RAD51 Inhibition Shows Antitumor Activity in Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
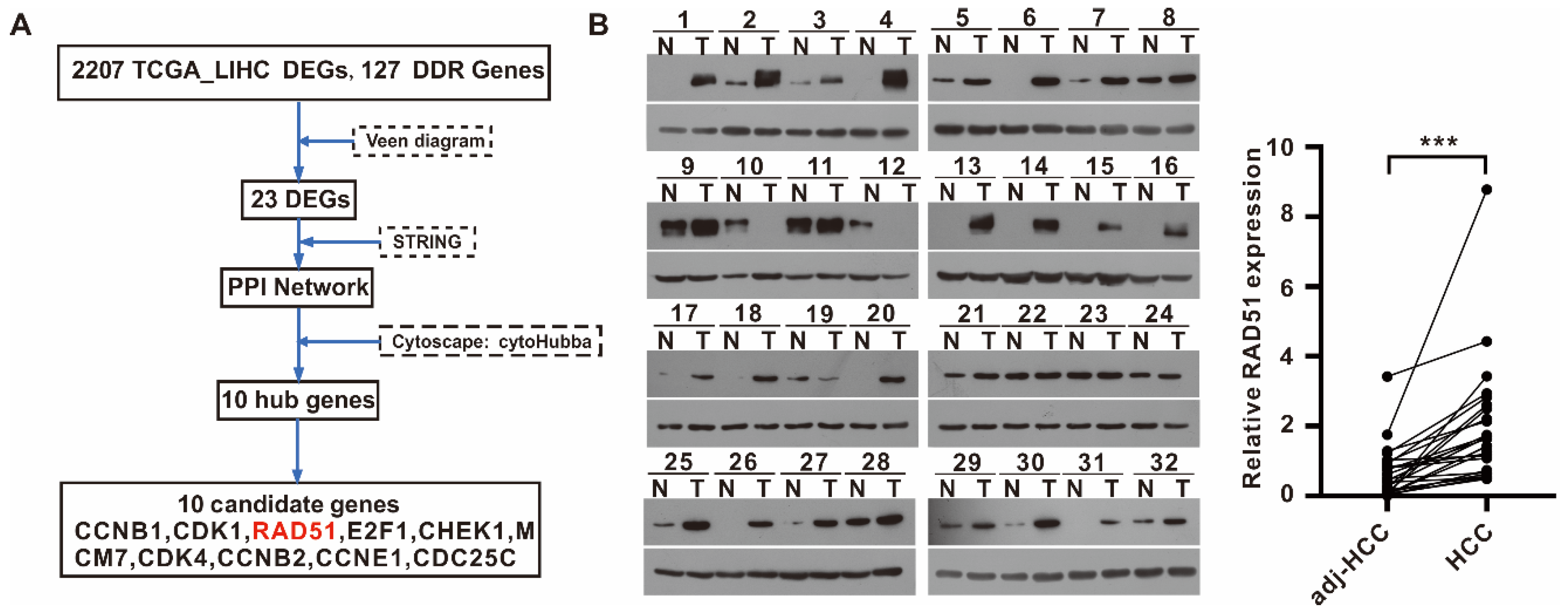
2.1. RAD51 Identified as a Potential DDR Target for HCC, and RAD51 Expression Was Upregulated in HCC
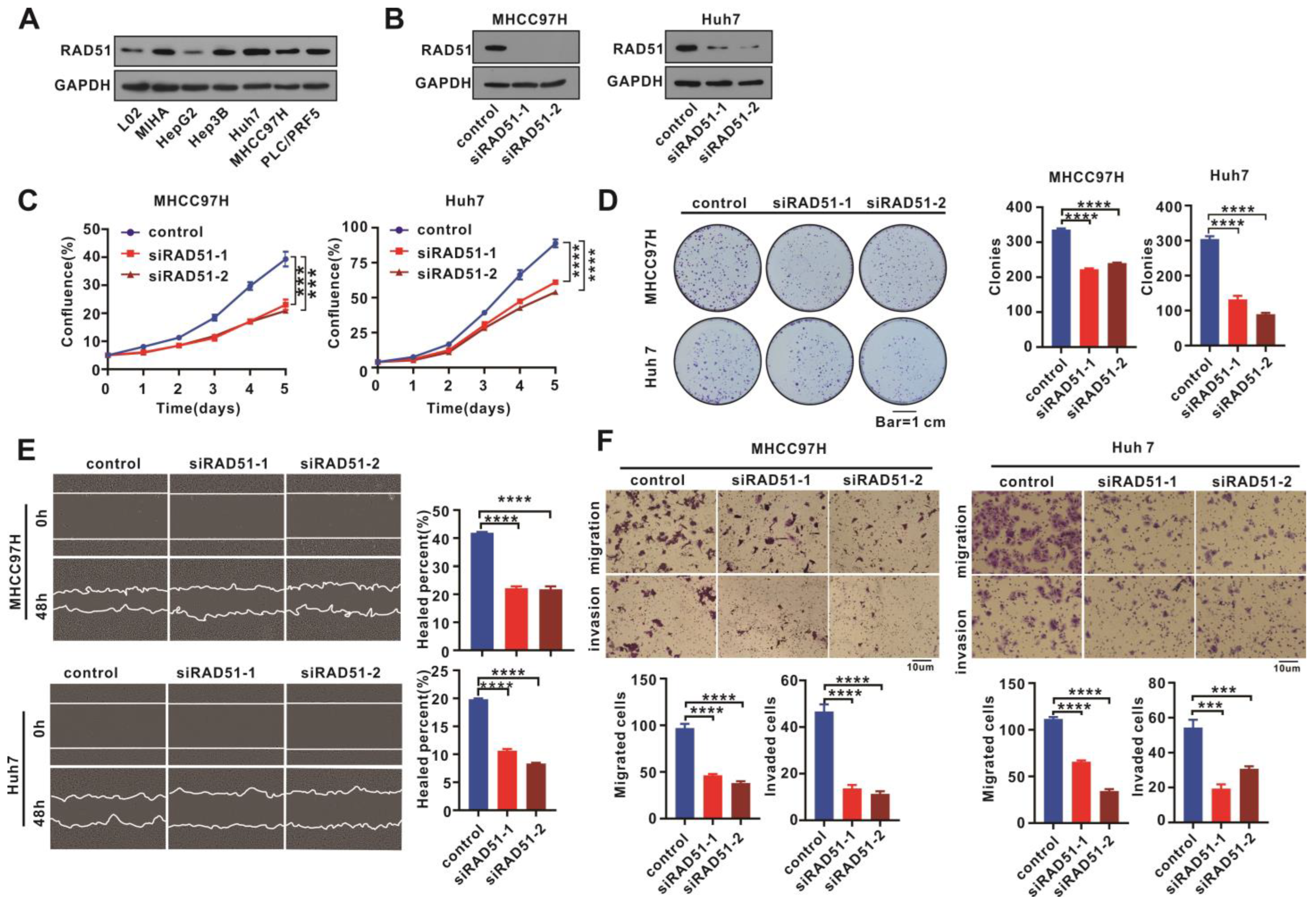
2.2. RAD51 Silencing Weakened Cell Proliferation, Migration, and Invasion
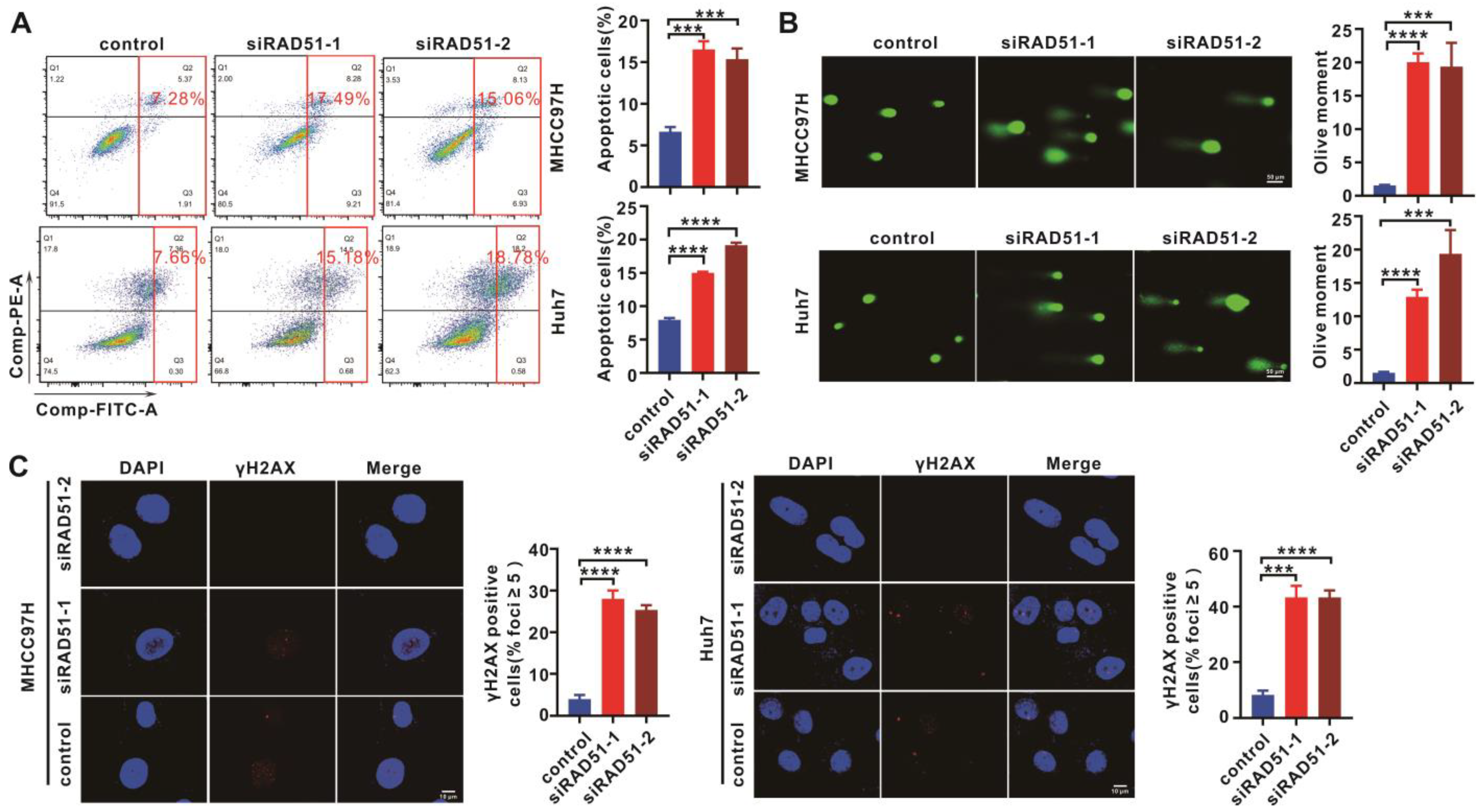
2.3. RAD51 Silencing Enhanced Cell Apoptosis and DNA Damage
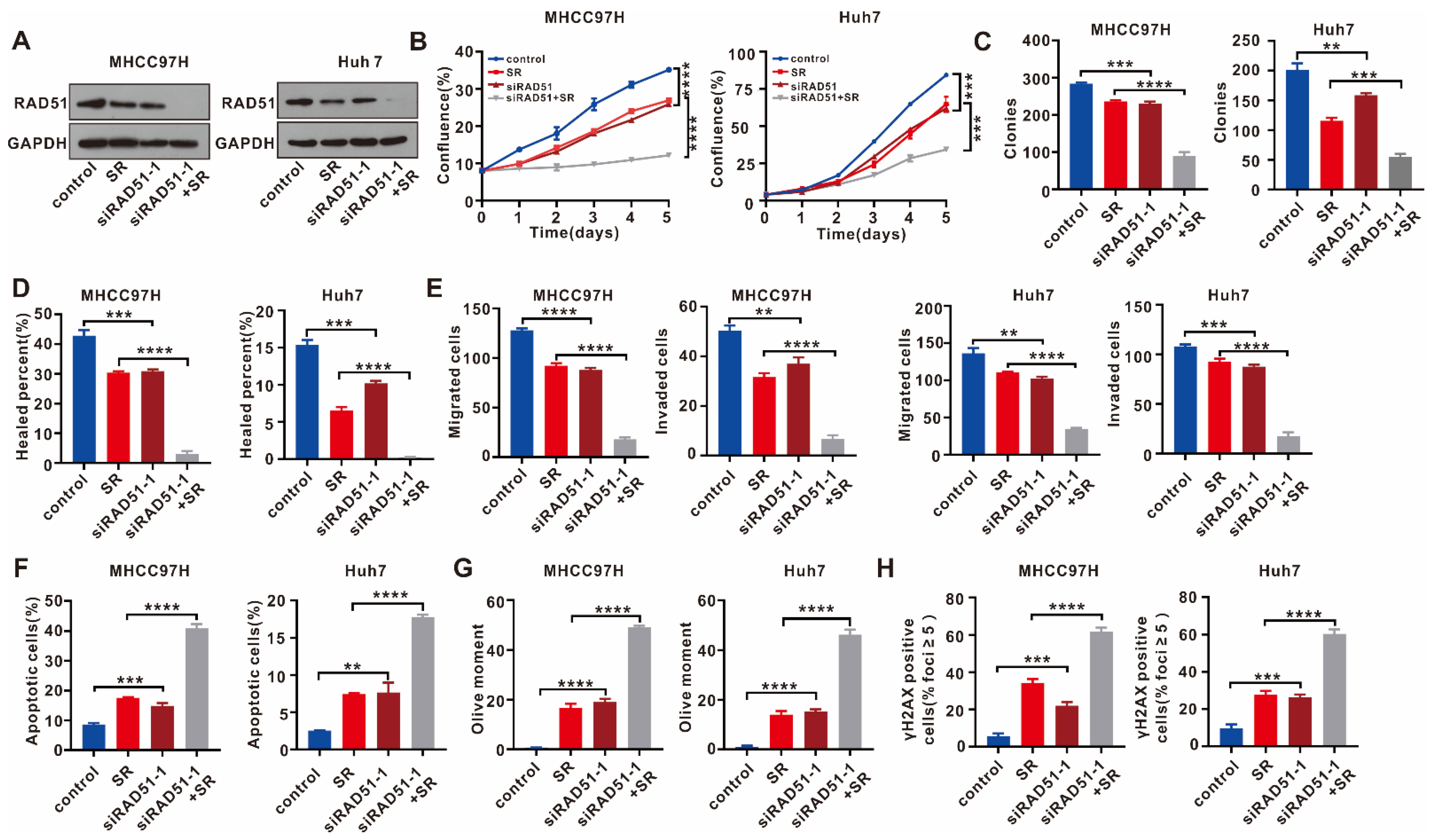
