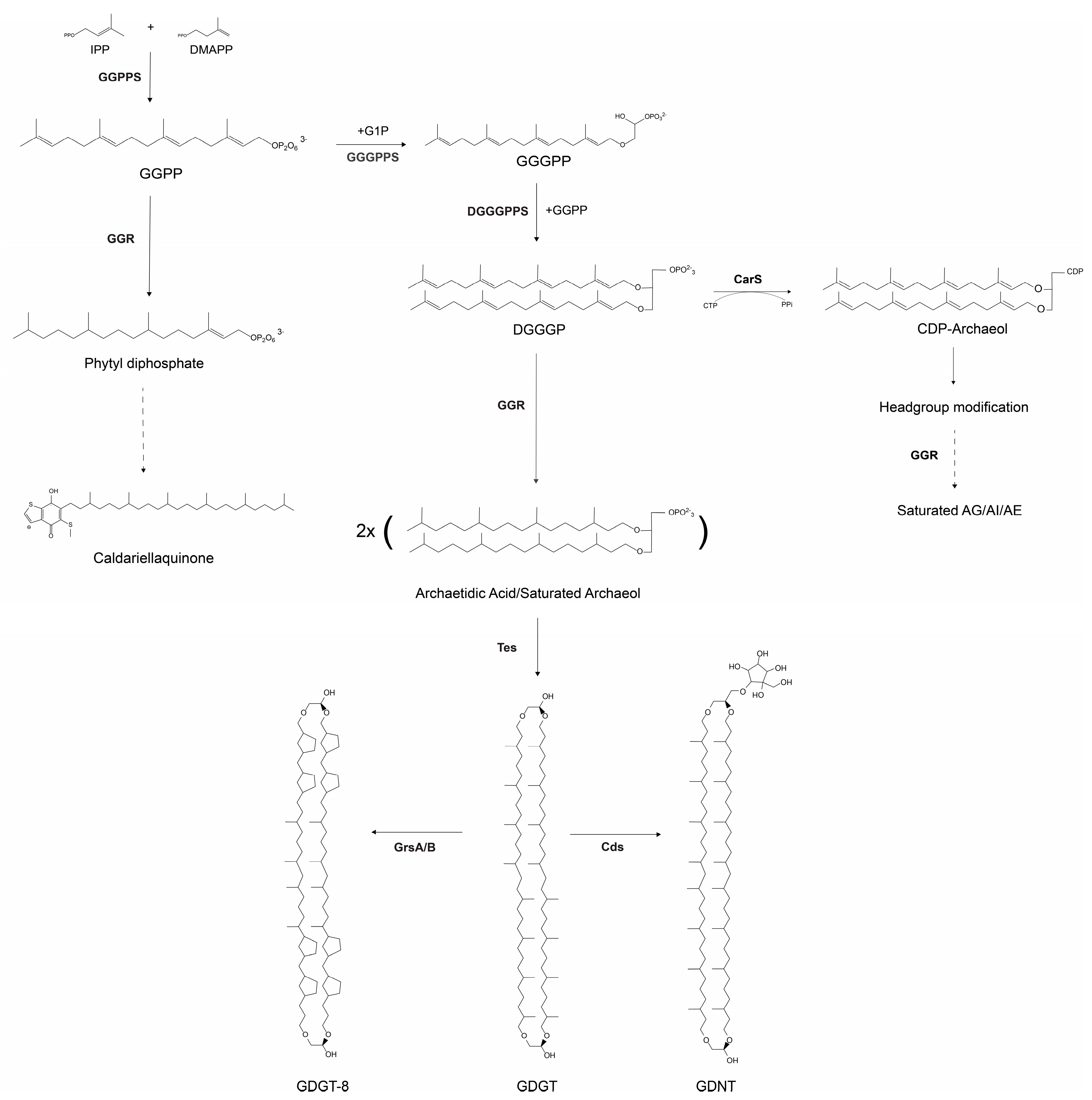
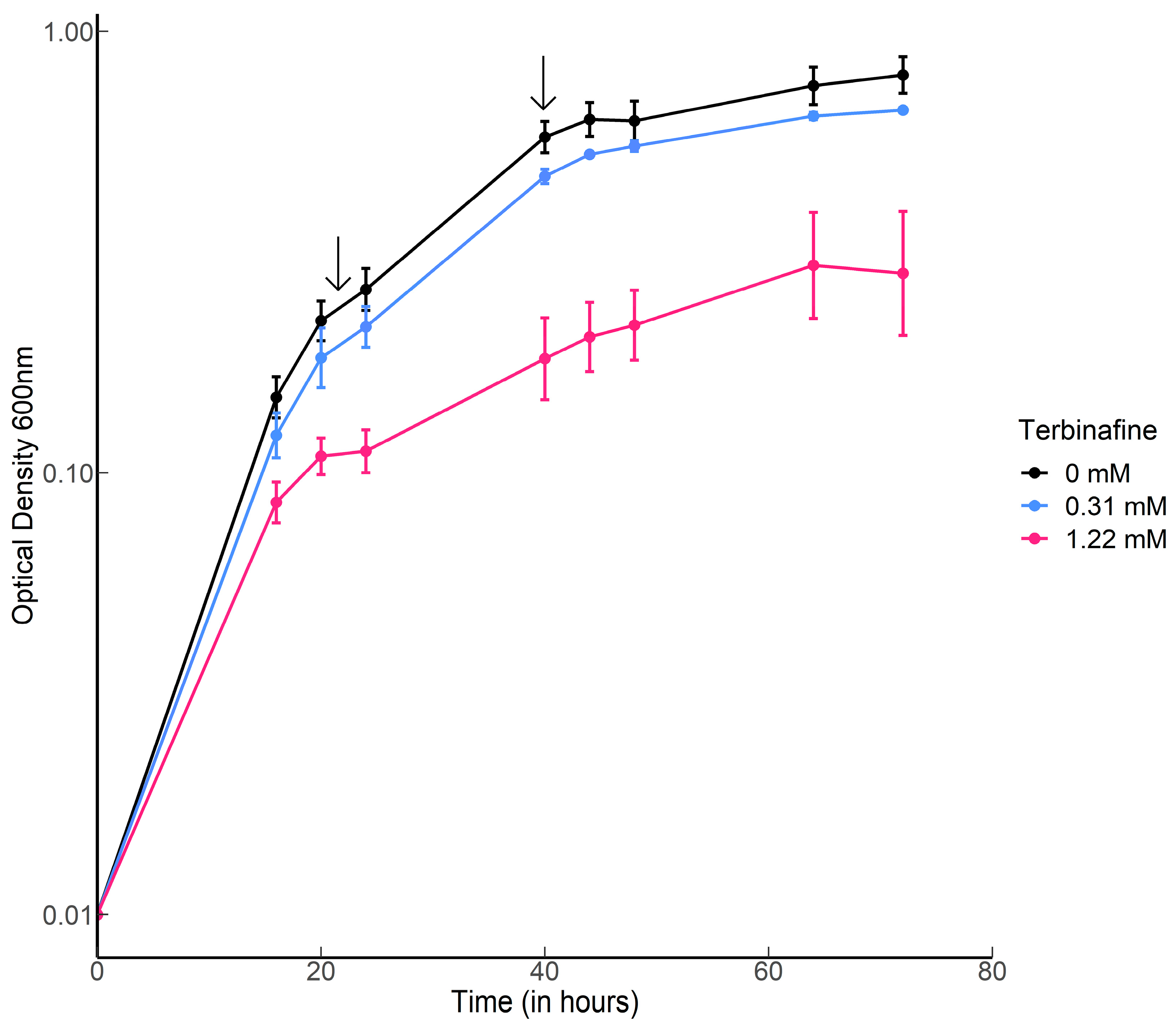
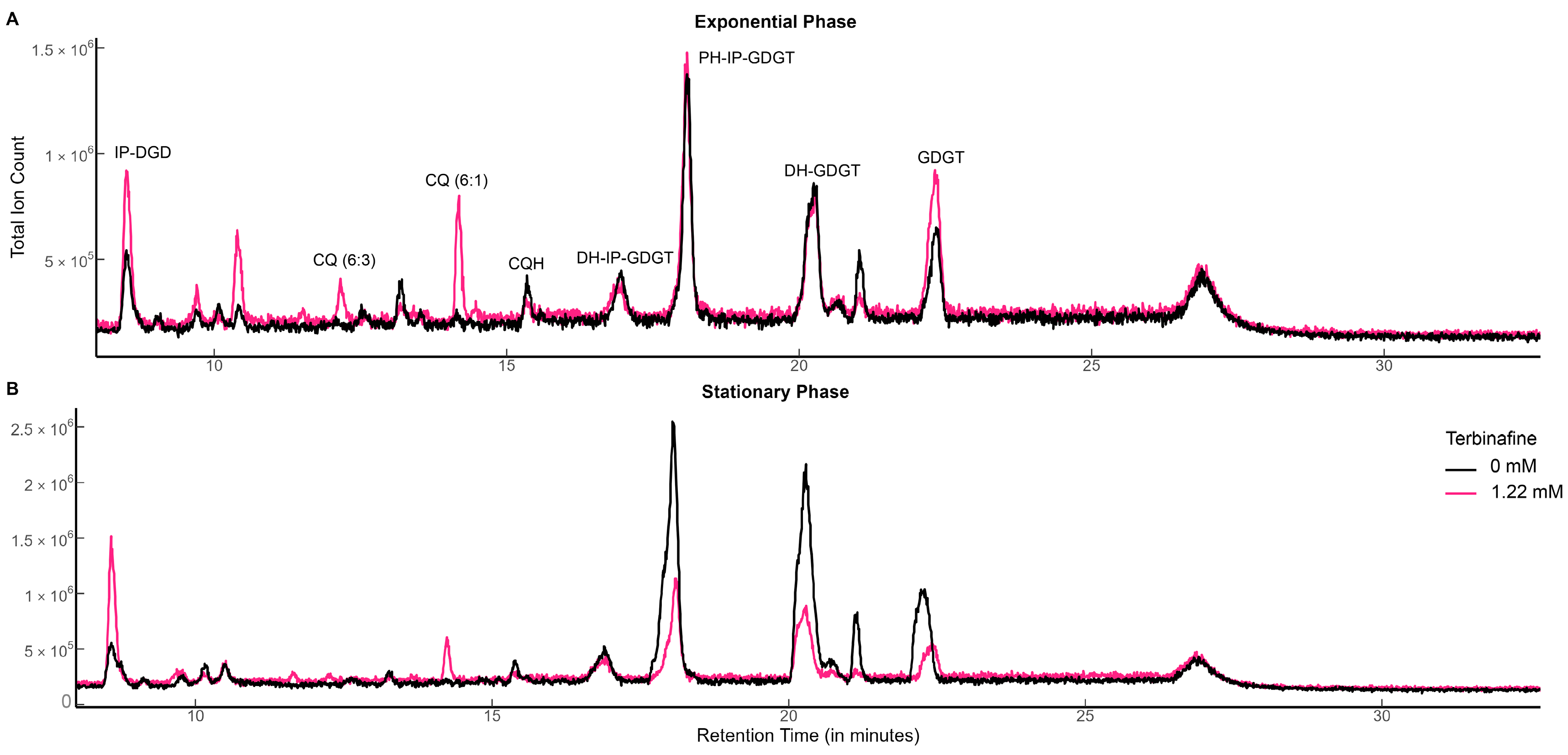
2.1. Terbinafine Causes the Accumulation of DGDs and Depletion of DGTs in S. acidocaldarius Membranes
2.2. Terbinafine Interferes with the Respiratory Complex in S. acidocaldarius
2.3. A Shift in the Saturation Levels of Caldariellaquinone
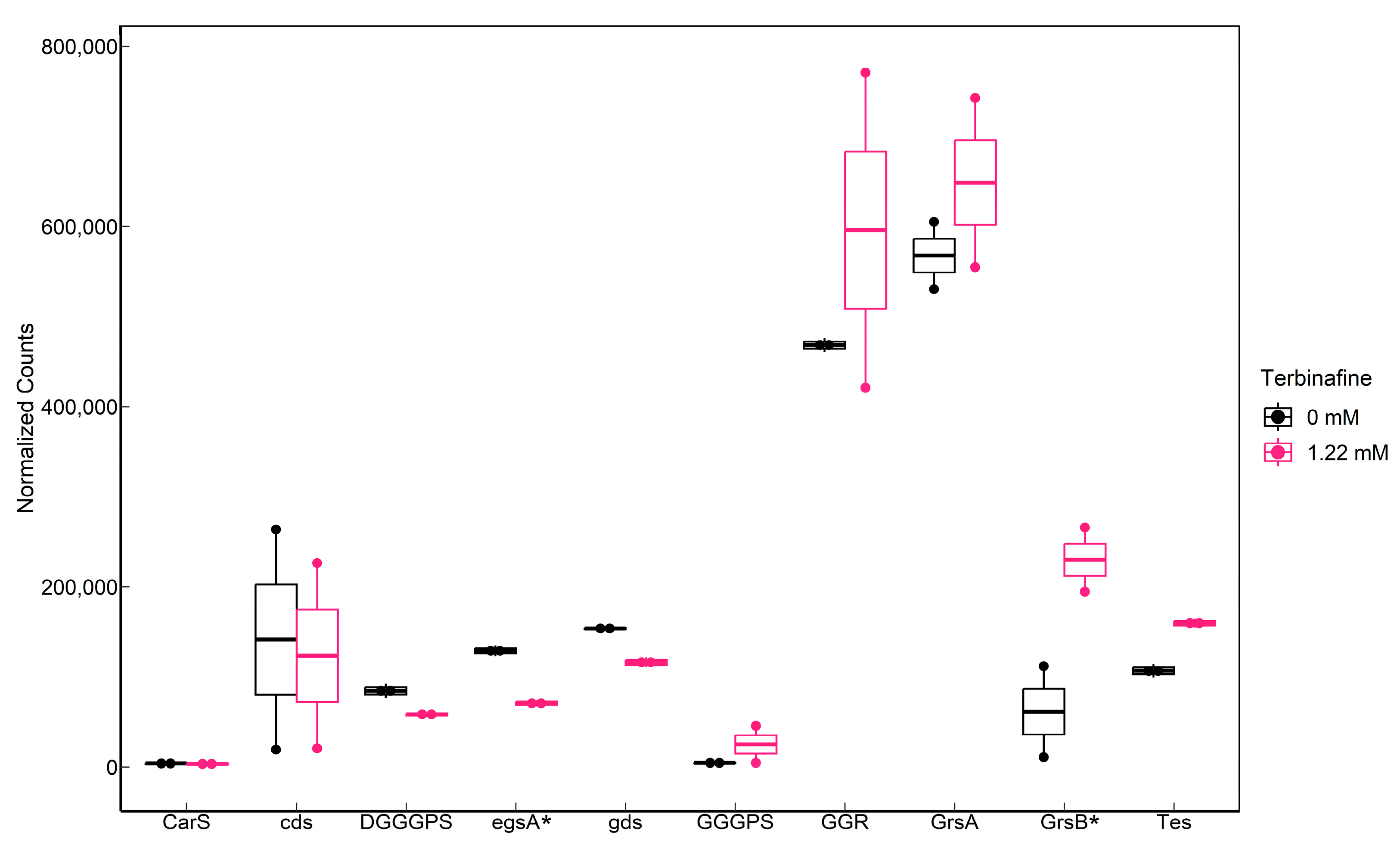
2.4. Influence of Terbinafine on the Expression of Phospholipid Biosynthesis Genes
2.5. Cell Envelope and Motility
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. RNA Isolation, Sequencing and Transcriptome Analysis
4.3. Lipid Extraction and Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jain, S.; Caforio, A.; Driessen, A.J.M. Biosynthesis of archaeal membrane ether lipids. Front. Microbiol. 2014, 5, 641. [Google Scholar] [CrossRef] [Green Version]
- Rastädter, K.; Wurm, D.J.; Spadiut, O.; Quehenberger, J. The cell membrane of Sulfolobus spp.—Homeoviscous adaption and biotechnological applications. Int. J. Mol. Sci. 2020, 21, 3935. [Google Scholar] [CrossRef]
- Koga, Y.; Ohga, M.; Tsujimura, M.; Morii, H.; Kawarabayasi, Y. Identification of sn-glycerol-1-phosphate dehydrogenase activity from genomic information on a hyperthermophilic archaeon, Sulfolobus tokodaii strain 7. Biosci. Biotechnol. Biochem. 2006, 70, 282–285. [Google Scholar] [CrossRef] [Green Version]
- Ren, S.; de Kok, N.A.W.; Gu, Y.; Yan, W.; Sun, Q.; Chen, Y.; He, J.; Tian, L.; Andringa, R.L.H.; Zhu, X.; et al. Structural and Functional Insights into an Archaeal Lipid Synthase. Cell Rep. 2020, 33, 108294. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Caforio, A.; Fodran, P.; Lolkema, J.S.; Minnaard, A.J.; Driessen, A.J.M. Identification of CDP-Archaeol Synthase, a Missing Link of Ether Lipid Biosynthesis in Archaea. Chem. Biol. 2014, 21, 1392–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, S.; Murakami, M.; Yoshimura, T.; Hemmi, H. Specific partial reduction of geranylgeranyl diphosphate by an enzyme from the thermoacidophilic archaeon Sulfolobus acidocaldarius yields a reactive prenyl donor, not a dead-end product. J. Bacteriol. 2008, 190, 3923–3929. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, D.; Fujihashi, M.; Iwata, Y.; Murakami, M.; Yoshimura, T.; Hemmi, H.; Miki, K. Structure and mutation analysis of archaeal geranylgeranyl reductase. J. Mol. Biol. 2011, 409, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Caforio, A.; Yang, Q.; Sun, B.; Yu, F.; Zhu, X.; Wang, J.; Dou, C.; Fu, Q.; Huang, N.; et al. Structural and mechanistic insights into the biosynthesis of CDP-archaeol in membranes. Cell Res. 2017, 27, 1378–1391. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, C.T.; Iwig, D.F.; Wang, B.; Cossu, M.; Metcalf, W.W.; Boal, A.K.; Booker, S.J. Discovery, structure and mechanism of a tetraether lipid synthase. Nature 2022, 609, 197–203. [Google Scholar] [CrossRef]
- Zeng, Z.; Chen, H.; Yang, H.; Chen, Y.; Yang, W.; Feng, X.; Pei, H.; Welander, P.V. Identification of a protein responsible for the synthesis of archaeal membrane-spanning GDGT lipids. Nat. Commun. 2022, 13, 1545. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.; Delago, A.; Nußbaum, P.; Meyer, B.H.; Albers, S.-V.; Eichler, J. Gene deletions leading to a reduction in the number of cyclopentane rings in Sulfolobus acidocaldarius tetraether lipids. FEMS Microbiol. Lett. 2018, 365, fnx250. [Google Scholar] [CrossRef]
- Yang, W.; Chen, H.; Chen, Y.; Chen, A.; Feng, X.; Zhao, B.; Zheng, F.; Fang, H.; Zhang, C.; Zeng, Z. Thermophilic archaeon orchestrates temporal expression of GDGT ring synthases in response to temperature and acidity stress. Environ. Microbiol. 2022, 25, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Liu, X.-L.; Wei, J.H.; Summons, R.E.; Welander, P.V. Calditol-linked membrane lipids are required for acid tolerance in Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA 2018, 115, 12932–12937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schouten, S.; Van Der Meer, M.T.J.; Hopmans, E.C.; Rijpstra, W.I.C.; Reysenbach, A.L.; Ward, D.M.; Damsté, J.S.S. Archaeal and bacterial glycerol dialkyl glycerol tetraether lipids in hot springs of Yellowstone National Park. Appl. Environ. Microbiol. 2007, 73, 6181–6191. [Google Scholar] [CrossRef] [Green Version]
