Effects of Drought and Flooding on Phytohormones and Abscisic Acid Gene Expression in Kiwifruit
Abstract
:1. Introduction
2. Results
2.1. Soil Moisture Content Increases with Flooding and Decreases with Drought
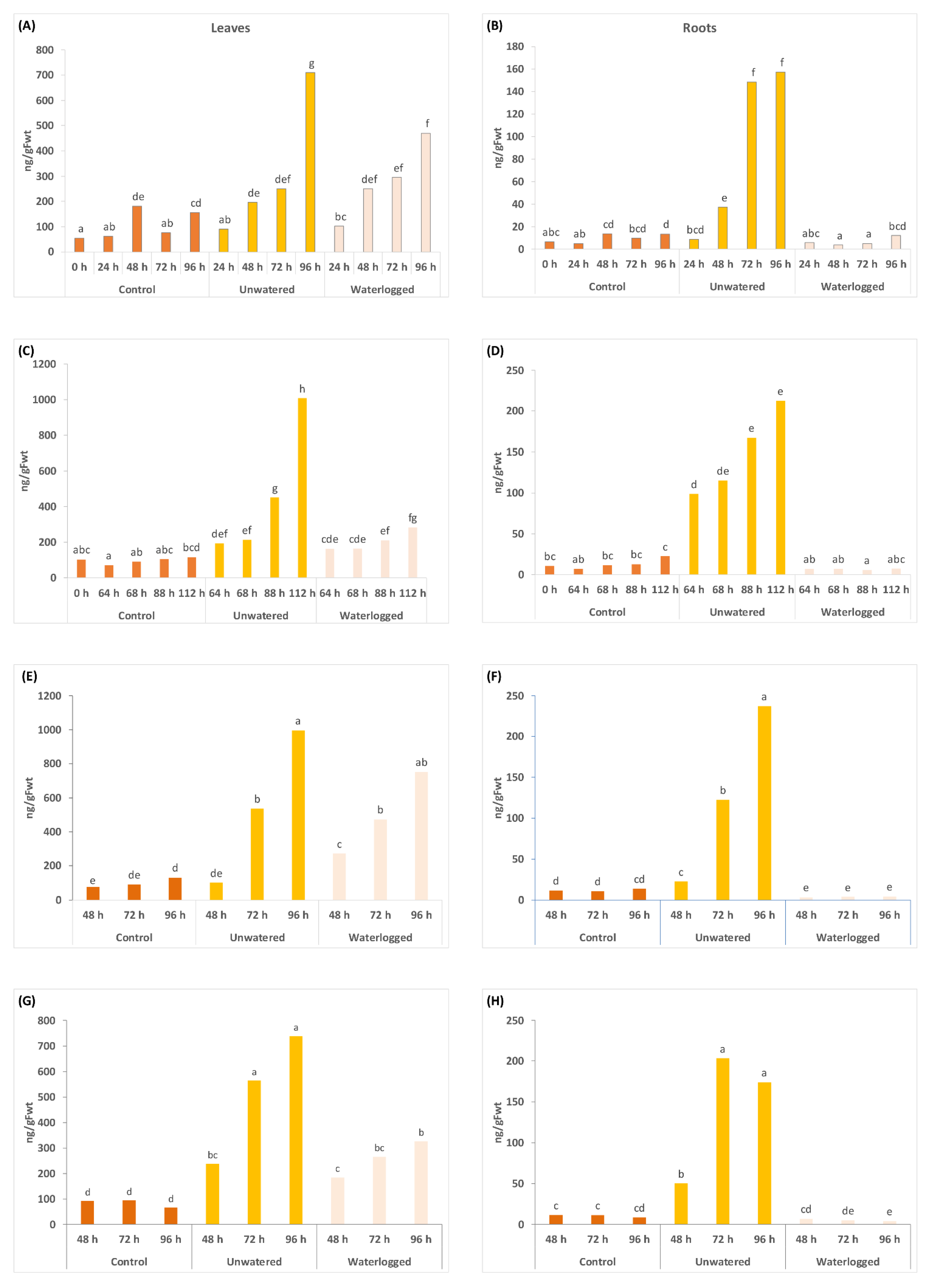
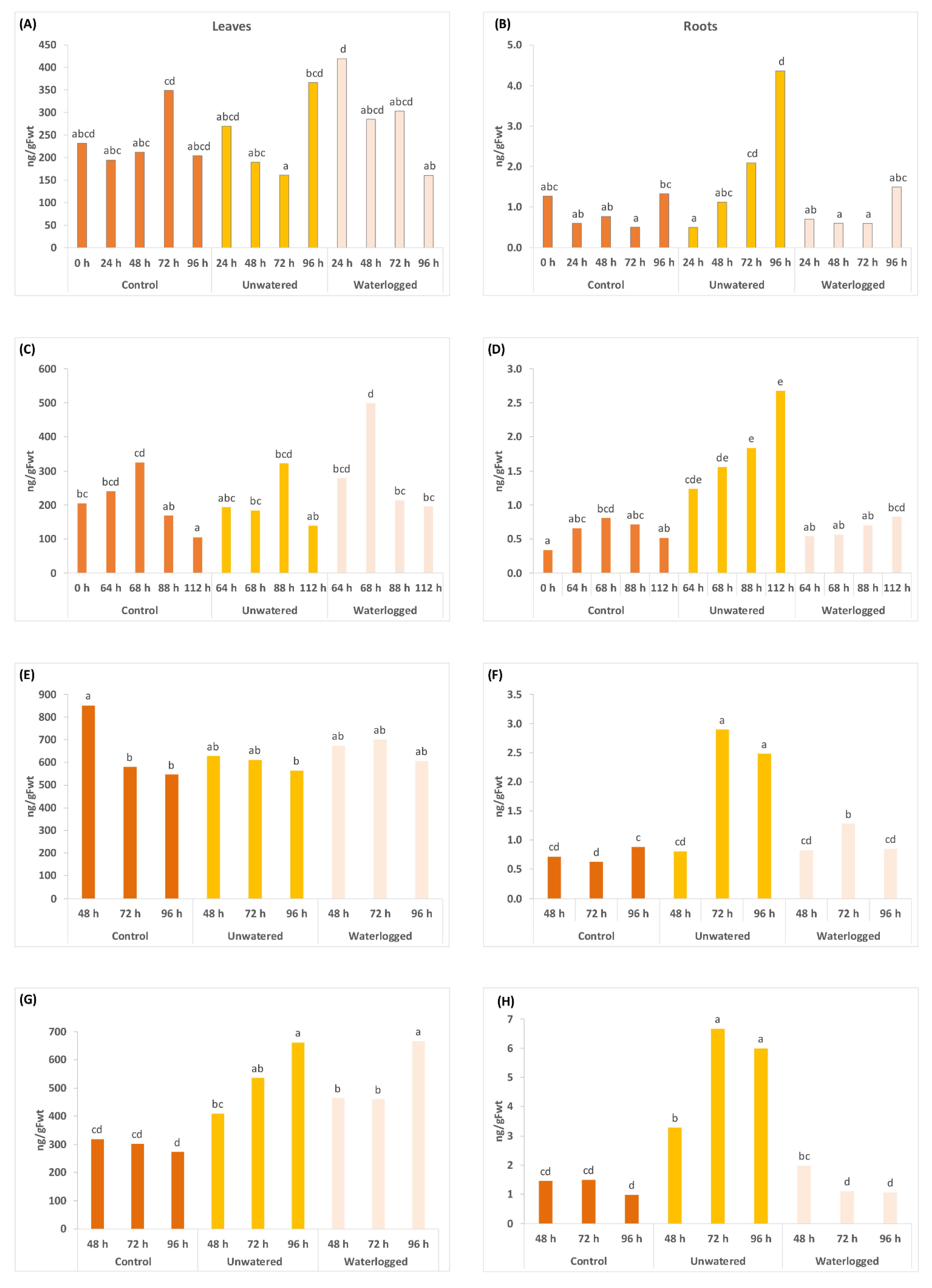
2.2. Water Stress Affects Concentrations of Phytohormones and Their Derivatives in a Tissue Specific Manner
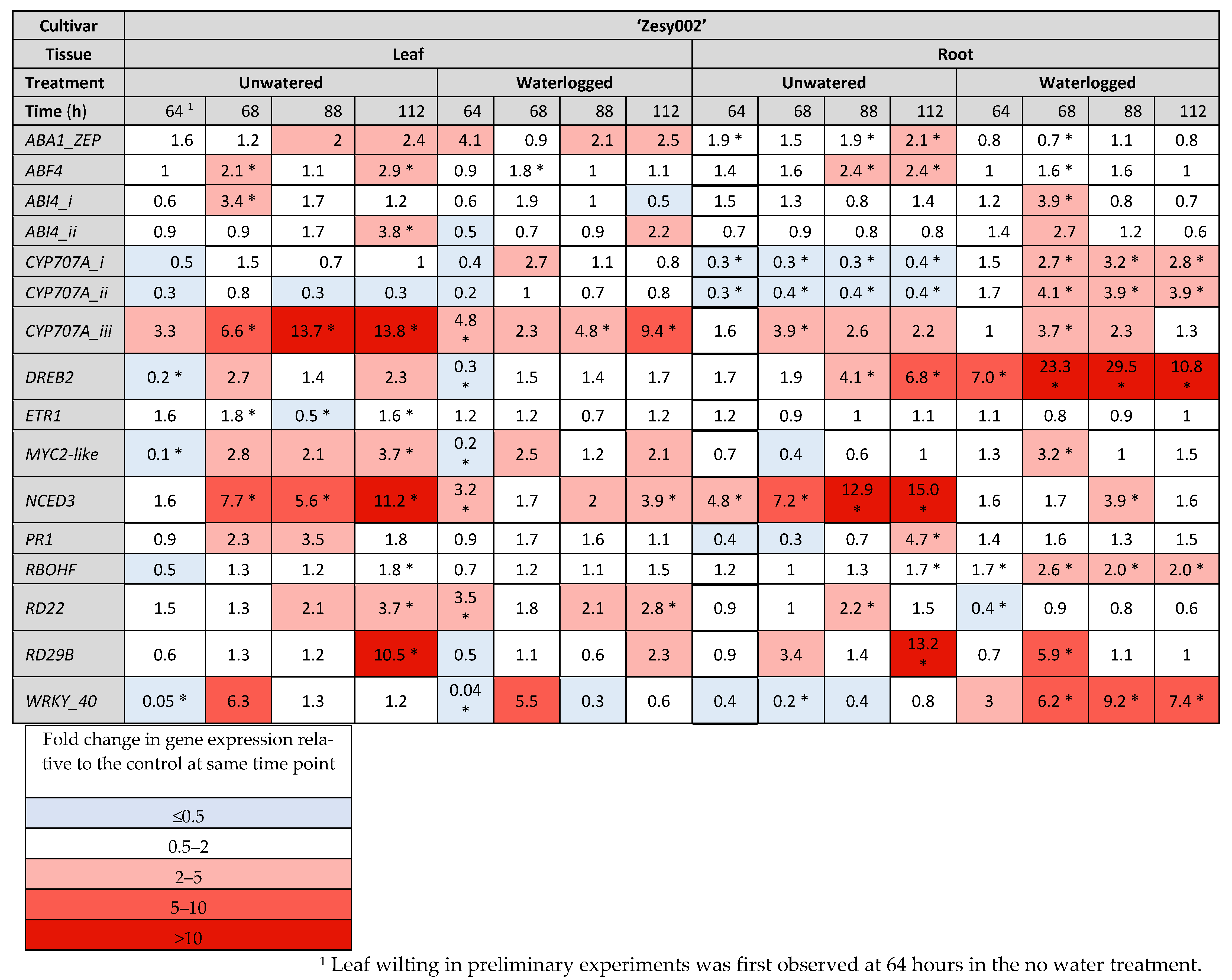
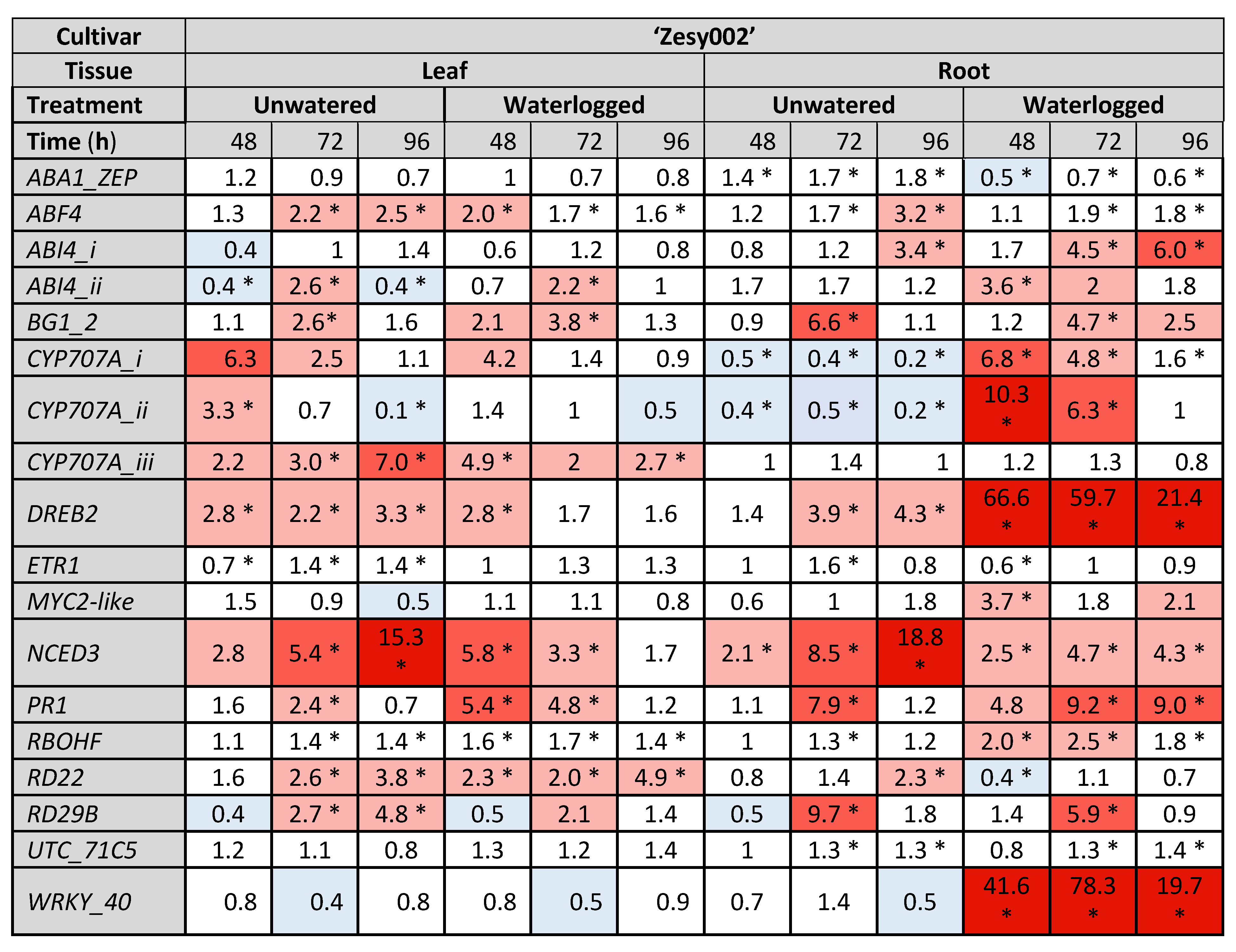
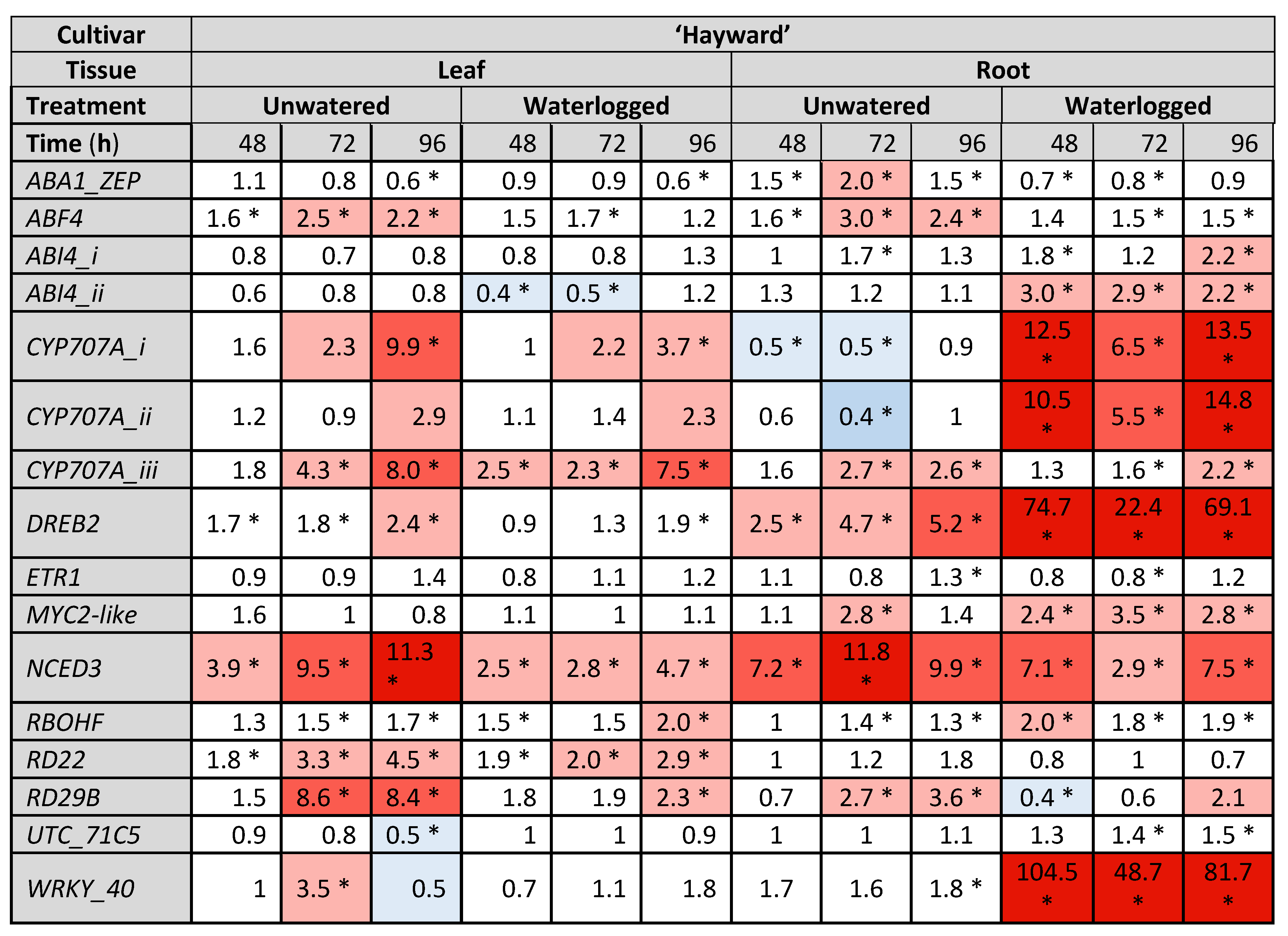
2.3. Gene Expresison Depends on the Type of Water Stress and the Tissue Sampled
3. Discussion
3.1. The Drought Response
3.2. Response to Flooding
3.3. Future Directions and Impact
4. Materials and Methods
4.1. Plant Material
4.2. Watering Conditions
4.3. Tissue Sampling for Phytohormone and Gene Expression Analysis
4.4. Phytohormone Analysis
4.4.1. Reagents
4.4.2. Phytohormone Extraction
4.4.3. LC-MS Analysis
4.5. Gene Expression Analysis
4.5.1. Sample Preparation/RNA Extraction
4.5.2. Gene Selection
4.5.3. Titration Analysis to Determine the Optimal RNA Input (ng)
4.5.4. Measurement of Gene Expression by PlexSet NanoString
4.5.5. Data Analysis
| Identification (NCBI Entry/ Acc#/Achn#) | Gene Name (Abbreviation) | Reference for Previously Identified Kiwifruit Genes | Reference for Kiwifruit Genes Identified in This Analysis (Published Orthologue 4) |
|---|---|---|---|
| FG499230/ Acc06864.1/Achn146991 | Pathogenesis-related protein family 1 (PR1) | [45] | - |
| Acc10322.1 | Zeaxanthin epoxidase (ABA1/ZEP) | [47] | - |
| Acc06947.1 | ABA-responsive element binding factor 4 (ABF4) | - | [51] |
| Acc03929.1 | β-1,3-Glucanase 1/2 (β-glucanase) | [52] | - |
| Acc01538.1 1 | Dehydration-responsive element-binding protein 2 (DREB2) | - | [23] |
| Acc33422.1 | Ethylene receptor 1 (ETR1) | - | [53] |
| Acc21754.1 | 9-cis-epoxycarotenoid dioxygenase 3 (NCED3) | - | [12] |
| Acc03466.1 | Responsive to desiccation 29B (RD29B) | - | [54] |
| Acc291141.1 | Responsive to desiccation 22 (RD22) | [47] | - |
| Acc28747.1 2 | Myelocytomatosis 2-like (MYC2-like) | [55] | - |
| Acc17926.1 | W-tryptophan, R-arginine, K-lysine, Y-tyrosine 40 (WRKY 40) | - | [23] |
| Acc22643.1 | Cytochrome P450, family 707, subfamily A, polypeptide i (CYP707A-i) | - | [12] |