2.4. RAD51 Silencing Displays a Synergistic Antitumor Effect with Sorafenib
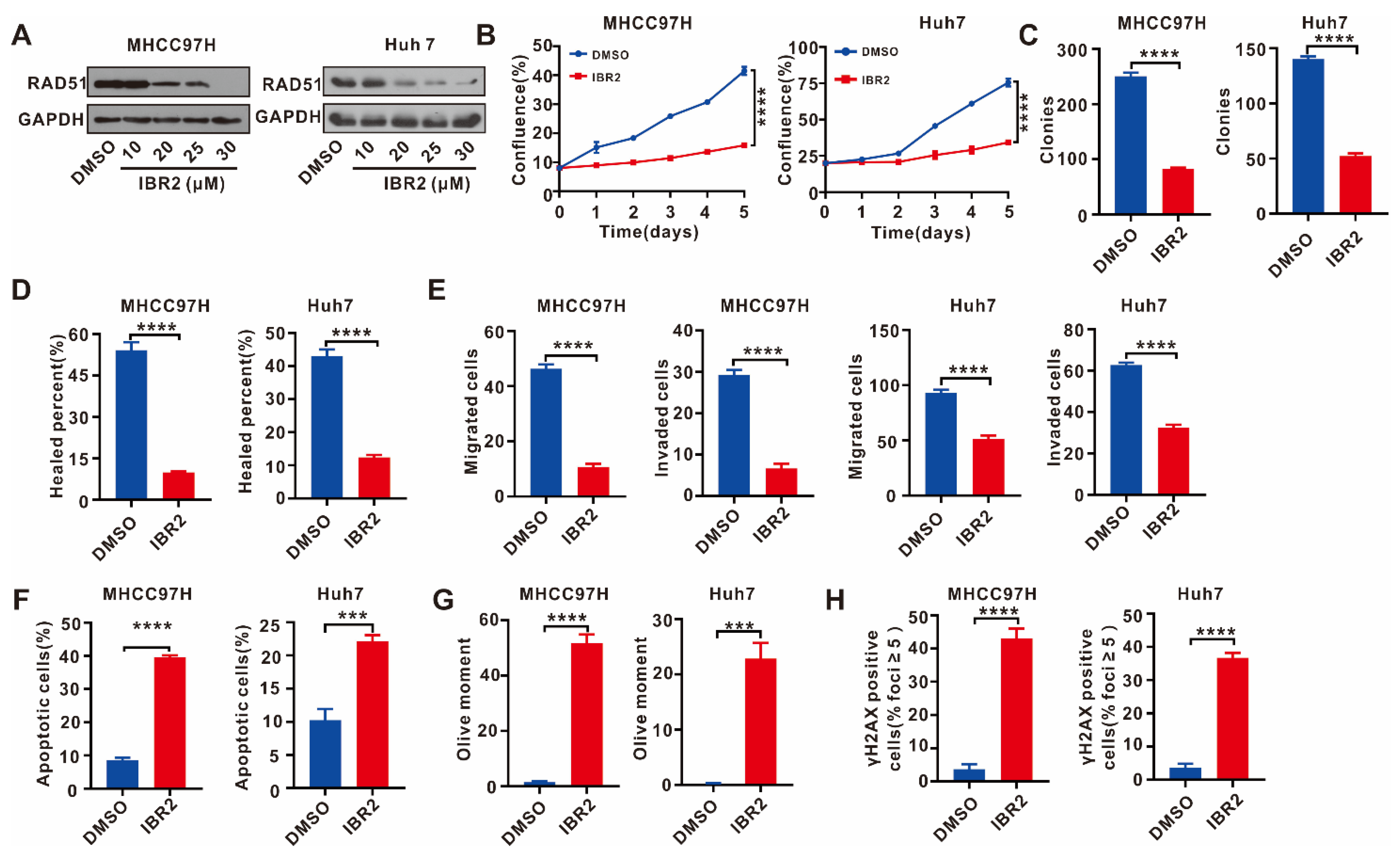
2.5. IBR2 Reduced Cell Proliferation, Migration, and Invasion and Increased Apoptosis and DNA Damage
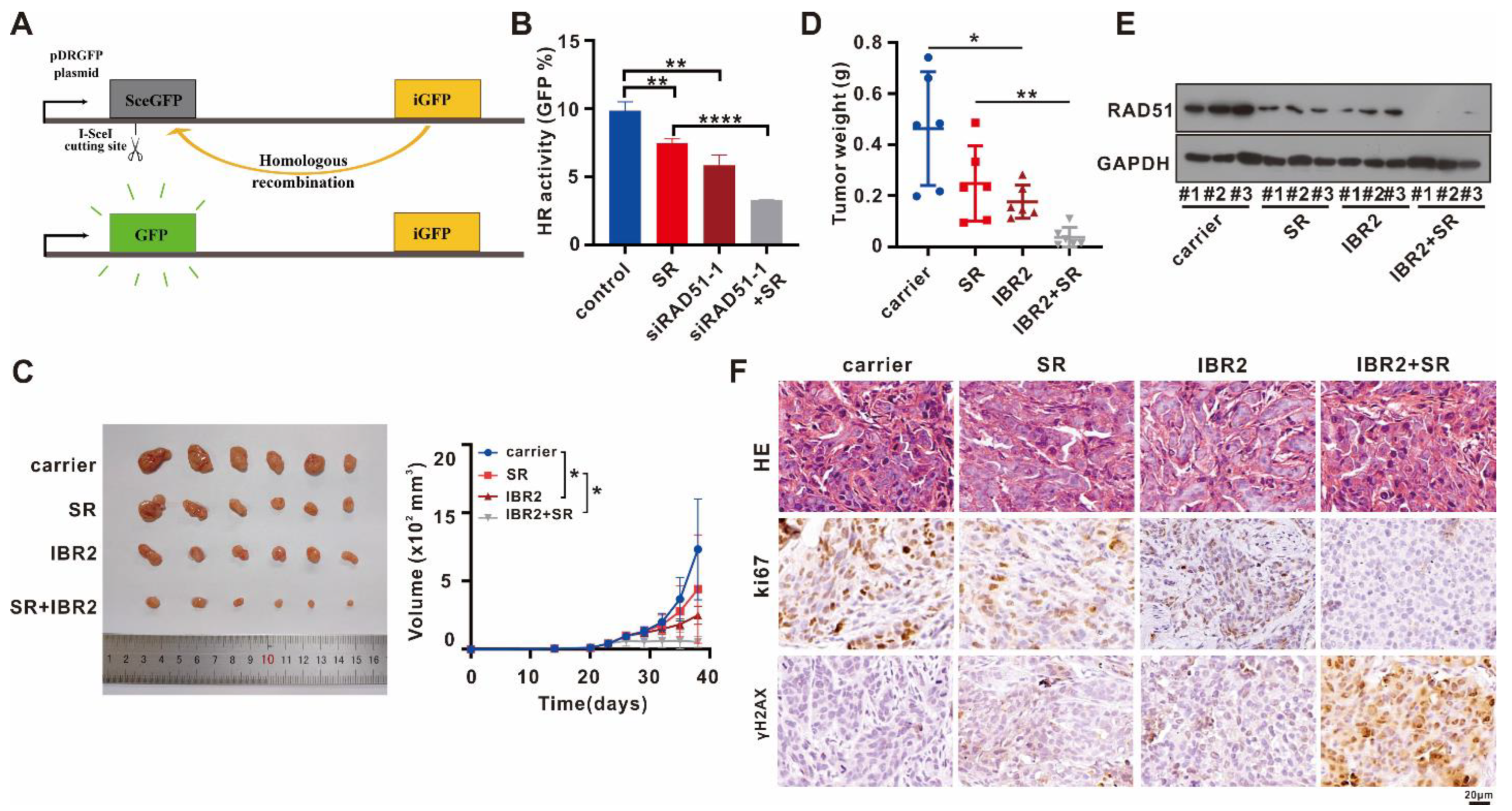
2.6. Targeting RAD51 Inhibited HR Activity In Vitro, Attenuated Tumor Growth In Vivo, and Exhibited a Synergistic Antitumor Effect with Sorafenib
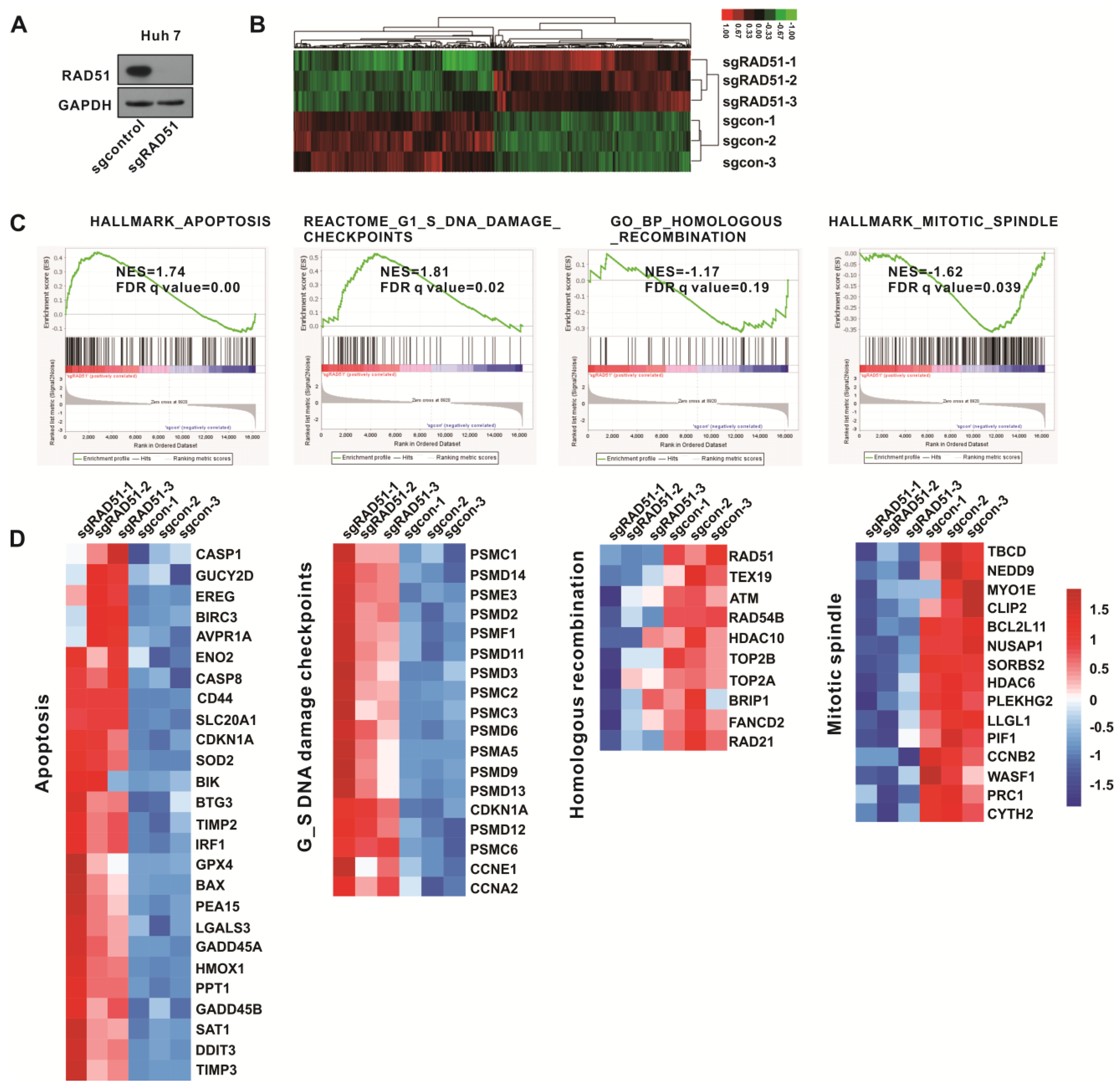
2.7. Gene Set Enrichment Analysis (GSEA) Confirmed the Antitumor Activity of Inactivated RAD51