- Jensen, S.M.; Neesgaard, V.L.; Skjoldbjerg, S.L.N.; Brandl, M.; Ejsing, C.S.; Treusch, A.H. The Effects of Temperature and Growth Phase on the Lipidomes of Sulfolobus islandicus and Sulfolobus tokodaii. Life 2015, 5, 1539–1566. [Google Scholar] [CrossRef] [Green Version]
- Tourte, M.; Schaeffer, P.; Grossi, V.; Oger, P.M. Membrane adaptation in the hyperthermophilic archaeon Pyrococcus furiosus relies upon a novel strategy involving glycerol monoalkyl glycerol tetraether lipids. Environ. Microbiol. 2022, 24, 2029–2046. [Google Scholar] [CrossRef]
- Quehenberger, J.; Pittenauer, E.; Allmaier, G.; Spadiut, O. The influence of the specific growth rate on the lipid composition of Sulfolobus acidocaldarius. Extremophiles 2020, 24, 413–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Driessen, A.J.M.; Albers, S. Membrane Adaptations of (Hyper)Thermophiles to High Temperatures. In Physiology and Biochemistry of Extremophiles; Gerday, C., Glansdorff, N., Eds.; ASM Press: Washington, DC, USA, 2007; pp. 104–116. [Google Scholar]
- Lo, S.-L.; Montague, C.E.; Chang, E.L. Purification of glycerol dialkyl nonitol tetraether from Sulfolobus acidocaldarius. J. Lipid Res. 1989, 30, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Castell, M.; Tourte, M.; Oger, P.M. In search for the membrane regulators of archaea. Int. J. Mol. Sci. 2019, 20, 4434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elling, F.J.; Becker, K.W.; Könneke, M.; Schröder, J.M.; Kellermann, M.Y.; Thomm, M.; Hinrichs, K.-U. Respiratory quinones in Archaea: Phylogenetic distribution and application as biomarkers in the marine environment. Environ. Microbiol. 2016, 18, 692–707. [Google Scholar] [CrossRef]
- Janssen, S.; Schäfer, G.; Anemüller, S.; Moll, R. A succinate dehydrogenase with novel structure and properties from the hyperthermophilic archaeon Sulfolobus acidocaldarius: Genetic and biophysical characterization. J. Bacteriol. 1997, 179, 5560–5569. [Google Scholar] [CrossRef] [Green Version]
- Reviews, F.M.; Femsre, E.; Sch, G.; Anemtiller, S. Electron transport and energy conservation in the archaebacterium Sulfolobus acidocaldarius. FEMS Microbiol. Lett. 1990, 75, 335–348. [Google Scholar]