| Acc28692.1 | Cytochrome P450, family 707, subfamily A, polypeptide ii (CYP707A-ii) | - | [12] |
| Acc29493.1 | Cytochrome P450, family 707, subfamily A, polypeptide iii (CYP707A-iii) | - | [12] |
| Acc02823.1 | Abscisic acid insensitive 4i (ABI4-i) | - | [12] |
| Acc01553.1 | Abscisic acid insensitive 4i (ABI4-ii) | - | [12] |
| Acc21776.1 3 | UDP-glycosyltransferase 71K1 (UTC-71C5) | - | [19] |
| Acc15636.1 | Respiratory burst oxidase homolog protein F (RBOHF) | [31] | - |
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adak, M.K.; Saha, I.; Dolui, D.; Hasanuzzaman, M. An updated overview of the physiological and molecular responses of rice to anoxia. Front. Biosci. 2021, 26, 1240–1255. [Google Scholar] [CrossRef]
- Ali, S.; Hayat, K.; Iqbal, A.; Xie, L. Implications of Abscisic Acid in the Drought Stress Tolerance of Plants. Agronomy 2020, 10, 1323. [Google Scholar] [CrossRef]
- Raza, A.; Mubarik, M.S.; Sharif, R.; Habib, M.; Jabeen, W.; Zhang, C.; Chen, H.; Chen, Z.; Siddique, K.H.M.; Zhuang, W.; et al. Developing drought-smart, ready-to-grow future crops. Plant Genome 2022, 37, e20279. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, J.; Hui, W.; Zhao, F.; Wang, P.; Su, C.; Gong, W. Physiology of Plant Responses to Water Stress and Related Genes: A Review. Forests 2022, 13, 324. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Sengupta, S.; Fritschi, F.B.; Azad, R.K.; Nechushtai, R.; Mittler, R. The impact of multifactorial stress combination on plant growth and survival. New Phytol. 2021, 230, 1034–1048. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, W.; Abou-Elwafa, S.F.; Shabala, S.; Xu, L. Understanding a Mechanistic Basis of ABA Involvement in Plant Adaptation to Soil Flooding: The Current Standing. Plants 2021, 10, 1982. [Google Scholar] [CrossRef] [PubMed]
- Scrimgeour, F.; Hughes, W.; Kumar, V. The Economic Contribution of Kiwifruit Industry Expansion to the Bay of Plenty, Northland and New Zealand Economies; Institute for Business Research, University of Waikato: Hamilton, New Zealand, 2017. [Google Scholar]
- The New Zealand Institute for Plant and Food Research. New Zealand Horticultural Exports 2021; The New Zealand Institute for Plant and Food Research: Auckland, New Zealand, 2021; p. 8. [Google Scholar]