3. Discussion
4. Materials and Methods
4.1. Bioinformatics Analysis
4.2. Clinical Patient Specimens
4.3. Cell Culture and siRNA Transfection
4.4. RNA Extraction and Quantitative Reverse Transcription (RT-qPCR)
4.5. siRNA
4.6. WB Assay
4.7. Antibodies and Reagents
4.8. Immunohistochemistry (IHC)
4.9. IncuCyte Cell Proliferation Assay
4.10. Colony Formation Assay
4.11. Wound Healing Assay
4.12. Transwell Cell Migration and Invasion Assays
4.13. Cell Apoptosis Analysis
4.14. Immunofluorescence Assay
4.15. Comet Assay
4.16. Plasmid DNA Construction and Lentivirus Packaging
4.17. HR Assays
4.18. RNA Sequencing
4.19. Animal Models
4.20. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lenvatinib versus Sorafenib in First-Line Treatment of Patients with Unresectable Hepatocellular Carcinoma: A Randomised Phase 3 Non-Inferiority Trial-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/29433850/ (accessed on 19 March 2023).
- Huang, A.; Yang, X.-R.; Chung, W.-Y.; Dennison, A.R.; Zhou, J. Targeted Therapy for Hepatocellular Carcinoma. Signal. Transduct. Target Ther. 2020, 5, 146. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Loss of Ribonuclease DIS3 Hampers Genome Integrity in Myeloma by Disrupting DNA:RNA Hybrid Metabolism|The EMBO Journal. Available online: https://www.embopress.org/doi/full/10.15252/embj.2021108040 (accessed on 19 March 2023).
- SMG8/SMG9 Heterodimer Loss Modulates SMG1 Kinase to Drive ATR Inhibitor Resistance | Cancer Research | American Association for Cancer Research. Available online: https://aacrjournals.org/cancerres/article/82/21/3962/709958/SMG8-SMG9-Heterodimer-Loss-Modulates-SMG1-Kinase (accessed on 19 March 2023).
- Jurkovicova, D.; Neophytou, C.M.; Gašparović, A.Č.; Gonçalves, A.C. DNA Damage Response in Cancer Therapy and Resistance: Challenges and Opportunities. Int. J. Mol. Sci. 2022, 23, 14672. [Google Scholar] [CrossRef]
- Therapeutic Opportunities within the DNA Damage Response-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/25709118/ (accessed on 19 March 2023).