- Whit, A.V.; Kingston, D.G.I.; Niehaus, W.G. Biosynthesis of Caldariellaquinone in Sulfolobus Acidocaldarius. Ph.D. Thesis, Virginia Tech, Blacksburg, VA, USA, 1991. [Google Scholar]
- Kon, T.; Nemoto, N.; Oshima, T.; Yamagishi, A. Effects of a squalene epoxidase inhibitor, terbinafine, on ether lipid biosyntheses in a thermoacidophilic archaeon, Thermoplasma acidophilum. J. Bacteriol. 2002, 184, 1395–1401. [Google Scholar] [CrossRef] [Green Version]
- Nowosielski, M.; Hoffmann, M.; Wyrwicz, L.S.; Stepniak, P.; Plewczynski, D.M.; Lazniewski, M.; Ginalski, K.; Rychlewski, L. Detailed mechanism of squalene epoxidase inhibition by terbinafine. J. Chem. Inf. Model. 2011, 51, 455–462. [Google Scholar] [CrossRef] [PubMed]
- De Kok, N.A.W.; Exterkate, M.; Andringa, R.L.H.; Minnaard, A.J.; Driessen, A.J.M. A versatile method to separate complex lipid mixtures using 1-butanol as eluent in a reverse-phase UHPLC-ESI-MS system. Chem. Phys. Lipids 2021, 240, 105125. [Google Scholar] [CrossRef]
- Stark, H.; Wolf, J.; Albersmeier, A.; Pham, T.K.; Hofmann, J.D.; Siebers, B.; Kalinowski, J.; Wright, P.C.; Neumann-Schaal, M.; Schomburg, D. Oxidative Stickland reactions in an obligate aerobic organism-amino acid catabolism in the Crenarchaeon Sulfolobus solfataricus. FEBS J. 2017, 284, 2078–2095. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, G.; Moll, R.; Schmidt, C.L. Respiratory enzymes from Sulfolobus acidocaldarius. Methods Enzymol. 2001, 331, 369–410. [Google Scholar] [CrossRef]
- Wakao, H.; Wakagi, T.; Oshima, T. Purification and properties of NADH dehydrogenase from a thermoacidophilic archaebacterium, Sulfolobus acidocaldarius. J. Biochem. 1987, 102, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Auernik, K.S.; Kelly, R.M. Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl. Environ. Microbiol. 2008, 74, 7723–7732. [Google Scholar] [CrossRef] [Green Version]
- Nicolaus, B.; Trincone, A.; Lama, L.; Palmieri, G.; Gambacorta, A. Quinone Composition in Sulfolobus solfataricus Grown under Different Conditions. Syst. Appl. Microbiol. 1992, 15, 18–20. [Google Scholar] [CrossRef]