- Reglinski, T.; Wurms, K.; Vanneste, J.; Ah Chee, A.; Yu, J.; Oldham, J.; Cornish, D.; Cooney, J.; Jensen, D.; Trower, T.; et al. Transient Changes in Defence Gene Expression and Phytohormone Content Induced by Acibenzolar-S-Methyl in Glasshouse and Orchard Grown Kiwifruit. Front. Agron. 2022, 3, 831172. [Google Scholar] [CrossRef]
- Bashar, K.K.; Tareq, M.Z.; Amin, M.R.; Honi, U.; Tahjib-Ul-Arif, M.; Abu Sadat, M.; Hossen, Q.M.M. Phytohormone-Mediated Stomatal Response, Escape and Quiescence Strategies in Plants under Flooding Stress. Agronomy 2019, 9, 43. [Google Scholar] [CrossRef]
- Todaka, D.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Seo, M.; Marion-Poll, A. Chapter Three—ABA-responsive gene expression in response to drought stress: Cellular regulation and long-distance signaling. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2019; Volume 92, pp. 83–113. [Google Scholar]
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Masondo, N.A.; Aremu, A.O.; Kulkarni, M.G.; Petřík, I.; Plačková, L.; Šubrtová, M.; Novák, O.; Grúz, J.; Doležal, K.; Strnad, M.; et al. How Do Different Watering Regimes Affect the Growth, Chlorophyll Fluorescence, Phytohormone, and Phenolic Acid Content of Greenhouse-Grown Ceratotheca triloba? J. Plant Growth Regul. 2019, 38, 385–399. [Google Scholar] [CrossRef]
- Arbona, V.; Zandalinas, S.I.; Manzi, M.; González-Guzmán, M.; Rodriguez, P.L.; Gómez-Cadenas, A. Depletion of abscisic acid levels in roots of flooded Carrizo citrange (Poncirus trifoliata L. Raf. × Citrus sinensis L. Osb.) plants is a stress-specific response associated to the differential expression of PYR/PYL/RCAR receptors. Plant Mol. Biol. 2017, 93, 623–640. [Google Scholar] [CrossRef] [PubMed]
- Puértolas, J.; Larsen, E.K.; Davies, W.J.; Dodd, I.C. Applying ‘drought’ to potted plants by maintaining suboptimal soil moisture improves plant water relations. J. Exp. Bot. 2017, 68, 2413–2424. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Cao, J.; He, J.; Chen, Q.; Li, X.; Yang, Y. Molecular Mechanism for the Regulation of ABA Homeostasis During Plant Development and Stress Responses. Int. J. Mol. Sci. 2018, 19, 3643. [Google Scholar] [CrossRef] [PubMed]