- O’Connor, M.J. Targeting the DNA Damage Response in Cancer. Mol. Cell 2015, 60, 547–560. [Google Scholar] [CrossRef]
- Anders, C.K.; Winer, E.P.; Ford, J.M.; Dent, R.; Silver, D.P.; Sledge, G.W.; Carey, L.A. Poly(ADP-Ribose) Polymerase Inhibition: “Targeted” Therapy for Triple-Negative Breast Cancer. Clin. Cancer Res. 2010, 16, 4702–4710. [Google Scholar] [CrossRef]
- Papadimitriou, M.; Mountzios, G.; Papadimitriou, C.A. The Role of PARP Inhibition in Triple-Negative Breast Cancer: Unraveling the Wide Spectrum of Synthetic Lethality. Cancer Treat. Rev. 2018, 67, 34–44. [Google Scholar] [CrossRef]
- Mateo, J.; Lord, C.J.; Serra, V.; Tutt, A.; Balmaña, J.; Castroviejo-Bermejo, M.; Cruz, C.; Oaknin, A.; Kaye, S.B.; de Bono, J.S. A Decade of Clinical Development of PARP Inhibitors in Perspective. Ann. Oncol. 2019, 30, 1437–1447. [Google Scholar] [CrossRef]
- Xu, H.; Xiong, C.; Chen, Y.; Zhang, C.; Bai, D. Identification of Rad51 as a Prognostic Biomarker Correlated with Immune Infiltration in Hepatocellular Carcinoma. Bioengineered 2021, 12, 2664–2675. [Google Scholar] [CrossRef]
- Zhu, J.; Zhou, L.; Wu, G.; Konig, H.; Lin, X.; Li, G.; Qiu, X.; Chen, C.; Hu, C.; Goldblatt, E.; et al. A Novel Small Molecule RAD51 Inactivator Overcomes Imatinib-resistance in Chronic Myeloid Leukaemia. EMBO Mol. Med. 2013, 5, 353–365. [Google Scholar] [CrossRef]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular Therapies and Precision Medicine for Hepatocellular Carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef]
- Gordan, J.D.; Kennedy, E.B.; Abou-Alfa, G.K.; Beg, M.S.; Brower, S.T.; Gade, T.P.; Goff, L.; Gupta, S.; Guy, J.; Harris, W.P.; et al. Systemic Therapy for Advanced Hepatocellular Carcinoma: ASCO Guideline. J. Clin. Oncol. 2020, 38, 4317–4345. [Google Scholar] [CrossRef]
- RAD51 Gene Family Structure and Function-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/32663049/ (accessed on 19 March 2023).
- Dynamics of DNA Double-Strand Breaks Revealed by Clustering of Damaged Chromosome Domains-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/14704429/ (accessed on 19 March 2023).
- DNA RECOMBINATION. Base Triplet Stepping by the Rad51/RecA Family of Recombinases-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/26315438/ (accessed on 19 March 2023).
- BRG1 Promotes the Repair of DNA Double-Strand Breaks by Facilitating the Replacement of RPA with RAD51-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/25395584/ (accessed on 19 March 2023).
- Deng, Y.; Guo, W.; Xu, N.; Li, F.; Li, J. CtBP1 Transactivates RAD51 and Confers Cisplatin Resistance to Breast Cancer Cells. Mol. Carcinog. 2020, 59, 512–519. [Google Scholar] [CrossRef]
- Welsh, J.W.; Ellsworth, R.K.; Kumar, R.; Fjerstad, K.; Martinez, J.; Nagel, R.B.; Eschbacher, J.; Stea, B. Rad51 Protein Expression and Survival in Patients with Glioblastoma Multiforme. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 1251–1255. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, N.; Yao, W.; Li, S.; Ren, Z. RAD51 Is a Potential Marker for Prognosis and Regulates Cell Proliferation in Pancreatic Cancer. Cancer Cell Int. 2019, 19, 356. [Google Scholar] [CrossRef]
- Qiao, G.-B.; Wu, Y.-L.; Yang, X.-N.; Zhong, W.-Z.; Xie, D.; Guan, X.-Y.; Fischer, D.; Kolberg, H.-C.; Kruger, S.; Stuerzbecher, H.-W. High-Level Expression of Rad51 Is an Independent Prognostic Marker of Survival in Non-Small-Cell Lung Cancer Patients. Br. J. Cancer 2005, 93, 137–143. [Google Scholar] [CrossRef]
- Mitra, A.; Jameson, C.; Barbachano, Y.; Sanchez, L.; Kote-Jarai, Z.; Peock, S.; Sodha, N.; Bancroft, E.; Fletcher, A.; Cooper, C.; et al. Overexpression of RAD51 Occurs in Aggressive Prostatic Cancer. Histopathology 2009, 55, 696–704. [Google Scholar] [CrossRef]
- Richardson, C.; Stark, J.M.; Ommundsen, M.; Jasin, M. Rad51 Overexpression Promotes Alternative Double-Strand Break Repair Pathways and Genome Instability. Oncogene 2004, 23, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Klein, H.L. The Consequences of Rad51 Overexpression for Normal and Tumor Cells. DNA Repair. 2008, 7, 686–693. [Google Scholar] [CrossRef]
- TRIM36 Enhances Lung Adenocarcinoma Radiosensitivity and Inhibits Tumorigenesis through Promoting RAD51 Ubiquitination and Antagonizing Hsa-MiR-376a-5p-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/36058131/ (accessed on 19 March 2023).
- Liu, Z.; Huang, J.; Jiang, Q.; Li, X.; Tang, X.; Chen, S.; Jiang, L.; Fu, G.; Liu, S. MiR-125a Attenuates the Malignant Biological Behaviors of Cervical Squamous Cell Carcinoma Cells through Rad51. Bioengineered 2022, 13, 8503–8514. [Google Scholar] [CrossRef]
- Targeting Rad51 as a Strategy for the Treatment of Melanoma Cells Resistant to MAPK Pathway Inhibition-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/32719412/ (accessed on 19 March 2023).
- Enhancing the Efficacy of Glycolytic Blockade in Cancer Cells via RAD51 Inhibition-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/30183475/ (accessed on 19 March 2023).