- Hemmi, H.; Takahashi, Y.; Shibuya, K.; Nakayama, T.; Nishino, T. Menaquinone-specific prenyl reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus. J. Bacteriol. 2005, 187, 1937–1944. [Google Scholar] [CrossRef] [Green Version]
- Hemmi, H.; Ikejiri, S.; Yamashita, S.; Nishino, T. Novel medium-chain prenyl diphosphate synthase from the thermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 2002, 184, 615–620. [Google Scholar] [CrossRef] [Green Version]
- Zhou, A.; Chiu, B.; Weber, Y.; Elling, F.; Cobban, A.; Pearson, A.; Leavitt, W. Energy flux controls tetraether lipid cyclization in Sulfolobus acidocaldarius. Environ. Microbiol. 2019, 22, 744623. [Google Scholar] [CrossRef] [PubMed]
- Albers, S.V.; Jarrell, K.F. The archaellum: How Archaea swim. Front. Microbiol. 2015, 6, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, A.; Hartung, S.; Van Der Does, C.; Tainer, J.A.; Albers, S.V. Archaeal flagellar ATPase motor shows ATP-dependent hexameric assembly and activity stimulation by specific lipid binding. Biochem. J. 2011, 437, 43–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lassak, K.; Peeters, E.; Wróbel, S.; Albers, S.-V. The one-component system ArnR: A membrane-bound activator of the crenarchaeal archaellum. Mol. Microbiol. 2013, 88, 125–139. [Google Scholar] [CrossRef]
- Hoffmann, L.; Anders, K.; Bischof, L.F.; Ye, X.; Reimann, J.; Khadouma, S.; Pham, T.K.; Van Der Does, C.; Wright, P.C.; Essen, L.O.; et al. Structure and interactions of the archaeal motility repression module ArnA-ArnB that modulates archaellum gene expression in Sulfolobus acidocaldarius. J. Biol. Chem. 2019, 294, 7460–7471. [Google Scholar] [CrossRef] [Green Version]
- Bischof, L.F.; Haurat, M.F.; Albers, S.V. Two membrane-bound transcription factors regulate expression of various type-IV-pili surface structures in Sulfolobus acidocaldarius. PeerJ 2019, 2019, e6459. [Google Scholar] [CrossRef]
- Lassak, K.; Neiner, T.; Ghosh, A.; Klingl, A.; Wirth, R.; Albers, S.-V. Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 2012, 83, 110–124. [Google Scholar] [CrossRef]