- Hewage, K.A.H.; Yang, J.; Wang, D.; Hao, G.; Yang, G.; Zhu, J. Chemical Manipulation of Abscisic Acid Signaling: A New Approach to Abiotic and Biotic Stress Management in Agriculture. Adv. Sci. 2020, 7, 2001265. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, L.; Chandrasekaran, U.; Luo, X.; Zheng, C.; Shu, K. ABA Biosynthesis and Signaling Cascades Under Hypoxia Stress. Front. Plant Sci. 2021, 12, 661228. [Google Scholar] [CrossRef]
- Liu, Z.; Yan, J.-P.; Li, D.-K.; Luo, Q.; Yan, Q.; Liu, Z.-B.; Ye, L.-M.; Wang, J.-M.; Li, X.-F.; Yang, Y. UDP-Glucosyltransferase71C5, a Major Glucosyltransferase, Mediates Abscisic Acid Homeostasis in Arabidopsis. Plant Physiol. 2015, 167, 1659–1670. [Google Scholar] [CrossRef]
- Urano, K.; Maruyama, K.; Jikumaru, Y.; Kamiya, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Analysis of plant hormone profiles in response to moderate dehydration stress. Plant J. 2017, 90, 17–36. [Google Scholar] [CrossRef]
- Kim, H.; Seomun, S.; Yoon, Y.; Jang, G. Jasmonic Acid in Plant Abiotic Stress Tolerance and Interaction with Abscisic Acid. Agronomy 2021, 11, 1886. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Singh, D.; Laxmi, A. Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant Sci. 2015, 6, 895. [Google Scholar] [CrossRef]
- Wurms, K.; Ah Chee, A.; Stannard, K.; Anderson, R.; Jensen, D.; Cooney, J.; Hedderley, D. Defence Responses Associated with Elicitor-Induced, Cultivar-Associated Resistance to Latania Scale in Kiwifruit. Plants 2021, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Damián-Zamacona, S.; Toledo-Ibelles, P.; Ibarra-Abundis, M.Z.; Uribe-Figueroa, L.; Hernández-Lemus, E.; Macedo-Alcibia, K.P.; Delgado–Coello, B.; Mas-Oliva, J.; Reyes-Grajeda, J.P. Early Transcriptomic Response to LDL and oxLDL in Human Vascular Smooth Muscle Cells. PLoS ONE 2016, 11, e0163924. [Google Scholar] [CrossRef] [PubMed]
- Meimoun, P.; Mordret, E.; Langlade, N.B.; Balzergue, S.; Arribat, S.; Bailly, C.; El-Maarouf-Bouteau, H. Is Gene Transcription Involved in Seed Dry After-Ripening? PLoS ONE 2014, 9, e86442. [Google Scholar] [CrossRef] [PubMed]
- Mosqueira, M.; Willmann, G.; Zeiger, U.; Khurana, T.S. Expression Profiling Reveals Novel Hypoxic Biomarkers in Peripheral Blood of Adult Mice Exposed to Chronic Hypoxia. PLoS ONE 2012, 7, e37497. [Google Scholar] [CrossRef] [PubMed]