- Flygare, J.; Fält, S.; Ottervald, J.; Castro, J.; Dackland, A.L.; Hellgren, D.; Wennborg, A. Effects of HsRad51 Overexpression on Cell Proliferation, Cell Cycle Progression, and Apoptosis. Exp. Cell Res. 2001, 268, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jin, R.; Zhao, J.; Liu, J.; Ying, H.; Yan, H.; Zhou, S.; Liang, Y.; Huang, D.; Liang, X.; et al. Potential Molecular, Cellular and Microenvironmental Mechanism of Sorafenib Resistance in Hepatocellular Carcinoma. Cancer Lett. 2015, 367, 1–11. [Google Scholar] [CrossRef]
- The DNA Damage Response: Ten Years after-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/18082599/ (accessed on 20 March 2023).
- Xu, R.; Yu, S.; Zhu, D.; Huang, X.; Xu, Y.; Lao, Y.; Tian, Y.; Zhang, J.; Tang, Z.; Zhang, Z.; et al. HCINAP Regulates the DNA-Damage Response and Mediates the Resistance of Acute Myelocytic Leukemia Cells to Therapy. Nat. Commun. 2019, 10, 3812. [Google Scholar] [CrossRef] [PubMed]
- Targeting DNA Flap Endonuclease 1 to Impede Breast Cancer Progression. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5161424/ (accessed on 20 March 2023).
- Wang, C.; Wang, H.; Lieftink, C.; du Chatinier, A.; Gao, D.; Jin, G.; Jin, H.; Beijersbergen, R.L.; Qin, W.; Bernards, R. CDK12 Inhibition Mediates DNA Damage and Is Synergistic with Sorafenib Treatment in Hepatocellular Carcinoma. Gut 2020, 69, 727–736. [Google Scholar] [CrossRef]
- Samadaei, M.; Senfter, D.; Madlener, S.; Uranowska, K.; Hafner, C.; Trauner, M.; Rohr-Udilova, N.; Pinter, M. Targeting DNA Repair to Enhance the Efficacy of Sorafenib in Hepatocellular Carcinoma. J. Cell Biochem. 2022, 123, 1663–1673. [Google Scholar] [CrossRef] [PubMed]
- Chiu, W.-C.; Fang, P.-T.; Lee, Y.-C.; Wang, Y.-Y.; Su, Y.-H.; Hu, S.C.-S.; Chen, Y.-K.; Tsui, Y.-T.; Kao, Y.-H.; Huang, M.-Y.; et al. DNA Repair Protein Rad51 Induces Tumor Growth and Metastasis in Esophageal Squamous Cell Carcinoma via a P38/Akt-Dependent Pathway. Ann. Surg. Oncol. 2020, 27, 2090–2101. [Google Scholar] [CrossRef]
- FindFoci: A Focus Detection Algorithm with Automated Parameter Training That Closely Matches Human Assignments, Reduces Human Inconsistencies and Increases Speed of Analysis-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/25478967/ (accessed on 16 March 2023).
- Gyori, B.M.; Venkatachalam, G.; Thiagarajan, P.S.; Hsu, D.; Clement, M.-V. OpenComet: An Automated Tool for Comet Assay Image Analysis. Redox. Biol. 2014, 2, 457–465. [Google Scholar] [CrossRef]
- BRCA2 Is Required for Homology-Directed Repair of Chromosomal Breaks-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/11239455/ (accessed on 19 March 2023).
| Catalog | Antibody/Regent | Antibody Species | MW (KD) | Company | Application and Radio |
|---|---|---|---|---|---|
| Ab133534 | RAD51 | R | 37 | Abcam | WB 1:10,000 IHC 1:200 |
| TA-08 | GAPDH | M | 37 | ZSGB-BIO | WB 1:2000 |
| 9718 | γH2AX | R | 15 | CST | IF IHC 1:250 |
| PA5-16785 | Ki67 | M | Invitrogen | IHC 1:200 | |
| Catalog | Regent | Company | |||
| HY-103710 S7397 WLA123a | IBR2 | MCE | |||
| Sorafenib | Selleck | ||||
| Comet assay kit | Wanlei |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, M.; Sha, Y.; Qiu, J.; Chen, Y.; Liu, L.; Luo, M.; Huang, A.; Xia, J. RAD51 Inhibition Shows Antitumor Activity in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 7905. https://doi.org/10.3390/ijms24097905
Pan M, Sha Y, Qiu J, Chen Y, Liu L, Luo M, Huang A, Xia J. RAD51 Inhibition Shows Antitumor Activity in Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2023; 24(9):7905. https://doi.org/10.3390/ijms24097905
Chicago/Turabian StylePan, Mingang, Yu Sha, Jianguo Qiu, Yunmeng Chen, Lele Liu, Muyu Luo, Ailong Huang, and Jie Xia. 2023. "RAD51 Inhibition Shows Antitumor Activity in Hepatocellular Carcinoma" International Journal of Molecular Sciences 24, no. 9: 7905. https://doi.org/10.3390/ijms24097905
APA StylePan, M., Sha, Y., Qiu, J., Chen, Y., Liu, L., Luo, M., Huang, A., & Xia, J. (2023). RAD51 Inhibition Shows Antitumor Activity in Hepatocellular Carcinoma. International Journal of Molecular Sciences, 24(9), 7905. https://doi.org/10.3390/ijms24097905