- Gambelli, L.; Meyer, B.H.; McLaren, M.; Sanders, K.; Quax, T.E.F.; Gold, V.A.M.; Albers, S.V.; Daum, B. Architecture and modular assembly of Sulfolobus S-layers revealed by electron cryotomography. Proc. Natl. Acad. Sci. USA 2019, 116, 25278–25286. [Google Scholar] [CrossRef] [Green Version]
- Bischof, L.F.; Haurat, M.F.; Hoffmann, L.; Albersmeier, A.; Wolf, J.; Neu, A.; Pham, T.K.; Albaum, S.P.; Jakobi, T.; Schouten, S.; et al. Early Response of Sulfolobus acidocaldarius to Nutrient Limitation. Front. Microbiol. 2019, 9, 3201. [Google Scholar] [CrossRef]
- Shimada, H.; Nemoto, N.; Shida, Y.; Oshima, T.; Yamagishi, A. Effects of pH and temperature on the composition of polar lipids in Thermoplasma acidophilum HO-62. J. Bacteriol. 2008, 190, 5404–5411. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, M.L. The multifunctional fungal ergosterol. mBio 2018, 9, e01755-18. [Google Scholar] [CrossRef] [Green Version]
- Rohmer, M. A Mevalonate-independent Route to Isopentenyl Diphosphate. In Comprehensive Natural Products Chemistry; Elsevier: Amsterdam, The Netherlands, 1999; pp. 45–67. [Google Scholar]
- Sáenz, J.P.; Sezgin, E.; Schwille, P.; Simons, K. Functional convergence of hopanoids and sterols in membrane ordering. Proc. Natl. Acad. Sci. USA 2012, 109, 14236–14240. [Google Scholar] [CrossRef] [Green Version]
- Collins, M.D.; Langworthy, T.A. Respiratory Quinone Composition of Some Acidophilic Bacteria. Syst. Appl. Microbiol. 1983, 4, 295–304. [Google Scholar] [CrossRef]
- Wang, K.; Sybers, D.; Maklad, H.R.; Lemmens, L.; Lewyllie, C.; Zhou, X.; Schult, F.; Bräsen, C.; Siebers, B.; Valegård, K.; et al. A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius. Nat. Commun. 2019, 10, 1542. [Google Scholar] [CrossRef] [Green Version]
- Nemoto, N.; Shida, Y.; Shimada, H.; Oshima, T.; Yamagishi, A. Characterization of the precursor of tetraether lipid biosynthesis in the thermoacidophilic archaeon Thermoplasma acidophilum. Extremophiles 2003, 7, 235–243. [Google Scholar] [CrossRef]
- Van de Vossenberg, J.L.C.M.; Driessen, A.J.M.; Konings, W.N. The essence of being extremophilic: The role of the unique archaeal membrane lipids. Extremophiles 1998, 2, 163–170. [Google Scholar] [CrossRef] [Green Version]