- Sharp, R.E.; Lenoble, M.E.; Else, M.A.; Thorne, E.T.; Gherardi, F. Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: Evidence for an interaction with ethylene. J. Exp. Bot. 2000, 51, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, L.; Liu, M.; Peng, D.; Wei, A.; Hou, B.; Lei, Y.; Li, X. TaFDL2-1A confers drought stress tolerance by promoting ABA biosynthesis, ABA responses, and ROS scavenging in transgenic wheat. Plant J. 2022, 112, 722–737. [Google Scholar] [CrossRef]
- Wang, C.; Chen, S.; Dong, Y.; Ren, R.; Chen, D.; Chen, X. Chloroplastic Os3BGlu6 contributes significantly to cellular ABA pools and impacts drought tolerance and photosynthesis in rice. New Phytol. 2020, 226, 1042–1054. [Google Scholar] [CrossRef]
- de Jong, H.; Reglinski, T.; Elmer, P.A.G.; Wurms, K.; Vanneste, J.L.; Guo, L.F.; Alavi, M. Integrated Use of Aureobasidium pullulans Strain CG163 and Acibenzolar-S-Methyl for Management of Bacterial Canker in Kiwifruit. Plants 2019, 8, 287. [Google Scholar] [CrossRef]
- Aires, E.S.; Ferraz, A.K.L.; Carvalho, B.L.; Teixeira, F.P.; Putti, F.F.; de Souza, E.P.; Rodrigues, J.D.; Ono, E.O. Foliar Application of Salicylic Acid to Mitigate Water Stress in Tomato. Plants 2022, 11, 1775. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell. Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Cisse, E.-H.M.; Zhang, J.; Li, D.-D.; Miao, L.-F.; Yin, L.-Y.; Yang, F. Exogenous ABA and IAA modulate physiological and hormonal adaptation strategies in Cleistocalyx operculatus and Syzygium jambos under long-term waterlogging conditions. BMC Plant Biol. 2022, 22, 20. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Zhou, W.; Chen, F.; Luo, X.; Yang, W. Abscisic Acid and Gibberellins Antagonistically Mediate Plant Development and Abiotic Stress Responses. Front. Plant Sci. 2018, 9, 416. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, A.R. The Genus Actinidia. In Kiwifruit Science and Management; Warrington, I.J., Weston, G.C., Eds.; Ray Richards Publisher New Zealand Society for Horticultural Science: Auckland, New Zealand, 1990; pp. 15–35. [Google Scholar]
- Chartzoulakis, K.; Noitsakis, B.; Therios, I. Photosynthesis, plant growth and carbon allocation in kiwi, cv hayward, as influenced by water deficits. Acta Hortic. 1993, 335, 227–234. [Google Scholar] [CrossRef]
- Green, S.R.; Clothier, B.E. Root water uptake by kiwifruit vines following partial wetting of the root zone. Plant Soil 1995, 173, 317–328. [Google Scholar] [CrossRef]