- Siliakus, M.F.; van der Oost, J.; Kengen, S.W.M. Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 2017, 21, 651–670. [Google Scholar] [CrossRef] [Green Version]
- Hanford, M.J.; Peeples, T.L. Archaeal Tetraether Lipids: Unique Structures and Applications. Appl. Biochem. Biotechnol. 2002, 97, 45–62. [Google Scholar] [CrossRef]
- Zhang, C.; Phillips, A.P.R.; Wipfler, R.L.; Olsen, G.J.; Whitaker, R.J. The essential genome of the crenarchaeal model Sulfolobus islandicus. Nat. Commun. 2018, 9, 4908. [Google Scholar] [CrossRef] [Green Version]
- Bzdrenga, J.; Hiblot, J.; Gotthard, G.; Champion, C.; Elias, M.; Chabriere, E. SacPox from the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius is a proficient lactonase. BMC Res. Notes 2014, 7, 333. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Archaeal clusters of orthologous genes (arCOGs): An update and application for analysis of shared features between thermococcales, methanococcales, and methanobacteriales. Life 2015, 5, 818–840. [Google Scholar] [CrossRef] [Green Version]
- Neumann-Schaal, M.; Koblitz, J.; Schomburg, D. MetaboMAPS: Pathway sharing and multi-omics data visualization in metabolic context. F1000Research 2020, 9, 288. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Gatto, L.; Gibb, S.; Rainer, J. MSnbase, Efficient and Elegant R-Based Processing and Visualization of Raw Mass Spectrometry Data. J. Proteome Res. 2021, 20, 1063–1069. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing 2022; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
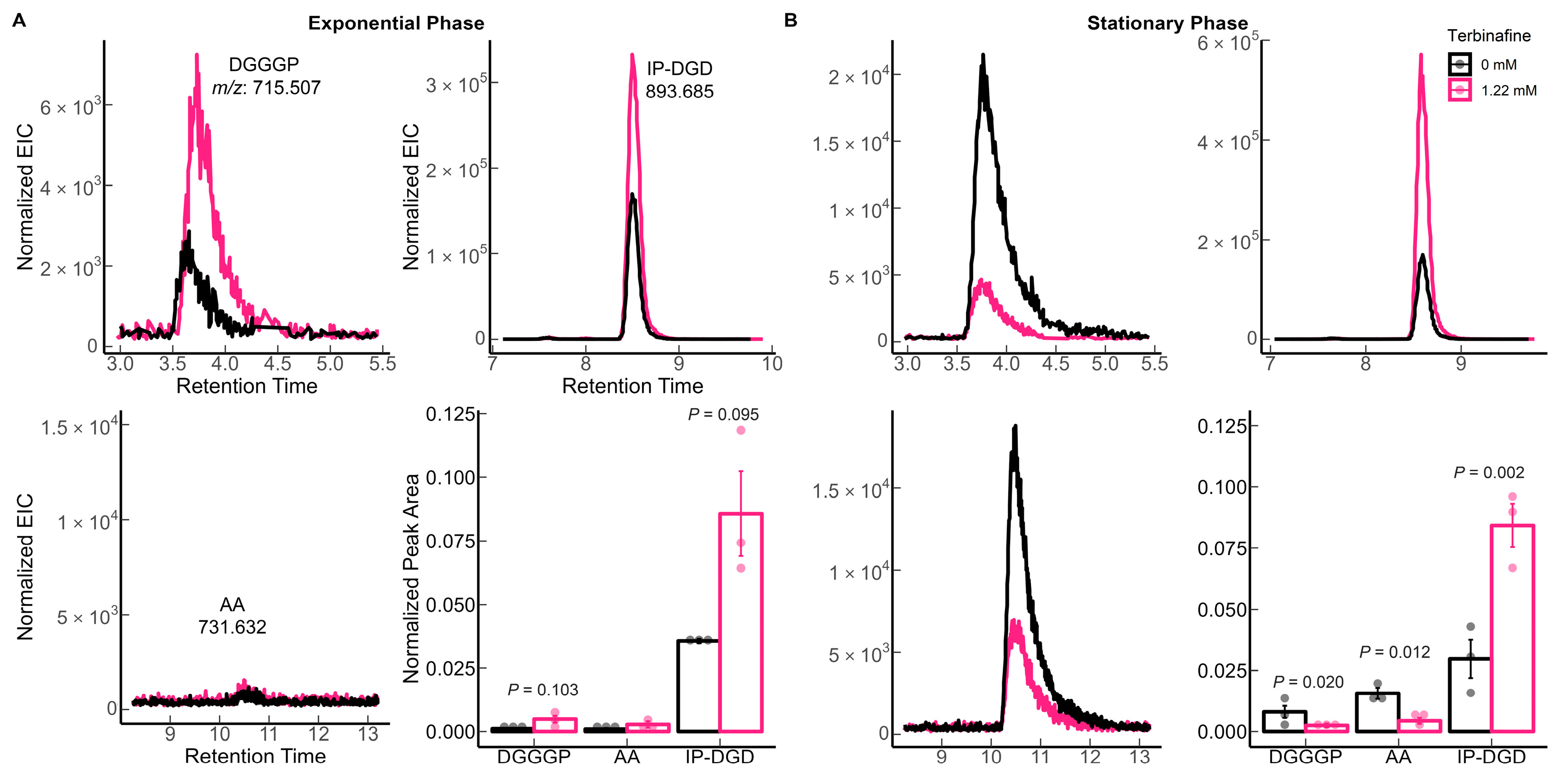
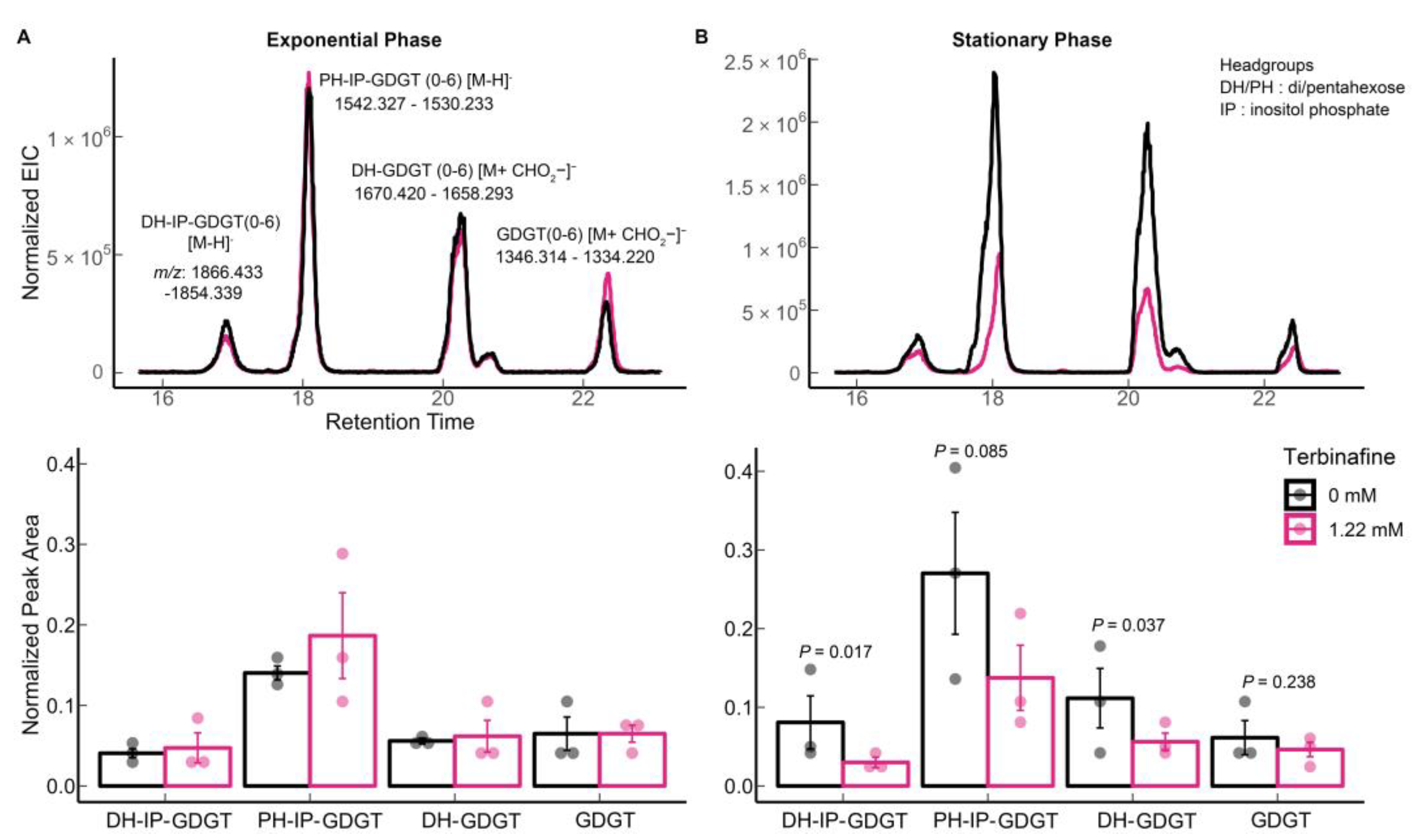
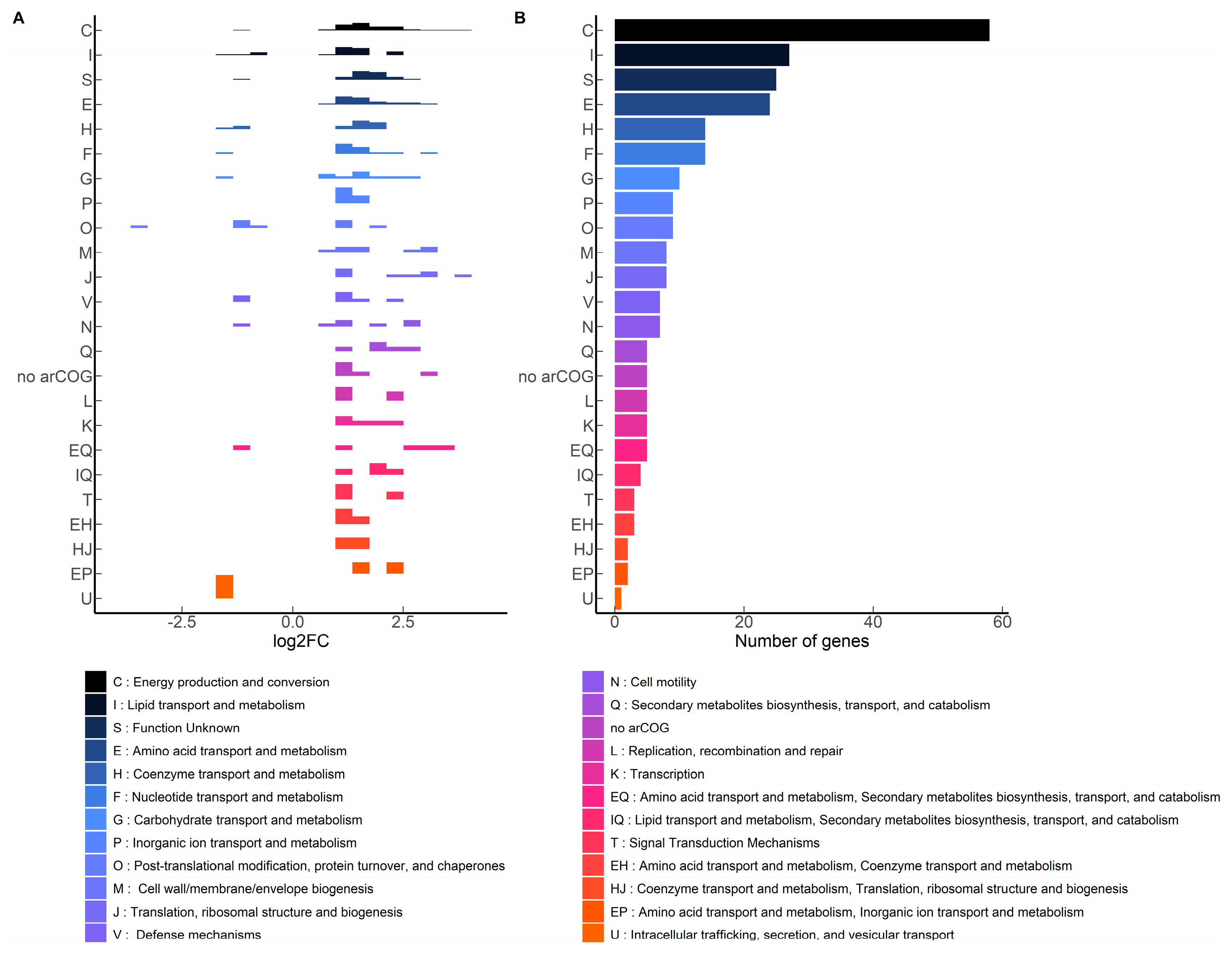
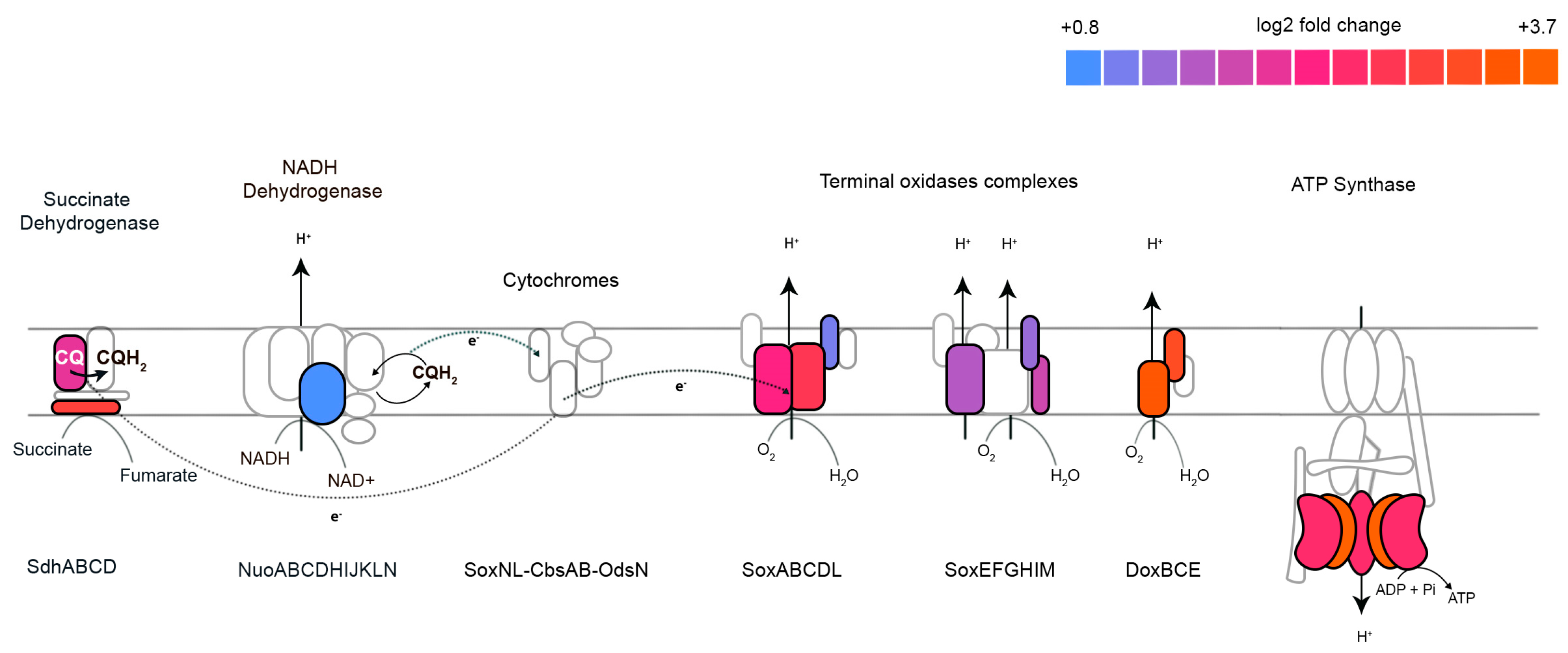

| Lipid Species | Theoretical m/z 1 | Observed m/z | Ppm Error |
|---|---|---|---|
| DOPG | 773.533 [M-H]− | 773.533 | 0.34 |
| DGGGP | 715.507 [M-H]− | 715.507 | −0.08 |
| AA | 731.632 [M-H]− | 731.633 | 0.56 |
| IP-DGD | 893.685 [M-H]− | 893.685 | 0.02 |
| CQH | 630.451 [M] | 630.451 | 0.22 |
| CQ (6:1) | 627.428 [M-H]− | 627.427 | −0.21 |
| CQ (6:2) | 625.412 [M-H]− | 625.412 | 0.03 |
| CQ (6:3) | 623.396 [M-H]− | 623.396 | −0.13 |