- Jo, Y.-S.; Cho, H.-S.; Park, M.-Y.; Bang, G.-P.; Kim, W.-S. Drought stress tolerance of Actinidia arguta and Actinidia eriantha. Acta Hortic. 2008, 773, 283–288. [Google Scholar] [CrossRef]
- Miller, S.A.; Smith, G.S.; Boldingh, H.L.; Johansson, A. Effects of Water Stress on Fruit Quality Attributes of Kiwifruit. Ann. Bot. 1998, 81, 73–81. [Google Scholar] [CrossRef]
- Grynkiewicz, G.; Achmatowicz, O.; Hennig, J.; Indulski, J.; Klessig, D.F. Synthesis and characterization of the salicylic-acid beta-d-glucopyranoside. Pol. J. Chem. 1993, 67, 1251–1254. [Google Scholar]
- Kramell, R.; Schmidt, J.; Schneider, G.; Sembdner, G.; Schreiber, K. Synthesis of n-(jasmonoyl)amino acid conjugates. Tetrahedron 1988, 44, 5791–5807. [Google Scholar] [CrossRef]
- Cai, W.-J.; Ye, T.-T.; Wang, Q.; Cai, B.-D.; Feng, Y.-Q. A rapid approach to investigate spatiotemporal distribution of phytohormones in rice. Plant Methods 2016, 12, 47. [Google Scholar] [CrossRef]
- Cellini, A.; Fiorentini, L.; Buriani, G.; Yu, J.; Donati, I.; Cornish, D.A.; Novak, B.; Costa, G.; Vanneste, J.L.; Spinelli, F. Elicitors of the salicylic acid pathway reduce incidence of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidae. Ann. Appl. Biol. 2014, 165, 441–453. [Google Scholar] [CrossRef]
- Petriccione, M.; Mastrobuoni, F.; Zampella, L.; Scortichini, M. Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 2015, 5, 16961. [Google Scholar] [CrossRef] [PubMed]
- Wurms, K.V.; Hardaker, A.J.; Ah Chee, A.; Bowen, J.; Phipps, J.; Taylor, J.; Jensen, D.; Cooney, J.; Wohlers, M.; Reglinski, T. Phytohormone and Putative Defense Gene Expression Differentiates the Response of ‘Hayward’ Kiwifruit to Psa and Pfm Infections. Front. Plant Sci. 2017, 8, 1366. [Google Scholar] [CrossRef]
- Herrero, J.; Muffato, M.; Beal, K.; Fitzgerald, S.; Gordon, L.; Pignatelli, M.; Vilella, A.J.; Searle, S.M.J.; Amode, R.; Brent, S.; et al. Ensembl comparative genomics resources. Database 2016, 2016, bav096. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Allen, J.; Amode, R.M.; Azov, A.G.; Barba, M.; Becerra, A.; Bhai, J.; Campbell, L.I.; Martinez, M.C.; Chakiachvili, M.; et al. Ensembl Genomes 2022: An expanding genome resource for non-vertebrates. Nucleic Acids Res. 2022, 50, D996–D1003. [Google Scholar] [CrossRef]
- Nguyen, N.T.T.; Vincens, P.; Roest Crollius, H.; Louis, A. Genomicus 2018: Karyotype evolutionary trees and on-the-fly synteny computing. Nucleic Acids Res. 2017, 46, D816–D822. [Google Scholar] [CrossRef]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.-Y.; Ahn, J.H. Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant 2019, 12, 489–505. [Google Scholar] [CrossRef]
- Wurms, K.; Cui, W.; Ah-Chee, A.; Rees-George, J.; Bublin, M.; Breiteneder, H. Down Regulation of Putative Defence-associated Transcripts Correlates with Ripe Rot Symptoms on Kiwifruit (Actinidia chinensis). J. Phytopathol. 2011, 159, 435–442. [Google Scholar] [CrossRef]
- Schaller, G.E.; Ladd, A.N.; Lanahan, M.B.; Spanbauer, J.M.; Bleecker, A.B. The Ethylene Response Mediator ETR1 from Arabidopsis Forms a Disulfide-linked Dimer. J. Biol. Chem. 1995, 270, 12526–12530. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Genet. Genom. 1993, 236, 331–340. [Google Scholar] [CrossRef]
- Stroud, E.A. Understanding the Delicate Balance between Microbial Pathogens and Insect Pests and Optimising Protection in Actinidia chinensis. Ph.D. Thesis, University of Auckland, Auckland, New Zealand, 2023. [Google Scholar]
- Lee, S.J.; Kang, J.Y.; Park, H.J.; Kim, M.D.; Bae, M.S.; Choi, H.I.; Kim, S.Y. DREB2C Interacts with ABF2, a bZIP Protein Regulating Abscisic Acid-Responsive Gene Expression, and Its Overexpression Affects Abscisic Acid Sensitivity. Plant Physiol. 2010, 153, 716–727. [Google Scholar] [CrossRef] [PubMed]

| Experiment | Treatment | Time (h) | |||
|---|---|---|---|---|---|
| 24 | 48 | 72 | 96 | ||
| 1 | Unwatered | 93 ± 2 | 83 ± 1.6 | 76 ± 2 | 72 ± 2.3 |
| Waterlogged | 108 ± 2 | 105 ± 1.8 | 110 ± 2.3 | 111 ± 2.8 | |
| p-value | <0.001 | <0.001 | <0.001 | <0.001 | |
| 2 | 64 | 68 | 88 | 112 | |
| Unwatered | 76 ± 1.6 | 74 ± 1.6 | 69 ± 1.5 | 70 ± 1.93 | |
| Waterlogged | 109 ± 1.9 | 109 ± 1.9 | 107 ± 1.8 | 117 ± 2.3 | |
| p-value | <0.001 | <0.001 | <0.001 | <0.001 | |
| 3 | 24 | 48 | 72 | 96 | |
| Unwatered | 91 ± 1.5 | 84 ± 1.6 | 76 ± 1.4 | 70 ± 1 | |
| Waterlogged | 110 ± 1.6 | 114 ± 1.9 | 116 ± 1.6 | 113 ± 1.2 | |
| p-value | <0.001 | <0.001 | <0.001 | <0.001 | |
| 4 | 24 | 48 | 72 | 96 | |
| Unwatered | 91 ± 2.3 | 81 ± 2.5 | 75 ± 2.8 | 70 ± 2.1 | |
| Waterlogged | 112 ± 2.5 | 109 ± 2.9 | 111 ± 3.3 | 111 ± 2.5 | |