| GDGT-0 | 1346.314 [M + CHO2−]− | 1346.310 | −2.99 |
| GDGT-1 | 1344.298 | 1344.293 | −3.33 |
| GDGT-2 | 1342.283 | 1342.277 | −3.78 |
| GDGT-3 | 1340.266 | 1340.263 | −3.04 |
| GDGT-4 | 1338.251 | 1338.244 | −4.75 |
| GDGT-5 | 1336.235 | 1336.234 | −1.36 |
| GDGT-6 | 1334.220 | 1334.219 | −0.79 |
| GDGT-7 | 1332.204 | 1332.204 | −0.32 |
| GDGT-8 | 1330.188 | 1330.188 | −0.67 |
| DH-GDGT(0) | 1670.419 [M + CHO2−]− | 1670.412 | −4.05 |
| DH-GDGT(1) | 1668.404 | 1668.398 | −3.60 |
| DH-GDGT(2) | 1666.388 | 1666.382 | −3.44 |
| DH-GDGT(3) | 1664.372 | 1664.368 | −2.77 |
| DH-GDGT(4) | 1662.357 | 1662.350 | −3.86 |
| DH-GDGT(5) | 1660.341 | 1660.340 | −0.61 |
| DH-GDGT(6) | 1658.293 | 1658.288 | −3.11 |
| DH-IP-GDGT(0) | 1866.433 [M-H]- | 1866.433 | 0.17 |
| DH-IP-GDGT(1) | 1864.417 | 1864.410 | −3.68 |
| DH-IP-GDGT(2) | 1862.402 | 1862.396 | −3.28 |
| DH-IP-GDGT(3) | 1860.386 | 1860.381 | −2.75 |
| DH-IP-GDGT(4) | 1858.370 | 1858.369 | −0.96 |
| DH-IP-GDGT(5) | 1856.355 | 1856.354 | −0.43 |
| DH-IP-GDGT(6) | 1854.339 | 1854.338 | −0.54 |
| PH-IP-GDGT(0) | 1542.327 [M-H]− | 1542.321 | −4.27 |
| PH-IP-GDGT(1) | 1540.312 | 1540.306 | −3.47 |
| PH-IP-GDGT(2) | 1538.296 | 1538.292 | −2.82 |
| PH-IP-GDGT(3) | 1536.280 | 1536.277 | −2.49 |
| PH-IP-GDGT(4) | 1534.265 | 1534.263 | −1.13 |
| PH-IP-GDGT(5) | 1532.249 | 1532.248 | −0.72 |
| PH-IP-GDGT(6) | 1530.233 | 1530.233 | −0.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, A.; de Kok, N.A.W.; Driessen, A.J.M. Membrane Adaptations and Cellular Responses of Sulfolobus acidocaldarius to the Allylamine Terbinafine. Int. J. Mol. Sci. 2023, 24, 7328. https://doi.org/10.3390/ijms24087328
Rao A, de Kok NAW, Driessen AJM. Membrane Adaptations and Cellular Responses of Sulfolobus acidocaldarius to the Allylamine Terbinafine. International Journal of Molecular Sciences. 2023; 24(8):7328. https://doi.org/10.3390/ijms24087328
Chicago/Turabian StyleRao, Alka, Niels A. W. de Kok, and Arnold J. M. Driessen. 2023. "Membrane Adaptations and Cellular Responses of Sulfolobus acidocaldarius to the Allylamine Terbinafine" International Journal of Molecular Sciences 24, no. 8: 7328. https://doi.org/10.3390/ijms24087328