| p-value | <0.001 | <0.001 | <0.001 | <0.001 | |
| Cultivar | Experiment Start Date (Experiment #) | Sampling Times after Experiment Start (h) | Experimental Rationale |
|---|---|---|---|
| ‘Zesy002’ | 30 November 2020 (# 1) | 24, 48, 72 and 96 | Pilot study to investigate hormone levels under water stress (and to help guide gene selection). |
| ‘Zesy002’ | 11 December 2020 (# 2) | 64 1, 68, 88 and 112 | The experiment was carried out over a longer period to induce more severe water stress and included both phytohormone and gene measurements. |
| ‘Zesy002’ | 15 March 2021 (# 3) | 48, 72 and 96 | Repeat of exp 1 trial setup (with sample times before and after plants appear visibly water stressed) but with full phytohormone and gene measurements. |
| ‘Hayward’ | 21 March 2021 (# 4) | 48, 72 and 96 | Identical design to exp 3, but in a different cultivar. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wurms, K.V.; Reglinski, T.; Buissink, P.; Ah Chee, A.; Fehlmann, C.; McDonald, S.; Cooney, J.; Jensen, D.; Hedderley, D.; McKenzie, C.; et al. Effects of Drought and Flooding on Phytohormones and Abscisic Acid Gene Expression in Kiwifruit. Int. J. Mol. Sci. 2023, 24, 7580. https://doi.org/10.3390/ijms24087580
Wurms KV, Reglinski T, Buissink P, Ah Chee A, Fehlmann C, McDonald S, Cooney J, Jensen D, Hedderley D, McKenzie C, et al. Effects of Drought and Flooding on Phytohormones and Abscisic Acid Gene Expression in Kiwifruit. International Journal of Molecular Sciences. 2023; 24(8):7580. https://doi.org/10.3390/ijms24087580
Chicago/Turabian StyleWurms, Kirstin V., Tony Reglinski, Poppy Buissink, Annette Ah Chee, Christina Fehlmann, Stella McDonald, Janine Cooney, Dwayne Jensen, Duncan Hedderley, Catherine McKenzie, and et al. 2023. "Effects of Drought and Flooding on Phytohormones and Abscisic Acid Gene Expression in Kiwifruit" International Journal of Molecular Sciences 24, no. 8: 7580. https://doi.org/10.3390/ijms24087580
APA StyleWurms, K. V., Reglinski, T., Buissink, P., Ah Chee, A., Fehlmann, C., McDonald, S., Cooney, J., Jensen, D., Hedderley, D., McKenzie, C., & Rikkerink, E. H. A. (2023). Effects of Drought and Flooding on Phytohormones and Abscisic Acid Gene Expression in Kiwifruit. International Journal of Molecular Sciences, 24(8), 7580. https://doi.org/10.3390/ijms24087580






